Abstract
Humans are protected against infection from most African trypanosomes by lipoprotein complexes present in serum that contain the trypanolytic pore-forming protein, Apolipoprotein L1 (APOL1). The human-infective trypanosomes, Trypanosoma brucei rhodesiense in East Africa and T. b. gambiense in West Africa have separately evolved mechanisms that allow them to resist APOL1-mediated lysis and cause human African trypanosomiasis, or sleeping sickness, in man. Recently, APOL1 variants were identified from a subset of Old World monkeys, that are able to lyse East African T. b. rhodesiense, by virtue of C-terminal polymorphisms in the APOL1 protein that hinder that parasite’s resistance mechanism. Such variants have been proposed as candidates for developing therapeutic alternatives to the unsatisfactory anti-trypanosomal drugs currently in use. Here we demonstrate the in vitro lytic ability of serum and purified recombinant protein of an APOL1 ortholog from the West African Guinea baboon (Papio papio), which is able to lyse examples of all sub-species of T. brucei including T. b. gambiense group 1 parasites, the most common agent of human African trypanosomiasis. The identification of a variant of APOL1 with trypanolytic ability for both human-infective T. brucei sub-species could be a candidate for universal APOL1-based therapeutic strategies, targeted against all pathogenic African trypanosomes.
Author Summary
African trypanosomes are protozoan parasites that affect both humans and animals in poor rural areas of sub-Saharan Africa, and are a major constraint on health and agricultural development. Disease control is principally dependent on the administration of drugs, which are old and largely unsatisfactory. Humans are naturally resistant to infection by most African trypanosomes species because of a lytic protein component in their blood, called APOL1. However, human-infective trypanosomes, T. b. rhodesiense in East Africa, and T. b. gambiense in West Africa, have evolved separate mechanisms to disarm this lytic protein and cause disease. Recently, variants of APOL1 were discovered in some primates that are able to kill the East African human disease-causing sub-species. These APOL1 variants form the basis of current attempts to create novel therapeutic interventions that can kill both animal and human-infective trypanosomes. In this study, we show that another variant of the same protein from a West African baboon species is able to kill, not only East African human-infective trypanosomes, but also the West African parasites, which causes the majority of human African trypanosomiasis cases. This new APOL1 variant could be a potential candidate for anti-trypanosomal therapies targeted at all pathogenic trypanosome species.
Introduction
African trypanosomes continue to exert a significant barrier to agricultural production and rural development across sub-Saharan Africa [1]. Due to a primate-specific innate trypanolytic mechanism, the majority of trypanosome species are unable to infect man. However, two sub-species of Trypanosoma brucei, T. b. rhodesiense and T. b. gambiense, have evolved distinct processes to resist this lysis and cause the debilitating and often fatal human form of African trypanosomiasis, known as sleeping sickness. The West African T. b. gambiense parasite typically causes a chronic disease profile, while the zoonotic T. b. rhodesiense sub-species, located in Eastern and Southern Africa, results in a more rapidly progressing, acute infection [2,3]. Seventy-million people over an area of 1.55 million km2 are at risk of contracting either of the two human-infective sub-species [4].
Current anti-trypanosomal drugs for medical and veterinary administration are largely unsatisfactory due to high toxicity, difficult treatment regimens, and emerging resistance [5–7]. Decades of drug development for African trypanosomiasis has produced safer refinements of existing therapies [7,8] and a number of promising novel drug candidates [9–11], but as yet no new anti-trypanosomal therapy has successfully passed phase III clinical trials. Furthermore, the adaptive immune response of vertebrates is rendered largely ineffective by the trypanosome’s ability to cyclically evade detection through variant surface glycoprotein (VSG)-mediated antigenic variation [12,13], placing a significant hurdle in the path of vaccine development. Broad-spectrum, safe, easily administered, and effective therapies to treat African trypanosomiasis are therefore still needed. The recent discovery of primate serum proteins that are able to kill both animal and human-infective trypanosomes is now offering opportunities for novel therapeutic approaches [14,15].
It has been known for over a century that the serum of humans and a small number of other Catarrhine primates are highly toxic to most African trypanosome species [16,17]. The molecular basis of this innate immunity in man has been elucidated and centres on two trypanolytic serum complexes, Trypanosome Lytic Factor 1 (TLF-1) [18,19] and TLF-2 [20,21], which share the same core protein components: haptoglobin-related protein (HPR) and apolipoprotein L1 (APOL1). HPR bound to haemoglobin mediates TLF-1 endocytosis via the haem-scavenging, haptoglobin-haemoglobin receptor (HpHbR) on the trypanosome’s surface [22–25]. Difficulty in purifying TLF-2 ex-vivo, has hindered discovery of exactly how this complex is bound and internalised by the parasite but it is known that it does not require HpHbR [21,26]. Despite differences in uptake, both TLF-1 and TLF-2 utilize the same lytic component in the form of the ionic channel-forming protein, APOL1 [22,27,28]. Following internalization, APOL1 undergoes a pH-dependant conformational change in the endolysosome pathway which releases it from the TLF complex [29,30], and promotes insertion into parasite membranes [31,32]. The exact mechanism of APOL1-mediated lysis that follows remains to be elucidated. In one recent model APOL1 insertion was found to disrupt both lysosomal and mitochondrial membranes, inducing an apoptosis-like cell death [33]. In contrast, an alternative model proposes that endosome recycling of APOL1 to the neutral environment of the parasite’s plasma membrane accelerates cation-selective channel activity and promotes lysis by osmotic swelling [34].
The Trypanosoma parasites responsible for animal trypanosomiasis are rapidly killed by this innate defence system, whereas the human sleeping sickness parasites, T. b. rhodesiense and T. b. gambiense, are able to resist lysis. In T. b. rhodesiense, resistance is effected by the VSG-derived, serum resistance associated (SRA) protein [35,36] which binds to the C-terminal domain of APOL1 in the endolysosome pathway preventing channel-mediated lysis [27,37–39], plausibly by impeding correct membrane insertion of APOL1 [34,40].
The mechanism of human serum resistance in T. b. gambiense has taken longer to unravel. T. b. gambiense typically grows to very low parasitemia and is difficult to adapt to laboratory models. An additional complicating factor is that T. b. gambiense shows two distinct "groups" that differ in genotype and phenotype [41–44]. The classic, clonal T. b. gambiense type [45], labelled “group 1” and found in West and Central Africa, is the predominant human-infective sub-species, responsible for 97% of all reported human cases [46]. T. b. gambiense group 1 strains are invariably resistant even after prolonged passage in laboratory rodents [42,47] and the mechanism underlying this resistance appears multifactorial, with at least three independent contributing components so far identified. Firstly the reduction of TLF-1 uptake through reduced expression and polymorphism of the HpHbR receptor that reduces binding affinity [48–50]; secondly, expression of a VSG-related T. b. gambiense-specific glycoprotein (TgsGP) which is essential, but not sufficient, for resistance [51] and which may increase resistance to APOL1 pore-mediated lysis by stiffening trypanosomal membranes [52]; and thirdly, faster APOL1 degradation has been proposed, through the action of cysteine peptidase [52,53]. A second, more virulent type of T. b. gambiense was identified in Cote d’Ivoire and Burkina Faso in the 1980’s [42,44] and defined as “group 2”, but has since virtually disappeared and may now be extinct. Studies of the limited number of group 2 strains that have been isolated indicate that these parasites are closely related to West Africa T. b. brucei [41,43,44,54] and exhibit a variable human serum resistance phenotype, in a manner superficially similar to T. b. rhodesiense [42,47,48]. Although the underlying resistance mechanism remains elusive it does not appear to involve a reduction in TLF-1 uptake [48] or the SRA [55] or TgsGP gene [56,57].
Unlike humans and gorillas [58,59], from which they diverged around 25 million years ago [60], several members of the Cercopithecidae (Old World monkey) family appear intrinsically resistant to T. b. rhodesiense [58,59,61]. Both serum and APOL1 from the East African baboon species, Papio hamadryas, has been demonstrated to effectively lyse human-infective T. b. rhodesiense [14,58]. This difference in innate immunity between Homo sapiens and P. hamadryas, has been pinpointed to the position of a single amino acid in the baboon APOL1 C-terminus which prevents the parasite’s SRA protein from binding and neutralising APOL1 lytic activity [62]. Furthermore, a nearly identical mutation has now also been detected in the C-terminus of APOL1 variants of some humans with African ancestry whose serum exhibits lytic activity against T. b. rhodesiense but not T. b. gambiense [63].
This led to the hypothesis that as T. b. gambiense is found only in West Africa, another variant of APOL1 may exist in some West African primates that is able to kill T. b. gambiense. In this study we examined the serum and APOL1 protein of a West African baboon species, Papio papio, suggested to be refractory to T. b. gambiense infection, with the ability to eliminate parasites in a laboratory infection [64]. Here we demonstrate that serum and recombinant protein from the P. papio APOL1 ortholog lyses representative strains of all sub-species of T. brucei in an in vitro assay system. The identification of an APOL1 variant with broad trypanolytic ability against T. brucei sub-species, including the most prevalent T. b. gambiense type, may provide a potential reagent for the development of universal APOL1-based therapeutic agents.
Methods
Trypanosoma brucei stocks
Representative bloodstream form cell lines were selected for each subspecies from a collection at the University of Glasgow and have been previously described. STIB247 is a T. b. brucei strain originally isolated from a hartebeest in Serengeti, Tanzania in 1971 [65]. The T. b. rhodesiense strain EATRO98 was isolated by the East African Trypanosomiasis Research Organization (EATRO) from a human in Nyanza, Kenya in 1961 [66]. T. b. gambiense group 2 strain STIB386 (MHOM/CI/78/TH114) was originally isolated in 1978 from an infected patient in Côte d'Ivoire [67]. ELIANE (MHOM/CI/52/ITMAP 2188) is a T. b. gambiense group 1 strain isolated from a human in Côte d’Ivoire in 1952 [68]. Additional T. b. gambiense group 1 strains tested were human isolates, PA (MHOM/CG/80/ITMAP1843/PA) from Republic of the Congo in 1975 [43], BIM (MHOM/CM/75/ITMAP1789/BIM) from Cameroon in 1975 [43], and TOBO (MHOM/CI/83/DAL596/TOBO) and S1/1/6 RI from Côte d'Ivoire in 1983 [69] and 2002 [70], respectively. All bloodstream form culture lines were maintained in vitro in modified HMI9 medium [71] supplemented by 1.5 mM glucose, 1 mM methyl cellulose, 250 μM adenosine, 150 μM guanosine and 20% foetal bovine serum (FBS). Expression of the SRA human serum resistance gene in T. b. rhodesiense EATRO98 was maintained under selection with 1% normal human serum. Ectopic expression of functional T. b. brucei HpHbR in ELIANE was previously generated using the tubulin-targeting TbbHpHbR pTub-phelo construct (strain ELIANE TbbHpHbR-/+) [51], and maintained under phleomycin selection. Bloodstream form isolates BIM and S1/1/6 RI were grown from stabilate in donor BALB/C mice (Harlan, United Kingdom) and trypanosomes purified from blood by differential centrifugation as previously described [72]. Cells were maintained as for bloodstream culture cells lines at 37°C in 5% CO2 for up to 24 hours until use. All animal procedures were carried out in accordance with the Animals (Scientific Procedures) Act of 1986. Subspecies classification for T. b. gambiense group 1 strains was confirmed by a positive PCR result for the T. b. gambiense specific glycoprotein (TgsGP) gene and T. b. rhodesiense by a positive PCR result for the subspecies-specific serum resistance-associated (SRA) gene, as previously described [48]. T. b. brucei and T. b. gambiense group 2 strains were confirmed by a combination of negative SRA/TgsGP PCR results, the human serum sensitivity phenotype and their microsatellite genetic profile [73].
Serum stocks
Sera Laboratories International, UK, provided pooled adult P. papio baboon serum derived from two individuals. Additional P. papio baboon serum, derived from a single adult male individual, was provided by Matrix Biologicals, UK. Normal human serum was obtained from a consented human donor and subject to appropriate ethical approval. The APOL1 protein levels in all serum samples are unquantified.
Serum resistance assays
Trypanosomes were diluted to 5 x 105 parasites per ml in modified HMI9, with the addition of human serum or P. papio serum serially diluted in foetal bovine serum (FBS), or FBS only, to a total concentration of 20%. Assays were performed in a final volume of 200 μl in a standard 96 well plate at 37°C in a CO2-equilibrated incubator. The number of viable motile trypanosomes was quantified at 24 hours by haemocytometer counts under the microscope in triplicate, for at least three independent experiments. The percentage viability of parasites in human or P. papio serum was normalised relative to the FBS control for each cell line to account for inherent differences in strain growth rate. Dose–response curves and IC50 values were determined using GraphPad Prism software (version 7.0).
Cloning and expression of recombinant APOL1
The H. sapiens (accession no. CCDS13926.1) or P. papio (accession no. KC197810) APOL1 open reading frame (ORF) was synthesised and supplied by GeneArt life technologies in an Invitrogen Gateway-compatible entry vector. The entry vector containing the APOL1 cDNA sequence, minus the N-terminal signal peptide (H. sapiens, residues 28–398; P. papio, residues 28–288) was cloned into pDest17 destination vector, which added an N-terminal 6xHis-tag, and transformed into BL21- AI competent E. coli. Protein expression was induced using 0.2% L-Arabinose for 16 hours at 37°C. Cells were lysed with urea lysis buffer (8 M urea, 20 mM Tris-HCl, 0.5 M NaCl, 5 mM imidazole, pH 8) and the cellular detritus removed by centrifugation at 5000g for 15 minutes. A small aliquot was removed for analysis by SDS-PAGE and Western blot with 1:5000 HRP-conjugated mouse anti-His antibody (Qiagen) and the remainder was used for protein purification under denaturing conditions. Denatured 6x His-tagged APOL1 protein was purified by passing the cell lysate through a gravity-flow Ni-Sepharose column (Gravitrap, GE Healthcare), and washing several times with urea lysis buffer pH 8 supplemented with increasing concentrations of Imidazole (5 mM-50 mM). Finally, bound protein was eluted with urea lysis buffer pH 8 containing 500mM imidazole. The eluate was dialyzed overnight against 20mM acetic acid and 0.05% tween and concentrated using 10,000 MW Vivaspin columns (Sartorius). Purity and concentration of the final purified protein was checked using a Qubit fluorometer (Thermo fisher) and SDS-PAGE (S1 Fig), then the concentration adjusted to 1 mg/ml and stored in aliquots at 4°C.
Recombinant APOL1 lysis assay
To assess survival in recombinant APOL1, trypanosomes were diluted to 5 x 105 parasites per ml in modified HMI9 containing 20% FBS and incubated with a dilution series of recombinant human or P. papio APOL1. The recombinant APOL1 was formulated in protein-free buffer (20mM acetic acid, 0.05% tween) and added in a volume of 10 μl to a final assay volume of 200 μl in a standard 96 well plate. A control containing an equivalent volume of protein-free buffer was also prepared. Assays were performed at 37°C in a CO2-equilibrated incubator, and the number of viable motile trypanosomes in each well was quantified at 24 hours by haemocytometer counts under the microscope in triplicate for at least three independent experiments. Cell counts in recombinant APOL1 were compared to control wells containing protein-free buffer only to determine percentage survival. In each assay, cells were incubated in 20% normal human serum as a positive control. Dose–response curves, IC50 values and one-way ANOVA were determined using GraphPad Prism software (version 7.0). Where indicated, trypanosomes were pre-incubated with 10 mM ammonium chloride (NH4Cl), a weak base, for 30 minutes at 37°C to reverse acidification of the endolysosome system prior to the addition of recombinant APOL1.
APOL1 localisation immunofluorescence assays
Samples for IFA were prepared as follows. All incubation steps unless stated otherwise were performed in a humidor at room temperature. Bloodstream form trypanosomes were diluted in HMI9 medium containing 20% FBS at a concentration of 106 parasites/ml and incubated with 50 μg/ml purified recombinant H. sapiens or P. papio APOL1 for two hours at 37°C. After this period, cells were washed once in serum-free HMI9 medium, and settled onto glass slides before fixing in 1% paraformaldehyde for 10 minutes. Samples were permeabilised using 0.1% Triton X-100 in PBS for 20 minutes then incubated in blocking solution (2% BSA in PBS) for 20 minutes. After washing three times in PBS, slides were incubated for 40 minutes with 1:500 mouse anti-p67 antibody (gift from Jay Bangs, Department of Microbiology and Immunology, University at Buffalo, NY, USA) in blocking solution. Washes were repeated and then primary antibody was detected using 1:1000 goat anti-mouse AlexaFluor594 secondary antibody (Life technologies) incubated for 40 minutes in blocking solution. To detect His-tagged APOL1 slides were washed three times in PBS and then incubated for 40 minutes with 1:500 AlexaFluor488 mouse anti-penta-His antibody (Molecular Probes, Invitrogen) in blocking solution. Following a final three washes the cells were treated with 50% glycerol, 0.1% DAPI, 2.5% 1, 4-diazabicyclo [2.2.2] octane (DABCO) in PBS, protected with a coverslip and sealed with acetone. Slides were imaged using the Deltavision Core system and SoftWorx package (Applied Precision) with standard filter sets (DAPI/FITC/Texas-Red and Light transmission). Approximately 30 serial sections through each trypanosome were taken for each filter. The images were composited and the brightness, contrast and color levels normalised between samples and exposures using the ImageJ software package (US National Institute of Health).
Ethics statement
The University of Glasgow ethical review board approved the use of human serum in this study. The human serum volunteer gave written informed consent.
Results
P. papio serum is lethal to T. b. rhodesiense and T. b. gambiense groups 1 and 2
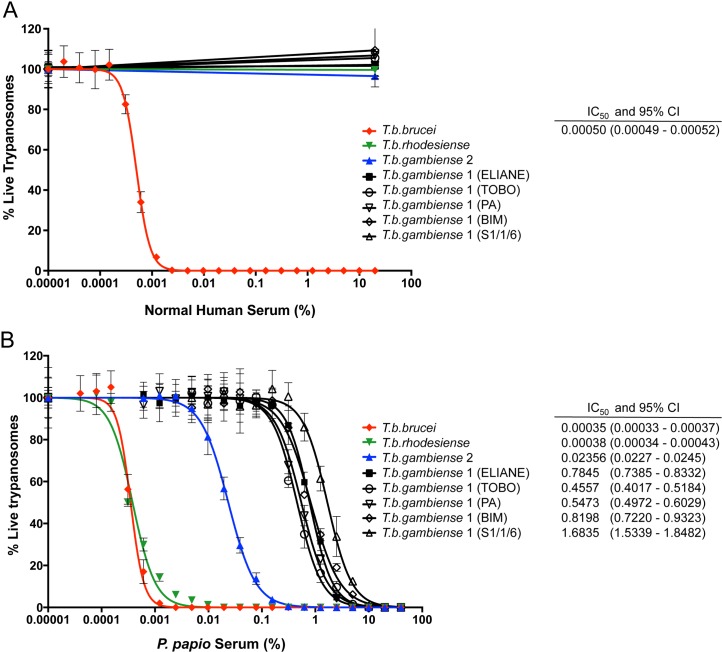
Trypanolytic activity against the human-infective East African T. b. rhodesiense sub-species has been demonstrated for sera from several members of the Cercopithecidae family, including baboons, mandrills and sooty mangabeys [14,37,58,59]. To date however, no primate has been identified with lytic activity against West African T. b. gambiense parasites. To determine the trypanolytic ability of serum from the West African Guinea baboon, P. papio, representative examples of the different T. brucei sub-species, were incubated for 24 hours in vitro, with a dilution series of P. papio or human serum. The strains selected included five different isolates of classic T. b. gambiense group 1, the cause of 97% of reported HAT cases [46], from a number of different disease foci in West Africa. As illustrated in Fig 1A, normal human serum efficiently lysed T. b. brucei bloodstream parasites (IC50; 0.0005%) in a 24 hour assay, but not strains of the human-infective T. b. rhodesiense or T. b. gambiense subspecies. In contrast, P. papio (pooled sera) was completely lytic to all tested strains, including both T. b. gambiense group 1 and 2 isolates, at concentrations ≥ 10% (Fig 1B). The sensitivity of T. b. brucei to P. papio pooled serum (IC50; 0.00035%) was comparable to that of T. b. rhodesiense (IC50; 0.00038%). T. b. gambiense group 1 and 2 strains however, were killed significantly less potently, with an IC50 approximately 70-fold (IC50; 0.024% serum, T. b. gambiense group 2) or 2000-fold (IC50; 0.46–1.68% serum, T. b. gambiense group 1) higher than that of the other sub-species, although still at a sub-physiological concentration. The trypanolytic activity of P. papio was also confirmed against a smaller collection of T. brucei strains using an alternative source of P. papio sera derived from a single male individual, which killed T. b. gambiense at a lower concentration > 2% (S2 Fig), presumably reflecting variation between individual animal samples.
Fig 1. Titration of the trypanolytic activity of Human (H. sapiens) and Guinea baboon (P. papio) sera against representative examples of the T. brucei sub-species.
The percentage of viable trypanosomes was determined following a 24-hour exposure to serial dilutions of (A) Human (H. sapiens) or (B) Guinea baboon (P. papio) sera. Representative T. brucei sub-species strains were tested: T. b. brucei (strain STIB247), T. b. rhodesiense (strain EATRO98), T. b. gambiense group 2 (strain STIB386), and T. b. gambiense group 1 (strains ELIANE, TOBO and S1/1/6 [Côte d'Ivoire], PA [Republic of the Congo], and BIM [Cameroon]). Mean percentage cell survival ± SD is expressed relative to FBS control, calculated from three independent experiments. Dose–response curves and IC50 values with 95% confidence intervals (CI) were determined using GraphPad Prism software version 7.
Lytic activity of P. papio recombinant APOL1
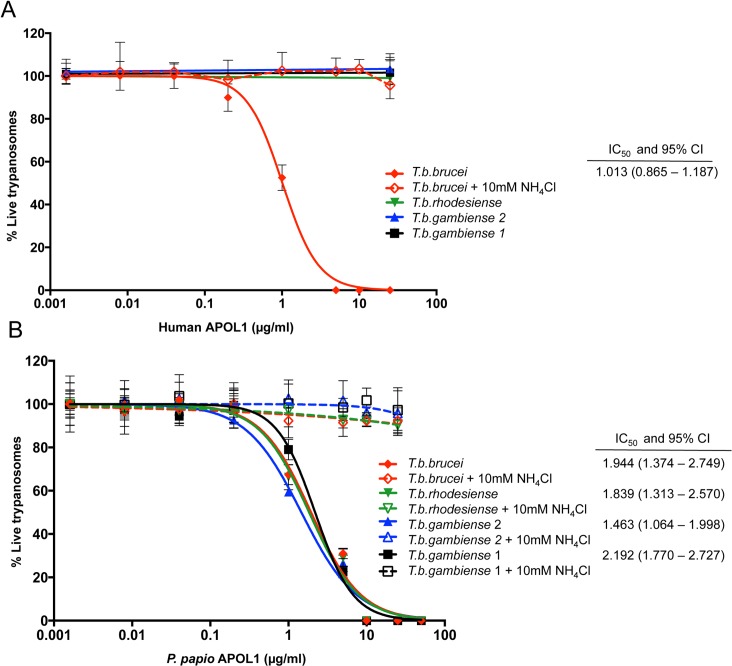
APOL1 has been demonstrated to be the lytic factor in normal human serum [22,27,28], and T. b. rhodesiense-lytic orthologs of APOL1 have now been identified in the serum of a number of Old World monkey species, including species of the Papio baboon genus [14,37,58]. Furthermore this lytic activity of Papio APOL1 against T. b. rhodesiense has been demonstrated to be the result of a single polymorphism [62]. We therefore hypothesize that the broad lytic ability of P. papio may be attributable to a functional variant of this protein. Sequenced APOL1 cDNA was used as a template for the production of recombinant variants of P. papio and human APOL1 protein (S3 Fig-Amino acid alignment). Representative strains of the different T. brucei sub-species were incubated in the presence of purified P. papio and human recombinant protein to determine if APOL1 alone had demonstrable trypanolytic ability. Titrated human recombinant APOL1 protein completely lysed T. b. brucei parasites after 24 hours (IC50; 1.013 μg/ml), at concentrations comparable to the physiological levels of APOL1 reported for normal human serum [74–76], but had no lytic effect on strains of the human serum resistant parasites, T. b. rhodesiense, T. b. gambiense group 1 or T. b. gambiense group 2 (Fig 2A). In contrast, recombinant P. papio APOL1 protein exhibited trypanolytic activity against representative strains of all T. brucei sub-species (Fig 2B with additional T. b. gambiense group 1 strains assays provided in S4 Fig). Furthermore, strains of all sub-species tested appeared equally susceptible to the effect of recombinant P. papio APOL1, with no significant difference in IC50 observed (one-way ANOVA, F (3, 24) = 1.741, p = 0.19). Notably, as has been observed for human APOL1, this lytic activity is inhibited by the addition of the acidotropic agent ammonium chloride to the assay (Fig 2A and 2B). Ammonium chloride is a weak base that raises endolysosomal pH, thereby preventing pH-dependant conformational changes to APOL1 that are predicted to be essential to efficient ion-channel mediated lysis [32,34,77]. This corresponding inhibition of APOL1-mediated lysis for both orthologs is further indicative of a conserved mechanism of action. In summary these assays demonstrate that the P. papio APOL1 ortholog in isolation exhibits trypanolytic ability against all tested examples of the human-infective T. brucei sub-species. Although there may be other, as yet uncharacterized factors that contribute to the lytic ability of P. papio serum, the APOL1 ortholog is a significant trypanolytic component.
Fig 2. Trypanolytic activity of recombinant APOL1.
The percentage of viable trypanosomes was determined following a 24-hour exposure to media containing serial dilutions of A) recombinant Human APOL1 protein and B) recombinant P. papio APOL1 protein. Representative T. brucei sub-species strains were tested: T. b. brucei (strain STIB247), T. b. rhodesiense (strain EATRO98), T. b. gambiense group 1 (strain ELIANE), and T. b. gambiense group 2 (strain STIB386). Additional assays were performed with different strains of T. b. gambiense group 1 and are provided in S4 Fig. The mean percentage cell survival ± SD, relative to protein-free control, was calculated from at least three independent experiments. Dose–response curves and IC50 values with 95% confidence intervals (CI) were determined using GraphPad Prism software version 7. APOL1-mediated lysis of each isolate was prevented by the inclusion of an acidotropic agent (10 mM NH4Cl) in the assay.
T. b. gambiense group 1 HpHbR reduces sensitivity to P. papio serum lysis
A reduced sensitivity to lysis was observed for both the predominant T. b. gambiense group 1 and minor group 2 strains, relative to T. b. brucei and T. b. rhodesiense, when incubated with P. papio serum, but not recombinant APOL1 protein. We postulated that for T. b. gambiense group 1, this difference might be the result of disparity in the rate of uptake of APOL1 versus APOL1–containing trypanolytic factors by these parasites. In normal human serum, HPR bound to haemoglobin, acts as ligand to facilitate TLF-1 uptake via the T. brucei HpHbR receptor [23,78]. However, a defining feature of T. b. gambiense group 1 strains is a decrease in TLF-1 internalisation as a result of reduced HpHbR expression and a conserved L210S substitution that reduces the binding affinity of HpHbR for its ligand [50,79]. Reduced TLF uptake via HpHbR contributes to the invariant human serum resistant phenotype of these parasites, although alone is insufficient to impart resistance to human serum [78] due to the existence of other speculated receptors for TLF-1 [80,81], and the additional TLF-2 particle in human serum for which the uptake mechanisms remain unknown [21,82,83]. In contrast, recombinant APOL1 is internalised by non-specific fluid phase endocytosis and trafficked through the endolysosome pathway, thus completely circumventing the HpHbR receptor [27,48].
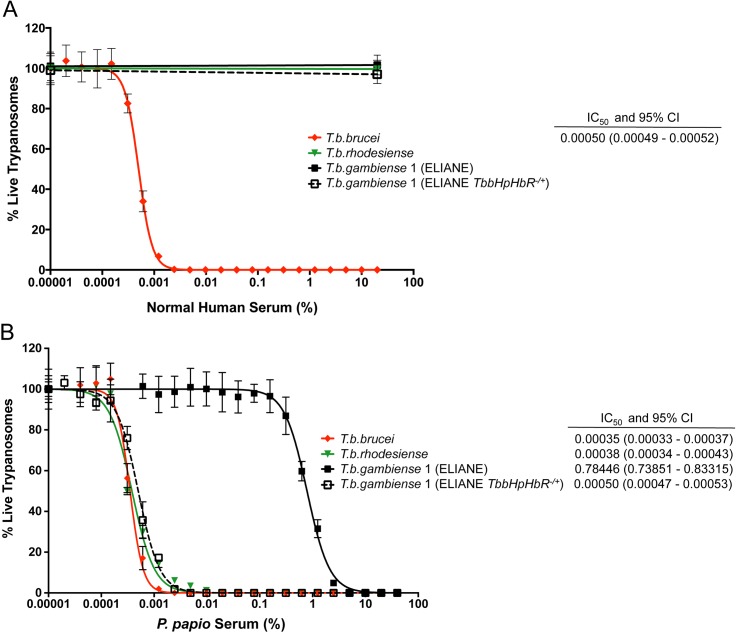
The number of molecules in the TLF complex and its exact structural composition in baboon serum is currently unresolved, but a representative baboon species, P. hamadryas, has been demonstrated to have similar constitutive components (HPR and APOL1) to human TLF [14]. As T. b. gambiense group 1 parasites have a reduced uptake of human TLF-1 but the other subspecies do not we postulated that a similar mechanism could reduce the uptake of P. papio TLF particles by T. b. gambiense group 1 strains, which is corrected by direct incubation in recombinant APOL1 protein. To investigate this we repeated the serum resistance assays using a T. b. gambiense ELIANE strain expressing a functional T. b. brucei HpHbR receptor (ELIANE TbbHpHbR -/+), that was previously generated by our laboratory and demonstrated to take up comparable amounts of TLF-1 to T. b. brucei [51]. As previously observed, expression of the functional T. b. brucei HpHbR receptor alone was insufficient to convert the phenotype of T. b. gambiense to human serum sensitivity and this clone (TbbHpHbR -/+ T. b. gambiense) retains full resistance to normal human serum (Fig 3A). However, it exhibits a 1000-fold increased sensitivity to P. papio serum (relative to the wild-type T. b. gambiense group 1 ELIANE strain), producing an IC50 value (0.0005%) comparable to that observed for the T. b. brucei and T. b. rhodesiense sub-species (Fig 3B and S2 Fig). Taken together, the serum and APOL1 assays indicate that diminished TLF uptake via the HpHbR receptor, rather than higher innate resistance to P. papio APOL1-mediated lysis underlies the increased resistance to P. papio serum observed for T. b. gambiense group 1 strains.
Fig 3. Expression of T. b. brucei HpHbR in T. b. gambiense group 1 increases sensitivity to P. papio serum lysis.
The percentage of viable trypanosomes following a 24-hour exposure to serial dilutions of A) human serum and B) P. papio serum was determined for T. b. gambiense (strain ELIANE) and T. b. gambiense expressing a functional T. b. brucei HpHbR receptor (strain ELIANE TbbHpHbR-/+) alongside representative T. brucei sub-species strains T. b. brucei (STIB247) and T. b. rhodesiense (EATRO98). Mean percentage cell survival ± SD is expressed, relative to FBS control. Dose–response curves and IC50 values with 95% confidence intervals (CI) were determined using GraphPad Prism software version 7.
In T. b. gambiense group 2, in contrast, an as yet uncharacterised HpHbR–independent mechanism/s determines human infectivity. T. b. gambiense group 2 strains, including the STIB386 isolate used in this study, have been shown to express the HpHbR gene at level comparable with T. b. brucei, with no demonstrable reduction in TLF-1 uptake [48]. Consequently, the reduced sensitivity to P. papio serum lysis, but not APOL1 protein, also observed for these HpHbR-functional parasites, further indicates that important differences exist in the cell biology of between T. b. gambiense group 2 and T. b. gambiense group 1 strains that determine sensitivity to these primate lytic factors.
Localisation of P. papio APOL1
Human recombinant APOL1 is taken up by fluid phase endocytosis and trafficked through the endocytic pathway to the endolysosome, the initial activation site of APOL1, in all T. brucei sub-species [27,48]. This results in lysis of T. b. brucei but not of T. b. rhodesiense or T. b. gambiense [48], which each possess mechanisms to resist the lytic effects of APOL1 [35,48,51,52]. To determine if P. papio APOL1 is localised through the parasite endolysosome pathway in a similar manner to that demonstrated for human APOL1, uptake of both recombinant proteins was compared in T. b. brucei and T. b. gambiense group 1 parasites using a fluorescent antibody to detect the His-tagged recombinant APOL1 protein. The cells were then examined by microscopy, in conjunction with the lysosomal marker p67. In order to achieve images of APOL1 uptake we used high concentrations of APOL1 (material and methods) to counteract possible degradation of APOL1 in the lysosome. Consistent with previous experiments of serum and APOL1 uptake in our laboratory [48,49,51], no lysosomal swelling was observed. As shown in Fig 4, both human and P. papio APOL1 are internalised by T. b. brucei and T. b. gambiense after a two hour incubation and are observed to co-localise with an antibody directed against the lysosomal membrane protein p67, indicative of the parasite endolysosome pathway [84,85]. These observations, in parallel with the ablation of lysis observed after co-incubation with acidotropic agent, ammonium chloride in APOL1 lysis assays, suggest that as previously demonstrated for human APOL1, exposure of the protein to the low pH of the endolysosomal pathway is also a requirement for trypanolytic activity of the baboon APOL1 ortholog.
Fig 4. Recombinant APOL1 uptake and localisation.
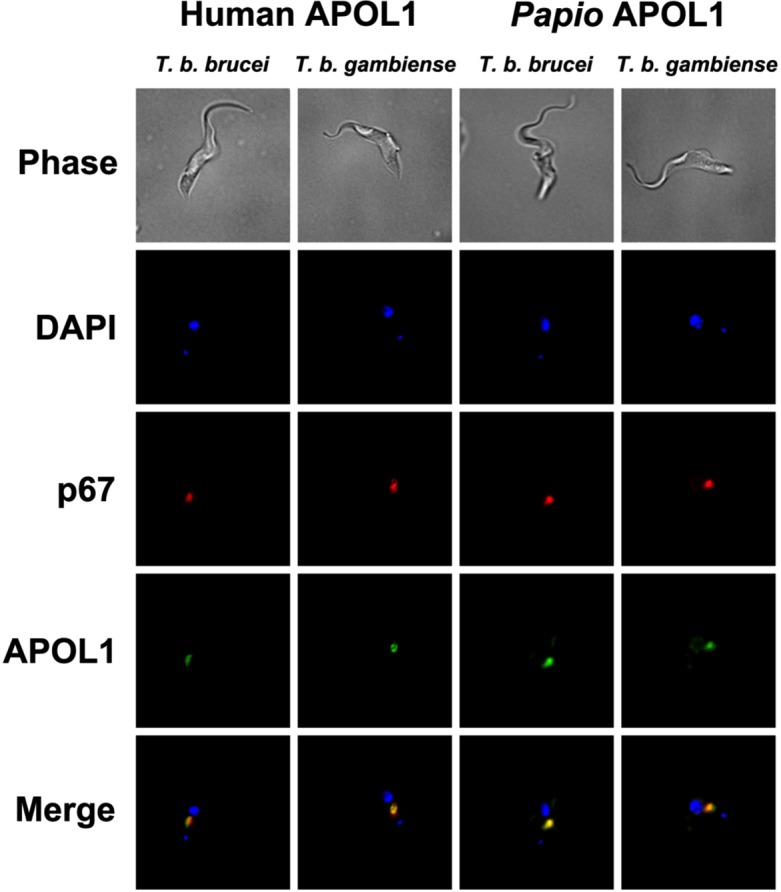
The localisation of Alexa488 (green) labelled anti-pentaHis antibody (APOL1), AlexaFluor594 (red) labelled anti-p67 (lysosomal membrane protein) and DAPI after a two-hour exposure to recombinant Human and P. papio APOL1 featuring an N-terminal 6xHis-tag. The panels represent human serum sensitive T. b. brucei, strain STIB247, and human serum resistant T. b. gambiense group 1, strain ELIANE.
Discussion
The ancient co-evolutionary engagement of African trypanosomes with their mammalian hosts has shaped an innate lytic molecule in man that protects from infection with most African trypanosomes. In response, the extensive antigenic repertoire of T. brucei [86] has provided a rich resource from which to evolve counter-measures to APOL1-mediated lysis on at least two occasions; SRA in T. b. rhodesiense in East Africa [35,36,87], and TgsGP in T. b. gambiense group 1 in West Africa [51,52,88]. In this study we present a novel APOL1 variant from a species of West African baboon that killed examples of all T. brucei sub-species, including T. b. rhodesiense, T. b. gambiense group 2, and T. b. gambiense group 1, the agent of most current cases of human African trypanosomiasis. The identification of such genetic variants, capable of killing both animal and human-infective parasites presents new opportunities for unconventional approaches to disease treatment and control, using APOL1-based biological therapies.
Previous studies have identified APOL1 orthologs in a subset of Old World monkeys [14,62], and an APOL1 variant with a key similarity in some humans with African ancestry [63], that encode proteins lytic to T. b. rhodesiense. In both variants, evidence suggests protection is mediated by the position of a single lysine residue in the C-terminal protein domain that obstructs coiled-coil interactions with SRA, thus allowing APOL1-directed lysis to proceed unimpeded [62]. Unfortunately in humans, the two amino acid deletion that alters the SRA-binding region in this APOL1-G2 variant come with an associated fitness cost: a 7–29-fold increased risk of developing a wide spectrum of kidney disorders in individuals carrying two copies of a variant allele [63,89–92]. The exact biological mechanism underlying this APOL1-associated nephropathy is not yet known but appears to be specific to the human variants. Engineered versions of the human APOL1 variant transiently expressed in a mouse model caused significant toxicity to the organ of expression (liver), which was not observed with baboon APOL1 or human APOL1 modified to introduce only the protective baboon lysine to the C-terminus [62]. This is an encouraging result, and such baboon-like APOL1 variants are now the focus of efforts to create suitable mechanisms of delivery, such as the conjugation of APOL1 protein to an antibody fragment targeted to parasite surface antigens [93] and an ambitious project to create targeted transgenic cattle expressing variant APOL1 [15].
These variants could be used to protect the reservoir host species from zoonotic T. b. rhodesiense sleeping sickness in addition to animal trypanosomiasis, which places severe restrictions on agricultural production and rural development in Sub-Saharan Africa [1]. Unfortunately, they will have a limited effect on the overall burden of human sleeping sickness. None of the APOL1 variants used in these experiments are able to kill the major human pathogen T. b. gambiense group 1 which places a population of 57 million people in West and Central Africa at risk of disease [4], less than 5% of whom are currently under surveillance [94]. Furthermore, there is a risk that the proposed interventions could result in the creation of a vacant ecological niche that increases the incidence of T. b. gambiense group 1 in domestic livestock through selective removal of susceptible competitor species such as T. b. brucei, T. congolense, T. vivax and T. b. rhodesiense.
We have addressed these concerns directly in this study by examining the serum of a West African baboon species P. papio that overlaps in distribution with that of T. b. gambiense, and which had been suggested to self-cure T. b. gambiense group 1 infection [64]. In that study primates infected with T. b. gambiense group 1 parasites exhibited a serological response that decreased throughout the course of the experiment and had no detectable parasitemia, consistent with an initial infection, followed by rapid parasite clearance and self-cure. In our study P. papio serum is able to lyse T. b. gambiense in 24 hours in vitro. The difference in timing of parasite killing between the in vivo and in vitro experiments, which could be due to a number of different factors such as parasite sequestration, is a well-recognised phenomenon. It is possible that parasites avoid lysis by residing in sites of low APOL1 concentrations (for example at the bite site in the skin) in the animal before eventually being cleared. This factor must be taken into account when attempting to develop APOL1-based therapies as in vitro assays do not always reflect the complexity of in vivo cell biology. The introduction of improved bioluminescent imaging to quantify parasite burden could be used to test in vivo for complete parasite clearance.
We have shown that the lytic effect of P. papio serum can be reproduced with an ortholog of the trypanolytic primate defence protein, APOL1, which demonstrates the uptake and localisation characteristics of other previously identified APOL1 proteins [27,48]. The trypanolytic action of this P. papio APOL1 variant against T. b. rhodesiense can be attributed to the C-terminal lysine mutation that is conserved among several members of the Cercopithecine subfamily that includes baboon, mandrills and mangabeys [62]. However the mechanism by which it counters T. b. gambiense, which has evolved multiple contributing mechanisms of human serum resistance, remains more elusive. All T. b. gambiense group 1 parasites share a mutated HpHbR with reduced affinity for one of the human APOL1-containing particles (TLF-1) via the HPR ligand [48–50], although a second particle, TLF2, appears to have alternative, as yet unresolved, mechanism(s) of internalisation [24,80,81,83]. The exact composition of TLF in baboon serum has not been clarified. However analysis of an HPR-affinity purified HDL sub-fraction from P. hamadryas baboon serum detected a TLF-equivalent particle that contains the same structural components as human TLF [14]. Furthermore, when transiently expressed in mice, all three components were required for maximum lytic activity against T. b. rhodesiense [14], suggesting HPR-HpHbR may play a role in uptake of baboon TLF. Here we show that T. b. gambiense group 1, although still fully susceptible to sub-physiological concentrations of P. papio serum, was 1000-fold less sensitive than T. b. brucei sub-species. This difference was ablated when functional T. b. brucei HpHbR was restored to the T. b. gambiense parasite, supporting a role for P. papio TLF uptake via both HpHbR-mediated endocytosis as well as unidentified alternative mechanisms, possible shared with those already proposed for human TLF [24,80,81,83].
Secondly, the TgsGP gene has been demonstrated to be essential for human serum resistance in T. b. gambiense group 1, as gene deletion renders the parasites sensitive to human serum lysis [51,52]. In contrast to the T. b. rhodesiense SRA protein, TgsGP and APOL1 do not appear to interact directly. Instead, TgsGP is proposed to bolster T. b. gambiense resistance to human APOL1 pore-forming activity through a process of plasma membrane stiffening [52]. A third mechanism by which T. b. gambiense might resist the actions of NHS, through enhanced APOL1 degradation within the endolysosomal system, has also been proposed [52]. Modulation of expression levels of the cysteine protease Cathepsin L and its inhibitor (ICP) has demonstrated an important role for cathepsin-mediated degradation of APOL1 in human serum resistance [53]. Difference in expression levels of these genes has not been detected in T. b. gambiense, however a lower pH is observed within the early endosomes that is predicted to accelerate their proteolytic activity relative to T. b. brucei [52]. Intriguingly, we observed equal sensitivity of all strains tested to P. papio APOL1-directed lysis, suggesting that the activity of TgsGP, and APOL1 degradation by cysteine peptidases, that effectively hinders human APOL1 in T. b. gambiense, poses no such barrier to the P. papio variant. This raises interesting questions about how exactly P. papio APOL1 is able to overcome these factors? Many details of the action of the TgsGP protein in particular remain cryptic. Despite its essential role in human serum resistance in T. b. gambiense, ectopic expression of T. b. gambiense TgsGP alone in T. b. brucei is insufficient to confer resistance to human serum [51,88]. There is evidently a role for other, as yet unidentified processes, in T. b. gambiense human serum resistance, which are absent or incomplete in T. b. brucei.
Sequence analysis has revealed that baboon and human APOL1 orthologs share only 58% amino acid sequence identity [14]. Despite this, in the recently elucidated example of baboon serum lysis of T. b. rhodesiense it was demonstrated that a single amino acid substitution conserved between baboon species is responsible for APOL1 evasion of SRA binding [62]. Uncovering the mechanism by which P. papio has developed its broad trypanolytic ability may offer further insights into the workings of T. b. gambiense human serum resistance, as well as aid in the design of an improved APOL1 therapy that could target all pathogenic trypanosomes across Sub-Saharan Africa. Such universal therapies that can treat both animal and human pathogens are particularly appropriate to the “one health” approach, currently advocated by WHO, FAO, and OIE, that integrates medical and veterinary health policy and research for addressing zoonotic diseases.
The Guinea baboon P. papio is found only in a limited area of western equatorial Africa, where its range overlaps with that of T. b. gambiense group 1. Five other baboons are represented in the Papio genus of which serum for only one, the east African P. cynocephalus (yellow baboon) has been previously tested against T. b. gambiense parasites, and was reported to be non–lytic [37]. Unfortunately APOL1 sequence is currently unavailable for comparative analysis with this species or the southern African P. ursinus (Chacma baboon) and P. kindae (Kinda baboon) species from Central Africa. Of the remaining Papio species, APOL1 sequences from cDNA have been successfully obtained for P. hamadryas (Hamadryas baboon) from North East Africa, and Central African P. anubis (Olive baboon) [62], the closest related species to P. papio in a recent phylogenetic study of mitochondrial DNA [95]. Amino acid alignments of P. papio APOL1 with these available sequences indicate ~98.5% identity to P. hamadryas and 93.5% to P. anubis (S3 Fig). A study in which C-terminal polymorphisms of P. anubis were incorporated into human recombinant APOL1 were observed to be lytic to T. b. rhodesiense but not T. b. gambiense [37], however full length APOL1 transcripts, unavailable at the time of the study, have not been tested. For P. hamadryas, serum and APOL1 have not yet been tested against T. b. gambiense, however a laboratory infection of two individual baboons with a strain of T. b. gambiense group 1 suggested hamadryas baboons to display a level of trypanotolerance to infection [64]. Future studies in which the sensitivity of T. brucei subspecies to serum and APOL1 from the other baboon species, followed by the construction of chimera mutants are now needed to help resolve the crucial polymorphisms responsible for T. b. gambiense lysis, as has been successful for T. b. rhodesiense.
Supporting Information
APOL1 variants were produced in E. coli, based on APOL1 cDNA sequence H. sapiens (accession no. CCDS13926.1) and P. papio (accession no. KC197810), minus the N-terminal signal peptide (H. sapiens, residues 28–398; P. papio, residues 28–288) and with the addition of an N-terminal 6xHis-tag. Proteins were purified using Ni-Sepharose under denaturing conditions and dialyzed against 20mM acetic acid and 0.05% tween. Purity and concentration of the final purified protein was checked using a Qubit fluorometer (Thermofisher) and SDS-PAGE (P = P. papio, H = human APOL1) stained with Brilliant blue G solution (Sigma-Aldrich) alongside SeeBlue Plus2 protein standards (Thermofisher). APOL1 concentration was adjusted to 1 mg/ml and stored in aliquots at 4°C.
(TIF)
To confirm the lytic ability of P. papio sera, the percentage of viable trypanosomes was determined following a 24-hour exposure to serial dilutions of an alternative Guinea baboon serum, sourced from an individual adult male (Matrix Biologicals, UK). Representative T. brucei sub-species strains were tested: T. b. brucei (strain STIB247), T. b. rhodesiense (strain EATRO98), T. b. gambiense group 1 (strain ELIANE) and T. b. gambiense group 1 expressing a functional T. b. brucei HpHbR receptor (ELIANE TbbHpHbR -/+). Mean percentage cell survival ± SD is expressed relative to FBS control, calculated from three independent experiments. Dose–response curves and IC50 values with 95% confidence intervals (CI) were determined using GraphPad Prism software version 7.
(TIF)
APOL1 amino acid sequence of the Old World monkey baboon species, Papio papio, was aligned with P. anubis, P. hamadryas and human (Homo sapiens) sequences. Dashes represent gaps introduced into the alignment by nucleotide deletions, and shading indicates amino acid differences. The position of the lysine (K) residue in the APOL1 C-terminus, implicated in resistance to T. b. rhodesiense, and present in several species of Old World monkey and the G2 APOL1 human variant is indicated (arrowhead).
(TIF)
The percentage of viable trypanosomes was determined following a 24-hour exposure to media containing serial dilutions of P. papio recombinant APOL1 protein. T. b. gambiense group 1 strains (ELIANE, TOBO and S1/1/6 [Côte d'Ivoire], PA [Republic of the Congo], and BIM [Cameroon]) were tested. The mean percentage cell survival ± SD, relative to protein-free control, was calculated from at least three independent experiments. Dose–response curves were determined using GraphPad Prism software version 7.
(TIF)
Acknowledgments
We gratefully acknowledge Jay Bangs for provision of anti-p67 antibody.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
AC PC CC NV WW are supported by a Wellcome Senior Fellowship Grant awarded to AM [095201/Z/10/Z]. The Wellcome Trust Centre for Molecular Parasitology is supported by core funding from the Wellcome Trust [085349]. The funders had no role in study design, data collection and analysis or the decision to publish.
References
- 1.Food and Agriculture Organization. On target against poverty: the Programme Against African Trypanosomiasis (PAAT) 1997–2007. Rome; 2008.
- 2.Kennedy PG. Clinical features, diagnosis, and treatment of human African trypanosomiasis (sleeping sickness). Lancet Neurol. 2013;12: 186–194. 10.1016/S1474-4422(12)70296-X [DOI] [PubMed] [Google Scholar]
- 3.Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. Elsevier; 2010;375: 148–159. 10.1016/S0140-6736(09)60829-1 [DOI] [PubMed] [Google Scholar]
- 4.Simarro PP, Cecchi G, Franco JR, Paone M, Diarra A, Ruiz-Postigo JA, et al. Estimating and mapping the population at risk of sleeping sickness. Ndung’u JM, editor. PLOS Negl Trop Dis. Public Library of Science; 2012;6: e1859 10.1371/journal.pntd.0001859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett MP, Vincent IM, Burchmore RJS, Kazibwe AJN, Matovu E. Drug resistance in human African trypanosomiasis. Future Microbiol. Future Medicine Ltd London, UK; 2011;6: 1037–1047. 10.2217/fmb.11.88 [DOI] [PubMed] [Google Scholar]
- 6.Delespaux V, Geysen D, Van den Bossche P, Geerts S. Molecular tools for the rapid detection of drug resistance in animal trypanosomes. Trends in Parasitology. Elsevier; 2008;24: 236–242. 10.1016/j.pt.2008.02.006 [DOI] [PubMed] [Google Scholar]
- 7.Simarro PP, Franco J, Diarra A, Postigo JAR, Jannin J. Update on field use of the available drugs for the chemotherapy of human African trypanosomiasis. Parasitology. Cambridge University Press; 2012;139: 842–846. 10.1017/S0031182012000169 [DOI] [PubMed] [Google Scholar]
- 8.Priotto G, Kasparian S, Mutombo W, Ngouama D, Ghorashian S, Arnold U, et al. Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. Lancet. Elsevier; 2009;374: 56–64. 10.1016/S0140-6736(09)61117-X [DOI] [PubMed] [Google Scholar]
- 9.Stich A, Ponte-Sucre A, Holzgrabe U. Do we need new drugs against human African trypanosomiasis? Lancet Infect Dis. Elsevier; 2013;13: 733–734. 10.1016/S1473-3099(13)70191-9 [DOI] [PubMed] [Google Scholar]
- 10.Nare B, Wring S, Bacchi C, Beaudet B, Bowling T, Brun R, et al. Discovery of novel orally bioavailable oxaborole 6-carboxamides that demonstrate cure in a murine model of late-stage central nervous system african trypanosomiasis. Antimicrob Agents Chemother. American Society for Microbiology; 2010;54: 4379–4388. 10.1128/AAC.00498-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brun R, Don R, Jacobs RT, Wang MZ, Barrett MP. Development of novel drugs for human African trypanosomiasis. Future Microbiol. Future Medicine Ltd London, UK; 2011;6: 677–691. 10.2217/fmb.11.44 [DOI] [PubMed] [Google Scholar]
- 12.Barry JD, McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv Parasitol. 2001;49: 1–70. [DOI] [PubMed] [Google Scholar]
- 13.Mugnier MR, Cross GAM, Papavasiliou FN. The in vivo dynamics of antigenic variation in Trypanosoma brucei. Science. American Association for the Advancement of Science; 2015;347: 1470–1473. 10.1126/science.aaa4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomson R, Molina-Portela P, Mott H, Carrington M, Raper J. Hydrodynamic gene delivery of baboon trypanosome lytic factor eliminates both animal and human-infective African trypanosomes. Proc Natl Acad Sci USA. National Acad Sciences; 2009;106: 19509–19514. 10.1073/pnas.0905669106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukeš J, Raper J. Prophylactic antiparasitic transgenesis for human parasitic disease? Mol Ther. Nature Publishing Group; 2010;18: 1745–1747. 10.1038/mt.2010.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hajduk SL, Hager KM, Esko JD. Human high density lipoprotein killing of African trypanosomes. Annu Rev Microbiol. Annual Reviews 4139 El Camino Way, P.O. Box 10139, Palo Alto, CA 94303–0139, USA; 1994;48: 139–162. 10.1146/annurev.mi.48.100194.001035 [DOI] [PubMed] [Google Scholar]
- 17.Laveran A, Mesnil F. Trypanosomes and Trypanosomiases. Paris: Masson et Cie; 1912. [Google Scholar]
- 18.Rifkin MR. Identification of the trypanocidal factor in normal human serum: high density lipoprotein. Proc Natl Acad Sci USA. National Academy of Sciences; 1978;75: 3450–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajduk SL, Moore DR, Vasudevacharya J, Siqueira H, Torri AF, Tytler EM, et al. Lysis of Trypanosoma brucei by a toxic subspecies of human high density lipoprotein. Journal of Biological Chemistry. 1989;264: 5210–5217. [PubMed] [Google Scholar]
- 20.Raper J, Portela MP, Lugli E, Frevert U, Tomlinson S. Trypanosome lytic factors: novel mediators of human innate immunity. Curr Opin Microbiol. 2001;4: 402–408. [DOI] [PubMed] [Google Scholar]
- 21.Tomlinson S, Jansen AM, Koudinov A, Ghiso JA, Choi-Miura NH, Rifkin MR, et al. High-density-lipoprotein-independent killing of Trypanosoma brucei by human serum. Molecular and Biochemical Parasitology. 1995;70: 131–138. [DOI] [PubMed] [Google Scholar]
- 22.Vanhollebeke B, Nielsen MJ, Watanabe Y, Truc P, Vanhamme L, Nakajima K, et al. Distinct roles of haptoglobin-related protein and apolipoprotein L-I in trypanolysis by human serum. Proc Natl Acad Sci USA. National Acad Sciences; 2007;104: 4118–4123. 10.1073/pnas.0609902104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Widener J, Nielsen MJ, Shiflett A, Moestrup SK, Hajduk S. Hemoglobin is a co-factor of human trypanosome lytic factor. PLOS Pathog. Public Library of Science; 2007;3: 1250–1261. 10.1371/journal.ppat.0030129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drain J, Bishop JR, Hajduk SL. Haptoglobin-related protein mediates trypanosome lytic factor binding to trypanosomes. Journal of Biological Chemistry. American Society for Biochemistry and Molecular Biology; 2001;276: 30254–30260. 10.1074/jbc.M010198200 [DOI] [PubMed] [Google Scholar]
- 25.Hager KM, Pierce MA, Moore DR, Tytler EM, Esko JD, Hajduk SL. Endocytosis of a cytotoxic human high density lipoprotein results in disruption of acidic intracellular vesicles and subsequent killing of African trypanosomes. J Cell Biol. The Rockefeller University Press; 1994;126: 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raper J, Nussenzweig V, Tomlinson S. The main lytic factor of Trypanosoma brucei brucei in normal human serum is not high density lipoprotein. J Exp Med. The Rockefeller University Press; 1996;183: 1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. Nature Publishing Group; 2003;422: 83–87. 10.1038/nature01461 [DOI] [PubMed] [Google Scholar]
- 28.Molina-Portela MP, Samanovic M, Raper J. Distinct roles of apolipoprotein components within the trypanosome lytic factor complex revealed in a novel transgenic mouse model. J Exp Med. 2008;205: 1721–1728. 10.1084/jem.20071463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanhollebeke B, Pays E. The function of apolipoproteins L. Cell Mol Life Sci. 2006;63: 1937–1944. 10.1007/s00018-006-6091-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrington JM, Howell S, Hajduk SL. Membrane permeabilization by trypanosome lytic factor, a cytolytic human high density lipoprotein. Journal of Biological Chemistry. American Society for Biochemistry and Molecular Biology; 2009;284: 13505–13512. 10.1074/jbc.M900151200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molina-Portela MDP, Lugli EB, Recio-Pinto E, Raper J. Trypanosome lytic factor, a subclass of high-density lipoprotein, forms cation-selective pores in membranes. Molecular and Biochemical Parasitology. 2005;144: 218–226. 10.1016/j.molbiopara.2005.08.018 [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, Nolan DP, Lins L, Homblé F, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. American Association for the Advancement of Science; 2005;309: 469–472. 10.1126/science.1114566 [DOI] [PubMed] [Google Scholar]
- 33.Vanwalleghem G, Fontaine F, Lecordier L, Tebabi P, Klewe K, Nolan DP, et al. Coupling of lysosomal and mitochondrial membrane permeabilization in trypanolysis by APOL1. Nat Commun. Nature Publishing Group; 2015;6: 8078 10.1038/ncomms9078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson R, Finkelstein A. Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers: relevance to trypanosome lysis. Proc Natl Acad Sci USA. National Acad Sciences; 2015;112: 2894–2899. 10.1073/pnas.1421953112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Xong H, Vanhamme L, Chamekh M, Chimfwembe CE, Van Den Abbeele J, Pays A, et al. A VSG Expression Site–Associated Gene Confers Resistance to Human Serum in Trypanosoma rhodesiense. Cell. 1998;95: 839–846. 10.1016/S0092-8674(00)81706-7 [DOI] [PubMed] [Google Scholar]
- 36.De Greef C, Imberechts H, Matthyssens G, Van Meirvenne N, Hamers R. A gene expressed only in serum-resistant variants of Trypanosoma brucei rhodesiense. Molecular and Biochemical Parasitology. 1989;36: 169–176. [DOI] [PubMed] [Google Scholar]
- 37.Lecordier L, Vanhollebeke B, Poelvoorde P, Tebabi P, Paturiaux-Hanocq F, Andris F, et al. C-terminal mutants of apolipoprotein L-I efficiently kill both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. Mansfield JM, editor. PLOS Pathog. Public Library of Science; 2009;5: e1000685 10.1371/journal.ppat.1000685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephens NA, Hajduk SL. Endosomal localization of the serum resistance-associated protein in African trypanosomes confers human infectivity. Eukaryotic Cell. 2011;10: 1023–1033. 10.1128/EC.05112-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bart J-M, Cordon-Obras C, Vidal I, Reed J, Perez-Pastrana E, Cuevas L, et al. Localization of serum resistance-associated protein in Trypanosoma brucei rhodesiense and transgenic Trypanosoma brucei brucei. Cell Microbiol. 2015;: n/a–n/a. 10.1111/cmi.12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Pérez-Morga D. The molecular arms race between African trypanosomes and humans. Nature Reviews Microbiology. Nature Publishing Group; 2014;12: 575–584. 10.1038/nrmicro3298 [DOI] [PubMed] [Google Scholar]
- 41.Gibson WC, de C Marshall TF, Godfrey DG. Numerical analysis of enzyme polymorphism: a new approach to the epidemiology and taxonomy of trypanosomes of the subgenus Trypanozoon. Adv Parasitol. 1980;18: 175–246. [DOI] [PubMed] [Google Scholar]
- 42.Mehlitz D, Zillmann U, Scott CM, Godfrey DG. Epidemiological studies on the animal reservoir of Gambiense sleeping sickness. Part III. Characterization of trypanozoon stocks by isoenzymes and sensitivity to human serum. Tropenmed Parasitol. 1982;33: 113–118. [PubMed] [Google Scholar]
- 43.Tait A, Babiker EA, Le Ray D. Enzyme variation in Trypanosoma brucei spp. I. Evidence for the sub-speciation of Trypanosoma brucei gambiense. Parasitology. 1984;89 (Pt 2): 311–326. [DOI] [PubMed] [Google Scholar]
- 44.Gibson WC. Will the real Trypanosoma b. gambiense please stand up. Parasitol Today (Regul Ed). 1986;2: 255–257. [DOI] [PubMed] [Google Scholar]
- 45.Weir W, Capewell P, Foth B, Clucas C, Pountain A, Steketee P, et al. Population genomics reveals the origin and asexual evolution of human infective trypanosomes. Elife. eLife Sciences Publications Limited; 2016;5: pdb.ip71. 10.7554/eLife.11473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simarro PP, Cecchi G, Paone M, Franco JR, Diarra A, Ruiz JA, et al. The Atlas of human African trypanosomiasis: a contribution to global mapping of neglected tropical diseases. Int J Health Geogr. BioMed Central Ltd; 2010;9: 57 10.1186/1476-072X-9-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zillmann U, Mehlitz D, Sachs R. Identity of Trypanozoon stocks isolated from man and a domestic dog in Liberia. Tropenmed Parasitol. 1984;35: 105–108. [PubMed] [Google Scholar]
- 48.Capewell P, Veitch NJ, Turner CMR, Raper J, Berriman M, Hajduk SL, et al. Differences between Trypanosoma brucei gambiense groups 1 and 2 in their resistance to killing by trypanolytic factor 1. Büscher P, editor. PLOS Negl Trop Dis. Public Library of Science; 2011;5: e1287 10.1371/journal.pntd.0001287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kieft R, Capewell P, Turner CMR, Veitch NJ, MacLeod A, Hajduk S. Mechanism of Trypanosoma brucei gambiense (group 1) resistance to human trypanosome lytic factor. Proc Natl Acad Sci USA. National Acad Sciences; 2010;107: 16137–16141. 10.1073/pnas.1007074107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeJesus E, Kieft R, Albright B, Stephens NA, Hajduk SL. A single amino acid substitution in the group 1 Trypanosoma brucei gambiense haptoglobin-hemoglobin receptor abolishes TLF-1 binding. Hill KL, editor. PLOS Pathog. Public Library of Science; 2013;9: e1003317 10.1371/journal.ppat.1003317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capewell P, Clucas C, DeJesus E, Kieft R, Hajduk S, Veitch N, et al. The TgsGP Gene Is Essential for Resistance to Human Serum in Trypanosoma brucei gambiense. Alsford S, editor. PLOS Pathog. Public Library of Science; 2013;9: e1003686 10.1371/journal.ppat.1003686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uzureau P, Uzureau S, Lecordier L, Fontaine F, Tebabi P, Homblé F, et al. Mechanism of Trypanosoma brucei gambiense resistance to human serum. Nature. Nature Publishing Group; 2013;501: 430–434. 10.1038/nature12516 [DOI] [PubMed] [Google Scholar]
- 53.Alsford S, Currier RB, Guerra-Assunção JA, Clark TG, Horn D. Cathepsin-L can resist lysis by human serum in Trypanosoma brucei brucei. Beverley SM, editor. PLOS Pathog. Public Library of Science; 2014;10: e1004130 10.1371/journal.ppat.1004130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balmer O, Beadell JS, Gibson W, Caccone A. Phylogeography and taxonomy of Trypanosoma brucei. Solano P, editor. PLoS Negl Trop Dis. 2011;5: e961 10.1371/journal.pntd.0000961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radwanska M, Chamekh M, Vanhamme L, Claes F, Magez S, Magnus E, et al. The serum resistance-associated gene as a diagnostic tool for the detection of Trypanosoma brucei rhodesiense. Am J Trop Med Hyg. 2002;67: 684–690. [DOI] [PubMed] [Google Scholar]
- 56.Gibson W, Nemetschke L, Ndung'u J. Conserved sequence of the TgsGP gene in Group 1 Trypanosoma brucei gambiense. Infect Genet Evol. 2010;10: 453–458. 10.1016/j.meegid.2010.03.005 [DOI] [PubMed] [Google Scholar]
- 57.Radwanska M, Claes F, Magez S, Magnus E, Pérez-Morga D, Pays E, et al. Novel primer sequences for polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am J Trop Med Hyg. 2002;67: 289–295. [DOI] [PubMed] [Google Scholar]
- 58.Lugli EB, Pouliot M, Portela MDPM, Loomis MR, Raper J. Characterization of primate trypanosome lytic factors. Molecular and Biochemical Parasitology. 2004;138: 9–20. 10.1016/j.molbiopara.2004.07.004 [DOI] [PubMed] [Google Scholar]
- 59.Poelvoorde P, Vanhamme L, Van Den Abbeele J, Switzer WM, Pays E. Distribution of apolipoprotein L-I and trypanosome lytic activity among primate sera. Molecular and Biochemical Parasitology. 2004;134: 155–157. [DOI] [PubMed] [Google Scholar]
- 60.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. Nature Publishing Group; 1998;392: 917–920. 10.1038/31927 [DOI] [PubMed] [Google Scholar]
- 61.Seed JR, Sechelski JB, Loomis MR. A survey for a trypanocidal factor in primate sera. J Protozool. 1990;37: 393–400. [DOI] [PubMed] [Google Scholar]
- 62.Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci USA. National Acad Sciences; 2014;111: E2130–9. 10.1073/pnas.1400699111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. American Association for the Advancement of Science; 2010;329: 841–845. 10.1126/science.1193032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kageruka P, Mangus E, Bajyana Songa E, Nantulya V, Jochems M, Hamers R, et al. Infectivity of Trypanosoma (Trypanozoon) brucei gambiense for baboons (Papio hamadryas, Papio papio). Ann Soc Belg Med Trop. 1991;71: 39–46. [PubMed] [Google Scholar]
- 65.Geigy R, Kauffmann M. Sleeping sickness survey in the Serengeti area (Tanzania) 1971. I. Examination of large mammals for trypanosomes. Acta Trop. 1973;30: 12–23. [PubMed] [Google Scholar]
- 66.Tait A, Barry JD, Wink R, Sanderson A, Crowe JS. Enzyme variation in T. brucei ssp. II. Evidence for T. b. rhodesiense being a set of variants of T. b. brucei. Parasitology. 1985;90 (Pt 1): 89–100. [DOI] [PubMed] [Google Scholar]
- 67.Felgner P, Brinkmann U, Zillmann U, Mehlitz D, Abu-Ishira S. Epidemiological studies on the animal reservoir of gambiense sleeping sickness. Part II. Parasitological and immunodiagnostic examination of the human population. Tropenmed Parasitol. 1981;32: 134–140. [PubMed] [Google Scholar]
- 68.O'Connell KM, Hutner SH, Fromentin H, Frank O, Baker H. Cryoprotectants for Crithidia fasciculata stored at -20 C, with notes on Trypanosoma gambiense and T. conorhini. J Protozool. 1968;15: 719–724. [DOI] [PubMed] [Google Scholar]
- 69.Barnes DA, Mottram J, Selkirk M, Agabian N. Two variant surface glycoprotein genes distinguish between different substrains of Trypanosoma brucei gambiense. Molecular and Biochemical Parasitology. 1989;34: 135–146. [DOI] [PubMed] [Google Scholar]
- 70.Holzmuller P, Biron DG, Courtois P, Koffi M, Bras-Gonçalves R, Daulouède S, et al. Virulence and pathogenicity patterns of Trypanosoma brucei gambiense field isolates in experimentally infected mouse: differences in host immune response modulation by secretome and proteomics. Microbes Infect. 2008;10: 79–86. 10.1016/j.micinf.2007.10.008 [DOI] [PubMed] [Google Scholar]
- 71.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75: 985–989. [PubMed] [Google Scholar]
- 72.Turner CMR, McLellan S, Lindergard LAG, Bisoni L, Tait A, MacLeod A. Human infectivity trait in Trypanosoma brucei: stability, heritability and relationship to sra expression. Parasitology. 2004;129: 445–454. [DOI] [PubMed] [Google Scholar]
- 73.MacLeod A, Tweedie A, McLellan S, Taylor S, Cooper A, Sweeney L, et al. Allelic segregation and independent assortment in T. brucei crosses: Proof that the genetic system is Mendelian and involves meiosis. Molecular and Biochemical Parasitology. 2005;143: 12–19. 10.1016/j.molbiopara.2005.04.009 [DOI] [PubMed] [Google Scholar]
- 74.Duchateau PN, Movsesyan I, Yamashita S, Sakai N, Hirano K, Schoenhaus SA, et al. Plasma apolipoprotein L concentrations correlate with plasma triglycerides and cholesterol levels in normolipidemic, hyperlipidemic, and diabetic subjects. J Lipid Res. 2000;41: 1231–1236. [PubMed] [Google Scholar]
- 75.Weckerle A, Snipes JA, Cheng D, Gebre AK, Reisz JA, Murea M, et al. Characterization of circulating APOL1 protein complexes in African Americans. J Lipid Res. American Society for Biochemistry and Molecular Biology; 2016;57: 120–130. 10.1194/jlr.M063453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bruggeman LA, O'Toole JF, Ross MD, Madhavan SM, Smurzynski M, Wu K, et al. Plasma apolipoprotein L1 levels do not correlate with CKD. J Am Soc Nephrol. American Society of Nephrology; 2014;25: 634–644. 10.1681/ASN.2013070700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pays E, Vanhollebeke B, Vanhamme L, Paturiaux-Hanocq F, Nolan DP, Pérez-Morga D. The trypanolytic factor of human serum. Nature Reviews Microbiology. Nature Publishing Group; 2006;4: 477–486. 10.1038/nrmicro1428 [DOI] [PubMed] [Google Scholar]
- 78.Vanhollebeke B, De Muylder G, Nielsen MJ, Pays A, Tebabi P, Dieu M, et al. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science. American Association for the Advancement of Science; 2008;320: 677–681. 10.1126/science.1156296 [DOI] [PubMed] [Google Scholar]
- 79.Higgins MK, Tkachenko O, Brown A, Reed J, Raper J, Carrington M. Structure of the trypanosome haptoglobin-hemoglobin receptor and implications for nutrient uptake and innate immunity. Proc Natl Acad Sci USA. National Acad Sciences; 2013;110: 1905–1910. 10.1073/pnas.1214943110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bullard W, Kieft R, Capewell P, Veitch NJ, MacLeod A, Hajduk SL. Haptoglobin-hemoglobin receptor independent killing of African trypanosomes by human serum and trypanosome lytic factors. Virulence. Taylor & Francis; 2012;3: 72–76. 10.4161/viru.3.1.18295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Green HP, Del Pilar Molina Portela M, St Jean EN, Lugli EB, Raper J. Evidence for a Trypanosoma brucei lipoprotein scavenger receptor. Journal of Biological Chemistry. American Society for Biochemistry and Molecular Biology; 2003;278: 422–427. 10.1074/jbc.M207215200 [DOI] [PubMed] [Google Scholar]
- 82.Raper J, Fung R, Ghiso J, Nussenzweig V, Tomlinson S. Characterization of a novel trypanosome lytic factor from human serum. Infect Immun. American Society for Microbiology; 1999;67: 1910–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vanhollebeke B, Pays E. The trypanolytic factor of human serum: many ways to enter the parasite, a single way to kill. Molecular Microbiology. Blackwell Publishing Ltd; 2010;76: 806–814. 10.1111/j.1365-2958.2010.07156.x [DOI] [PubMed] [Google Scholar]
- 84.Alexander DL, Schwartz KJ, Balber AE, Bangs JD. Developmentally regulated trafficking of the lysosomal membrane protein p67 in Trypanosoma brucei. J Cell Sci. 2002;115: 3253–3263. [DOI] [PubMed] [Google Scholar]
- 85.Kelley RJ, Alexander DL, Cowan C, Balber AE, Bangs JD. Molecular cloning of p67, a lysosomal membrane glycoprotein from Trypanosoma brucei. Molecular and Biochemical Parasitology. 1999;98: 17–28. [DOI] [PubMed] [Google Scholar]
- 86.Marcello L, Menon S, Ward P, Wilkes JM, Jones NG, Carrington M, et al. VSGdb: a database for trypanosome variant surface glycoproteins, a large and diverse family of coiled coil proteins. BMC Bioinformatics. BioMed Central; 2007;8: 143 10.1186/1471-2105-8-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gibson W, Ferris V. Conservation of the genomic location of the human serum resistance associated gene in Trypanosoma brucei rhodesiense. Molecular and Biochemical Parasitology. 2003;130: 159–162. [DOI] [PubMed] [Google Scholar]
- 88.Berberof M, Pérez-Morga D, Pays E. A receptor-like flagellar pocket glycoprotein specific to Trypanosoma brucei gambiense. Molecular and Biochemical Parasitology. 2001;113: 127–138. [DOI] [PubMed] [Google Scholar]
- 89.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. American Society of Nephrology; 2011;22: 2129–2137. 10.1681/ASN.2011040388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kasembeli AN, Duarte R, Ramsay M, Mosiane P, Dickens C, Dix-Peek T, et al. APOL1 Risk Variants Are Strongly Associated with HIV-Associated Nephropathy in Black South Africans. J Am Soc Nephrol. American Society of Nephrology; 2015;: ASN.2014050469. 10.1681/ASN.2014050469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;66: 390–396. 10.1002/art.38220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Larsen CP, Beggs ML, Saeed M, Walker PD. Apolipoprotein L1 risk variants associate with systemic lupus erythematosus-associated collapsing glomerulopathy. J Am Soc Nephrol. American Society of Nephrology; 2013;24: 722–725. 10.1681/ASN.2012121180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baral TN, Magez S, Stijlemans B, Conrath K, Vanhollebeke B, Pays E, et al. Experimental therapy of African trypanosomiasis with a nanobody-conjugated human trypanolytic factor. Nat Med. 2006;12: 580–584. 10.1038/nm1395 [DOI] [PubMed] [Google Scholar]
- 94.Simarro PP, Cecchi G, Franco JR, Paone M, Diarra A, Ruiz Postigo JA, et al. Mapping the capacities of fixed health facilities to cover people at risk of gambiense human African trypanosomiasis. Int J Health Geogr. BioMed Central; 2014;13: 4 10.1186/1476-072X-13-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zinner D, Wertheimer J, Liedigk R, Groeneveld LF, Roos C. Baboon phylogeny as inferred from complete mitochondrial genomes. Am J Phys Anthropol. Wiley Subscription Services, Inc., A Wiley Company; 2013;150: 133–140. 10.1002/ajpa.22185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APOL1 variants were produced in E. coli, based on APOL1 cDNA sequence H. sapiens (accession no. CCDS13926.1) and P. papio (accession no. KC197810), minus the N-terminal signal peptide (H. sapiens, residues 28–398; P. papio, residues 28–288) and with the addition of an N-terminal 6xHis-tag. Proteins were purified using Ni-Sepharose under denaturing conditions and dialyzed against 20mM acetic acid and 0.05% tween. Purity and concentration of the final purified protein was checked using a Qubit fluorometer (Thermofisher) and SDS-PAGE (P = P. papio, H = human APOL1) stained with Brilliant blue G solution (Sigma-Aldrich) alongside SeeBlue Plus2 protein standards (Thermofisher). APOL1 concentration was adjusted to 1 mg/ml and stored in aliquots at 4°C.
(TIF)
To confirm the lytic ability of P. papio sera, the percentage of viable trypanosomes was determined following a 24-hour exposure to serial dilutions of an alternative Guinea baboon serum, sourced from an individual adult male (Matrix Biologicals, UK). Representative T. brucei sub-species strains were tested: T. b. brucei (strain STIB247), T. b. rhodesiense (strain EATRO98), T. b. gambiense group 1 (strain ELIANE) and T. b. gambiense group 1 expressing a functional T. b. brucei HpHbR receptor (ELIANE TbbHpHbR -/+). Mean percentage cell survival ± SD is expressed relative to FBS control, calculated from three independent experiments. Dose–response curves and IC50 values with 95% confidence intervals (CI) were determined using GraphPad Prism software version 7.
(TIF)
APOL1 amino acid sequence of the Old World monkey baboon species, Papio papio, was aligned with P. anubis, P. hamadryas and human (Homo sapiens) sequences. Dashes represent gaps introduced into the alignment by nucleotide deletions, and shading indicates amino acid differences. The position of the lysine (K) residue in the APOL1 C-terminus, implicated in resistance to T. b. rhodesiense, and present in several species of Old World monkey and the G2 APOL1 human variant is indicated (arrowhead).
(TIF)
The percentage of viable trypanosomes was determined following a 24-hour exposure to media containing serial dilutions of P. papio recombinant APOL1 protein. T. b. gambiense group 1 strains (ELIANE, TOBO and S1/1/6 [Côte d'Ivoire], PA [Republic of the Congo], and BIM [Cameroon]) were tested. The mean percentage cell survival ± SD, relative to protein-free control, was calculated from at least three independent experiments. Dose–response curves were determined using GraphPad Prism software version 7.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.