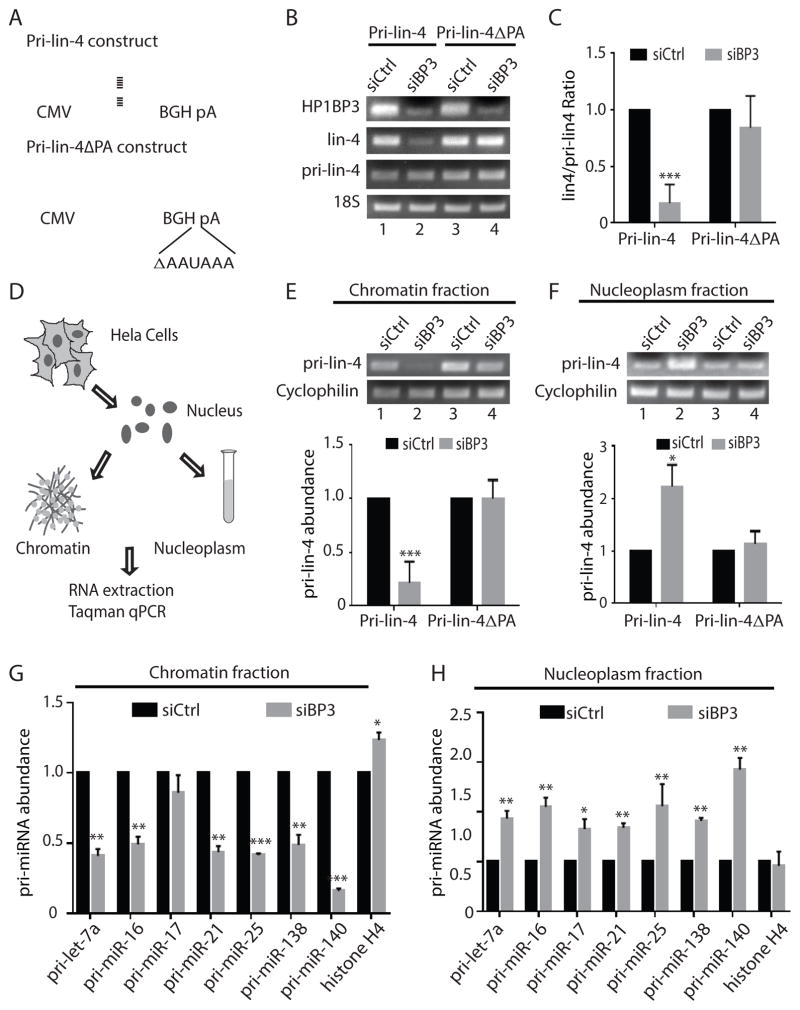
SUMMARY
Recent studies suggest that the Microprocessor (Drosha-DGCR8) complex can be recruited to chromatin to catalyze co-transcriptional processing of primary microRNAs (pri-miRNAs) in mammalian cells. However, the molecular mechanism of co-transcriptional miRNA processing is poorly understood. Here, we find that HP1BP3, a histone H1-like chromatin protein, specifically associates with the Microprocessor and promotes global miRNA biogenesis in human cells. Chromatin immunoprecipitation (ChIP) studies reveal genome-wide co-localization of HP1BP3 and Drosha and HP1BP3-dependent Drosha binding to actively transcribed miRNA loci. Moreover, HP1BP3 specifically binds endogenous pri-miRNAs and facilitates the Drosha/pri-miRNA association in vivo. Knockdown of HP1BP3 compromises pri-miRNA processing by causing premature release of pri-miRNAs from the chromatin. Taken together, these studies suggest that HP1BP3 promotes co-transcriptional miRNA processing via chromatin retention of nascent pri-miRNA transcripts. This work significantly expands the functional repertoire of the H1 family of proteins and suggests the existence of chromatin retention factors for widespread co-transcriptional miRNA processing.
Keywords: HP1BP3, histone H1, Drosha-DGCR8, pri-miRNA, co-transcriptional processing
Graphical Abstract
INTRODUCTION
MicroRNAs (miRNAs) are a family of ~22-nucleotide (nt) cellular RNAs that govern diverse physiological and pathological processes in eukaryotic organisms (Bartel, 2004; He and Hannon, 2004). Typically, miRNA biogenesis is catalyzed sequentially by two RNase III enzymes, Drosha and Dicer (Kim et al., 2009). In the nucleus, primary miRNA (pri-miRNA) transcripts are processed by the Microprocessor (Drosha-DGCR8) complex into ~70-nt stem-loop precursor miRNAs (pre-miRNAs) (Denli et al., 2004; Gregory et al., 2004; Han et al., 2004; Landthaler et al., 2004; Lee et al., 2003). DGCR8 is a double-stranded RNA (dsRNA)-binding protein that is essential for Drosha processing of pri-miRNA (Gregory et al., 2004; Han et al., 2004; Landthaler et al., 2004). In the cytoplasm, pre-miRNAs are further cleaved by the Dicer-TRBP/PACT complex into mature miRNAs (Chendrimada et al., 2005; Haase et al., 2005; Hutvagner et al., 2004; Lee et al., 2006; Paroo et al., 2009), which are then assembled into the miRNA-induced silencing complexes (miRISC) (Hutvagner and Zamore, 2002; Liu et al., 2004; Maniataki and Mourelatos, 2005; Yoda et al., 2010). In mammalian miRISC, miRNA guides Argonaute (Ago) and associated proteins to effect target silencing through translational inhibition and mRNA decay (Carthew and Sontheimer, 2009; Djuranovic et al., 2012; Guo et al., 2010; Iwakawa and Tomari, 2015).
In eukaryotic cells, the processing, e.g. capping, splicing, and 3′ end formation, of precursor messenger RNA (pre-mRNA) is closely coupled to its transcription by RNA polymerase II (Pol II) (Bentley, 2014). Co-transcriptional processing is thought to enhance the efficiency and accuracy of pre-mRNA maturation. Accumulating studies have suggested that the processing of pri-miRNAs can also occur co-transcriptionally. In mammals, many pri-miRNAs reside in the introns of other protein coding transcripts and intronic miRNAs are processed before splicing that occurs co-transcriptionally (Kim and Kim, 2007; Morlando et al., 2008). Chromatin immunoprecipitation (ChIP) reveals that Drosha-DGCR8 complex associates with several actively transcribed miRNA loci (Ballarino et al., 2009; Morlando et al., 2008). Moreover, co-transcriptional miRNA processing is also supported by native elongating transcript sequencing (mNET-seq) studies (Nojima et al., 2015). On the other hand, artificial retention of pri-miRNA transcripts on the chromatin, e.g. by blocking 3′ end formation, greatly enhances miRNA production in vivo (Pawlicki and Steitz, 2008). Both the kinetics and efficiency of pri-miRNA processing are enhanced when coupled to pri-miRNA transcription by Pol II in the nuclear extract of HeLa cells (Yin et al., 2015). However, the molecular mechanisms of co-transcriptional miRNA processing are poorly understood. It is also uncertain whether specific chromatin factors regulate co-transcriptional miRNA processing.
Histone H1 plays an essential role in the establishment and maintenance of higher order chromatin structure. The nucleosome-the basic unit of chromatin-consists of ~147 base pairs of DNA wrapping around an octamer comprised of two copies of core histones H2A, H2B, H3 and H4 (Kornberg, 1974). Linker histone H1 binds to the nucleosomes as well as the DNA linking nucleosomes to facilitate the compaction of chromatin from the “beads on a string” configuration into a 30 nm chromatin fiber (Hergeth and Schneider, 2015). Intriguingly, the H1 family is the fastest evolving among all histone families, which has expanded from single H1 in yeast or fly to eleven H1 subtypes in mammals. These H1 subtypes exhibit different developmental and tissue-specific expression patterns and may have redundant as well as specific functions (Hergeth and Schneider, 2015).
The heterochromatin protein 1 binding protein 3 (HP1BP3), also known as HP1-BP74, has recently been identified as a histone H1-related protein that also binds to the nucleosomes and protect linker DNA from degradation by micrococcal nuclease (Garfinkel et al., 2015b; Hayashihara et al., 2010). Here, we find that HP1BP3, but not canonical H1 variants, specifically associates with the Microprocessor and promotes global miRNA biogenesis in human cells. ChIP analysis reveals co-localization of HP1BP3/Drosha across the genome and HP1BP3-dependent Drosha binding to actively transcribed miRNA loci. Furthermore, we demonstrate that HP1BP3 binds both DNA and pri-miRNA and enhances co-transcriptional miRNA processing via chromatin retention of nascent pri-miRNA transcripts. Our study not only expands the functional repertoire of the H1 family of proteins, but also suggests the existence of a class of chromatin retention factors for widespread co-transcriptional miRNA processing.
RESULTS
Histone H1 binds pri-miRNA and promotes miRNA biogenesis in Drosophila cells
We purified Drosophila histone H1 as a potent pri-miRNA binding protein from the nuclear extract of Schnider 2 (S2) cells by biochemical fractionation (Figure S1A and S1B). Accordingly, recombinant fly H1, but not core histone H2A, H3 or H4, efficiently bound pri-miRNA in vitro (Figure S1C and S1D). RNA immunoprecipitation (RIP) showed that Flag-H1 could specifically bind endogenous pri-miRNA transcripts in vivo (Figure S1E). Knockdown of H1 expression by RNA interference (RNAi) did not reduce pri-miRNA expression, but decreased miRNA levels in S2 cells (Figure S1F and S1G), suggesting that histone H1 is probably involved in miRNA biogenesis in Drosophila cells.
HP1BP3 is involved in miRNA biogenesis in human cells
Because it was difficult to dissect the specific role of single fly H1 in miRNA processing from its other essential functions, we decided to test our hypothesis in the human system, which includes seven somatic H1 (H1.1, H1.2, H1.3, H1.4, H1.5, H1.0, H1x), three testis H1 (H1t, H1T2, HILS1), and one ovary H1 (H1oo). Additionally, HP1BP3 can be viewed as the twelfth H1 variant because of its high sequence and functional similarity to classical H1 proteins (Garfinkel et al., 2015b; Hayashihara et al., 2010). The H1 family of histones typically consists of a central winged helix globular domain (GD) surrounded by a variable amino (N)-terminal tail and a carboxyl (C)-terminal lysine rich tail (Hergeth and Schneider, 2015). By contrast, HP1BP3 contains three GD-repeats flanked by lysine rich sequences. Sequencing alignment and phylogenetic tree analysis suggests that the three GDs of HP1BP3 are more closely related to the GD of fly H1 or H1oo than other human H1 variants (Figure S2A, Figure 1A).
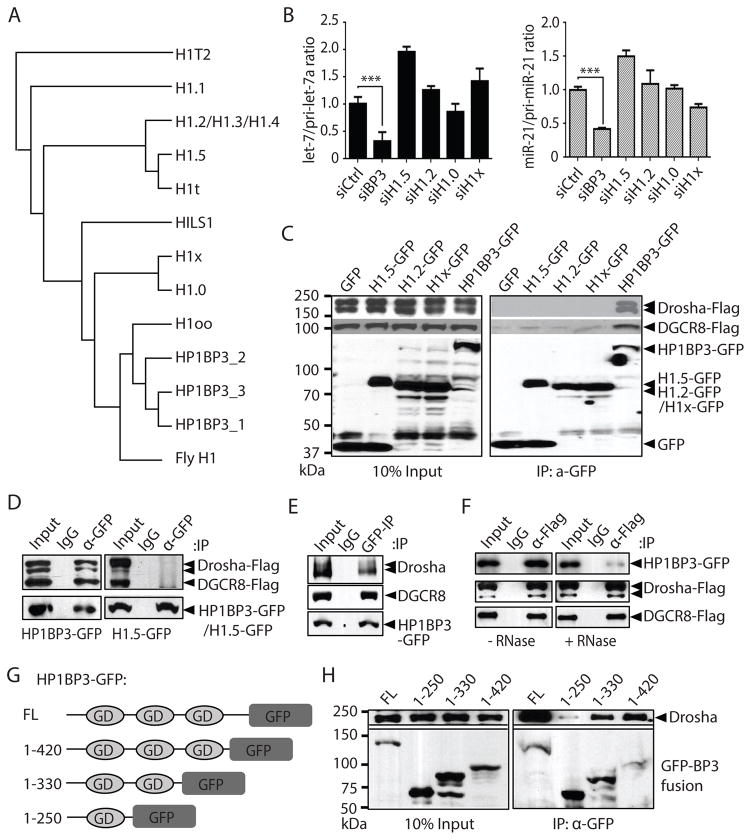
Figure 1. HP1BP3 specifically associates with the microprocessor and promotes miRNA biogenesis.
(A) A phylogenetic tree showing that the relative evolutionary distance among HP1BP3, eleven human H1 variants and fly H1 based on sequence homology of the central globular domains.
(B) Following siRNA-mediated knockdown of HP1BP3, H1.0, H1.2, H1.5, or H1x in HeLa cells, the ratio of let-7a/pri-let-7a or miR-21/pri-miR-21 was measured by Taqman qPCR, respectively (triplicate samples, data shown as mean ± SD). Paired t-test was used for statistical analysis (*** p< 0.001).
(C) HeLa cells were co-transfected with constructs expressing GFP-tagged HP1BP3, H1.2, H1.5 or H1x and Flag-tagged Drosha-DGCR8 complex followed by co-IP using anti-GFP antibody. Western blotting (WB) was performed with anti-Flag and anti-GFP antibodies, respectively.
(D) HP1BP3-GFP (left) or H1.5-GFP (right) BAC transgenic HeLa cells were transfected with Flag-tagged Drosha-Flag and DGCR8-Flag constructs followed by co-IP using anti-GFP antibody. Western blotting was performed with anti-Flag and anti-GFP antibodies.
(E) From HP1BP3-GFP BAC transgenic HeLa cells, co-IP of HP1BP3-GFP was performed with anti-GFP antibody followed by Western blotting to detect HP1BP3-GFP and endogenous Drosha or DGCR8 proteins with corresponding antibodies.
(F) HP1BP3-GFP BAC transgenic HeLa cells were transfected with the Drosha-Flag and DGCR8-Flag constructs. Cell lysates were untreated or treated with RNase A followed by co-IP with anti-Flag antibody and western blotting was performed with anti-GFP and anti-Flag antibodies.
(G) A schematic diagram of various truncated HP1BP3-GFP constructs.
(H) After co-transfection of HeLa cells with Drosha-Flag, DGCR8-Flag, and GFP-tagged HP1BP3 constructs, co-IP was performed with anti-GFP antibody followed by Western blotting with anti-Flag and anti-GFP antibodies.
To identify human ortholog of fly H1 in miRNA biogenesis, we performed siRNA-mediated knockdown of HP1BP3, H1.0, H1.2, H1.5 or H1x in HeLa cells. The efficiency of target mRNA knockdown was measured by quantitative RT-PCR (Figure S2B). Consistent with phylogenetic tree analysis, knockdown of HP1BP3, but not other H1 variants, significantly reduced the ratio of mature let-7a and miR-21 relative to pri-let-7a and pri-miR-21, respectively (Figure 1B). A similar result was obtained when HP1BP3 was depleted in human osteosarcoma (U2OS) cells (Figure S2C and S2D). These studies suggest that HP1BP3 is a histone H1-like protein that is specifically involved in miRNA processing in human cells.
HP1BP3 specifically associates with the Microprocessor
After co-transfecting HeLa cells with constructs expressing Flag-tagged Drosha-DGCR8 complex and GFP-tagged HP1BP3, H1.0, H1.2 or H1.5, co-immunoprecipitation (IP) studies showed that Drosha-DGCR8 complex specifically associated with HP1BP3, but not other H1 variants (Figure 1C). Furthermore, we constructed bacterial artificial chromosome (BAC) transgenic HeLa cell lines that stably expressed a C-terminal GFP-tagged Drosha, HP1BP3 or H1.5 protein (Figure S2E). The use of BACs that harbor large (>150 kb) genomic regions encompassing all exons, introns and regulatory regions allows for near physiological expression of transgenes as previously demonstrated (Kittler et al., 2005; Kittler et al., 2013; Poser et al., 2008). All three GFP-tagged proteins were localized in the nucleus and, as expected for chromatin factors, both H1.5-GFP and HP1BP3-GFP lighted up the mitotic chromosomes (Figure S2F). Reciprocal co-IP experiments indicated that HP1BP3-GFP, but not H1.5-GFP, specifically associated with Flag-tagged and endogenous Drosha-DGCR8 complex in the BAC transgenic cells (Figure 1D–1F). This association was sensitive to RNase treatment (Figure 1F), suggesting that HP1BP3 associates with the Microprocessor in a RNA-dependent manner.
We generated a series of truncated HP1BP3-GFP constructs to investigate whether the three GD domains of HP1BP3 are important for its association with the Microprocessor in vivo. Co-IP experiments showed that Flag-tagged Drosha-DGCR8 complex exhibited weak, medium and strong association, respectively, with GFP-tagged HP1BP3-1GD (1-250), 2GD (1-330) and 3GD (1-420) fusion proteins (Figure 1G and 1H). These results suggest that the three GDs of HP1BP3 function cooperatively to mediate its interaction with the Microprocessor in vivo.
HP1BP3 is involved in the processing, not the transcription, of pri-miRNAs
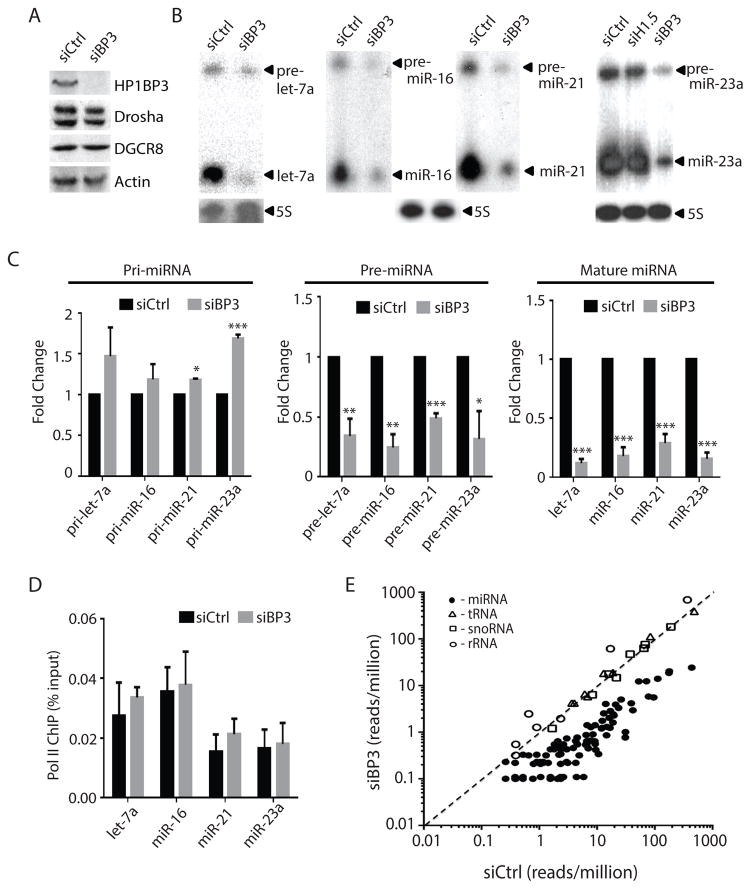
To investigate whether HP1BP3 affected pri-miRNA processing, we compared the levels of pri-, pre-, and mature miRNAs for let-7a, miR-16, miR-21 and miR-23a between the control siRNA (siCtrl) and HP1BP3 siRNA (siBP3) treated HeLa cells (Figure 2A–2C). Consistently, both pre-miRNAs and mature miRNAs were significantly reduced, but pri-miRNA levels remained unchanged or increased upon depletion of HP1BP3 (Figure 2B and 2C). This miRNA defect in the HP1BP3-depleted cells could be rescued by transfection of a siRNA-resistant HP1BP3 construct (Figure S3), confirming that this was not an off-target effect. Additionally, HP1BP3 depletion did not affect Drosha or DGCR8 expression, suggesting that the miRNA reduction was not due to insufficient level of Microprocessor (Figure 2A). Neither did HP1BP3 depletion affect Pol II occupancy at the promoters of miRNA genes (Figure 2D). Taken together, these results suggest that HP1BP3 is specifically involved in the processing, not the transcription, of pri-miRNAs.
Figure 2. HP1BP3 promotes pri-miRNA processing and global miRNA biogenesis.
(A) Western blotting was performed using the corresponding antibodies to detect HP1BP3, Drosha, DGCR8, and Actin proteins in the control and HP1BP3-depleted HeLa cell extracts.
(B) Northern blotting was performed to compare the levels of let-7a, miR-16, miR-21, miR-23a between the control and HP1BP3-depleted HeLa cells. 5S RNA was used as a loading control.
(C) Quantitative analysis of the expression fold change of pri-miRNA (by Taqman PCR), pre-miRNA and miRNA (by Northern blotting) between the control and HP1BP3-depleted HeLa cells (triplicate samples, data shown as mean ± SD). Unpaired t-test was used for statistical analysis (*** p< 0.001, ** p <0.01, * p<0.05).
(D) Pol II ChIP analysis comparing the Pol II occupancy at the promoter regions of let-7a, miR-16, miR-21, and miR-23a genes between the control and HP1BP3-depleted HeLa cells (triplicate samples, data shown as mean ± SD).
(E) Small RNA sequencing was performed to compare global miRNA expression profile between the control and HP1BP3-depleted HeLa cells. Each black circle represents an annotated miRNA expressed in HeLa cells. Each white triangle, square, and circle represents a selected tRNA, snoRNA, and rRNA, respectively. Diagonal line marks the 1:1 ratio of siCtrl and siBP3 samples.
HP1BP3 promotes global miRNA biogenesis
We performed massively parallel sequencing of 15 to 40-nt small RNAs (sRNA-Seq) to compare global miRNA expression profile between the control and HP1BP3-depleted HeLa cells (Figure 2E). We analyzed two independent libraries for the siCtrl and siBP3 samples, respectively. Sequencing reads were mapped to unique sites in the human genome (hg19) using Bowtie (v.2.2.5) (Langmead et al., 2009). Whereas the majority of small RNAs represent fragments of rRNA, tRNA and snoRNA, ~0.2% of the reads correspond to bona fide miRNAs according to a 523 re-annotated human miRNA list (Fromm et al., 2015). Although the abundance of most of small RNAs remained unchanged, 148 of 164 of expressed miRNAs were reduced, of which 68 miRNAs showed >2 fold reduction in the HP1BP3-depleted cells (Figure 2E and Table S1). The remaining 16 miRNAs showed a similar trend, but the results were inconclusive due to low counts. Thus, HP1BP3 promotes global miRNA biogenesis in human cells.
Genome-wide co-localization of HP1BP3 & Drosha at active miRNA loci
To determine whether HP1BP3 was involved in co-transcriptional miRNA processing, we took advantage of our BAC transgenic cell lines to compare the chromatin binding maps of HP1BP3-GFP, H1.5-GFP and Drosha-GFP by ChIP and massively parallel sequencing (ChIP-Seq). As reported for somatic H1 variants (Millan-Arino et al., 2014), H1.5-GFP showed a basal level of mostly uniform binding throughout the genome (Figure S4A). By contrast, HP1BP3-GFP was enriched at 9,149 islands across the genome, suggesting a different chromatin binding mode from that of H1.5-GFP. Similarly, Drosha-GFP was enriched at 5,814 islands, including many non-miRNA loci, which was consistent with previous reports that Drosha could also cleave mRNA with long hairpins and regulate other biological processes (Gromak et al., 2013; Han et al., 2009; Kadener et al., 2009; Knuckles et al., 2012).
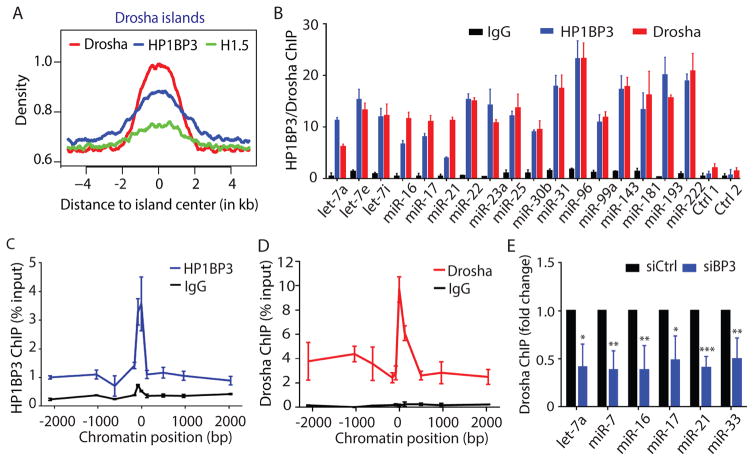
We used 5,000 base pair (bp) regions flanking the center of islands to generate density plots showing Drosha, HP1BP3 and H1.5 ChIP signals across all Drosha islands (Figure 3A). The analysis revealed a significant overlap in the overall binding sites between HP1BP3 and Drosha, but not between H1.5 and Drosha (odds ratio=1.32, p value=2.97e-5). A similar result was obtained when we compared the chromatin binding maps of HP1BP3-GFP and H1.5-GFP with endogenous Drosha protein using a ChIP-grade anti-Drosha antibody (data not shown). The genome-wide chromatin co-localization of HP1BP3 and Drosha suggests an important functional link between these two proteins.
Figure 3. Genome-wide co-localization of HP1BP3 and Drosha at active miRNA loci.
(A) Density map showing the enrichment of Drosha, HP1BP3, and H1.5 cumulative ChIP signal across all Drosha islands. X-axis represents the distance to island center. Y-axis represents per base read coverage.
(B) Individual ChIP analysis showing chromatin co-localization of HP1BP3-GFP and Drosha at the stem-loop regions of seventeen active miRNA loci in HeLa cells (triplicate samples, data shown as mean ± SD).
(C, D) Comprehensive ChIP analysis comparing chromatin co-localization of HP1BP3-GFP (C) and Drosha-GFP (D) in a 4 kb region surrounding the stem-loop region of the let-7a-1 locus.
(E) Individual ChIP analysis comparing chromatin binding of Drosha at six active miRNA loci between the control and HP1BP3-depleted HeLa cells (quadruplet samples, data shown as mean ± SD). Paired t-test was used for statistical analysis (*** p< 0.001, ** p <0.01, * p<0.05).
Previous studies have examined chromatin binding of Drosha or DGCR8 proteins at a few miRNA genes (Gromak et al., 2013; Morlando et al., 2008; Pawlicki and Steitz, 2008). Thus, it is uncertain whether all pri-miRNAs undergo co-transcriptional miRNA processing. In our ChIP-Seq data, HP1BP3-GFP bound to 42 expressed miRNA loci, and Drosha/Drosha-GFP bound to 18 expressed miRNA loci (Table S2), which was consistent with previous finding that Drosha preferentially bound to actively transcribed miRNA loci (Morlando et al., 2008). We confirmed that HP1BP3 also preferred to bind to active (e.g. let-7a, miR-16, miR-17, miR-21), but not silent (e.g. miR-1, miR-9) miRNA loci (Figure S4B). Importantly, HP1BP3 also bound to thirteen (~72%) of eighteen Drosha-bound miRNA loci (Table S2), indicative of a significant overlap of Drosha/HP1BP3 binding at active miRNA loci.
We suspected that our ChIP-Seq analysis underestimated the number of Drosha or HP1BP3-bound miRNA loci. Thus, we expanded individual ChIP analysis to examine the binding of HP1BP3-GFP and Drosha at the stem-loop regions of seventeen miRNA loci that were expressed at high or moderate levels (Figure 3B). Without an exception, HP1BP3 and Drosha showed co-binding to all seventeen miRNA loci (Figure 3B). By choosing one miRNA to represent every expressed polycistronic miRNA gene (Figure S4C), our combined ChIP-Seq and ChIP analyses indicated genome-wide co-localization of HP1BP3 and Drosha at a total of 42 actively transcribed miRNA loci (Table S2). These results, together with our finding that HP1BP3 promotes global miRNA biogenesis (Figure 2E), suggest that HP1BP3 plays a key role in co-transcriptional processing of most if not all of pri-miRNA transcripts.
Furthermore, we performed extensive ChIP analysis to compare the chromatin binding patterns of HP1BP3-GFP and Drosha at the let-7a-1 miRNA locus. Interestingly, both HP1BP3-GFP and Drosha showed a single chromatin-binding peak at the stem-loop region of let-7a-1 locus (Figure 3C and 3D). Furthermore, knockdown of HP1BP3 expression resulted in a significant reduction in Drosha ChIP signals at all six miRNA loci that we examined in HeLa cells (Figure 3E). These observations suggest that HP1BP3 may help recruit or retain the Microprocessor at active miRNA loci for co-transcriptional pri-miRNA processing.
HP1BP3 exhibits a specific pri-miRNA binding activity
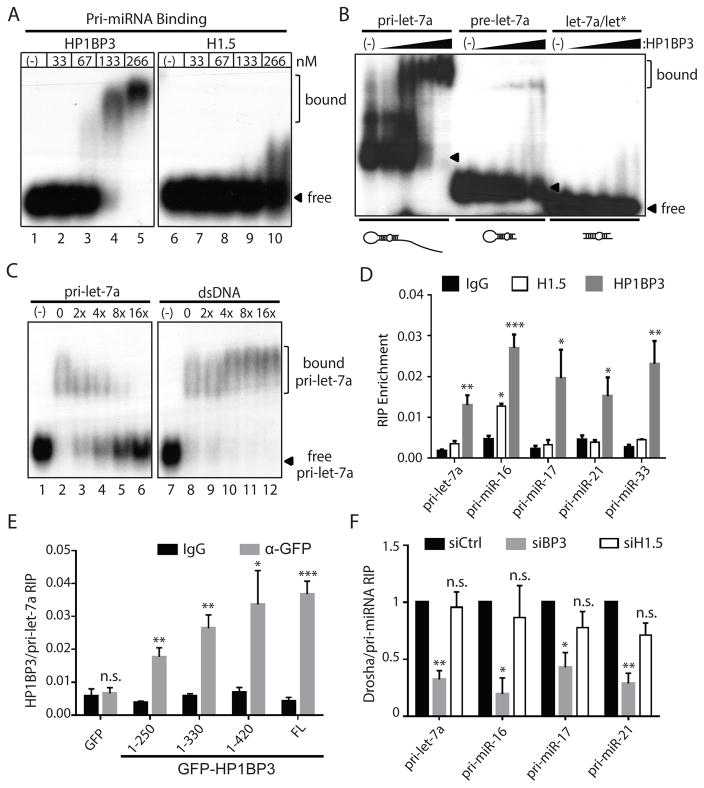
As previously reported (Hayashihara et al., 2010), His6-tagged HP1BP3 and H1.5 recombinant proteins exhibited double-stranded DNA (dsDNA) binding activity (Figure S5A and S5B). Unexpectedly, we found that recombinant HP1BP3, but not H1.5, also exhibited a specific pri-miRNA-binding activity in vitro (Figure 4A). Whereas pre-miRNA typically consists of a terminal loop and a ~22 bp imperfect double-stranded stem with 2-nt 3′ overhang, the corresponding pri-miRNA carries a longer (e.g. ~33 bp) stem and two single-stranded tails. Recombinant HP1BP3 efficiently bound to pri-miRNA and, to a lesser extent, pre-miRNA, but not duplex miRNA (Figure 4B), suggesting that HP1BP3 probably recognize the shared stem-loop structure of pri-/pre-miRNA. Neither pre-miRNA nor miRNA duplex could compete off binding of radiolabeled pri-miRNA by HP1BP3 (Figure S5C). Thus, HP1BP3 displayed a higher affinity for pri-miRNA than for pre-miRNA, suggesting that the single-stranded tails of pri-miRNA (absent in pre-miRNA) could also contribute critically to HP1BP3 binding.
Figure 4. HP1BP3 specifically binds pri-miRNA and promotes the Drosha/pri-miRNA association in vivo.
(A) Native gel-shift assays were performed by incubating 5′ radiolabeled pri-let-7a RNA in buffer alone or with increasing concentration (33, 67, 133, and 266nM) of recombinant HP1BP3 (lanes 1–5) and H1.5 (lanes 6–10). Arrowhead marks free pri-let-7a RNA substrate and bracket corresponds to the HP1BP3-bound pri-let-7a RNA.
(B) Native gel-shift assays were performed by incubating 5′ radiolabeled pri-let-7a, pre-let-7a, and let7a/let* duplex RNA in buffer alone, or with increasing concentration (38nM, 75nM, 150nM, and 300nM) of recombinant HP1BP3. Arrowhead marks free RNA substrate, whereas bracket corresponds to HP1BP3-bound pri-miRNA or pre-miRNA.
(C) Native gel-shift assays were performed by using excess non-radiolabeled pri-let-7a RNA or dsDNA to compete with binding of radiolabeled pri-let-7a RNA by recombinant HP1BP3.
(D) RNA immuneprecipitation (RIP) assays were performed to measure in vivo association of H1.5-GFP and HP1BP3-GFP with endogenous pri-miRNA transcripts (triplicate experiments, data shown as mean ± SD). Paired t-test was used for statistical analysis (*** p< 0.001, ** p <0.01, * p<0.05).
(E) RIP assays were performed to measure in vivo association of full-length and various truncated HP1BP3-GFP constructs with endogenous pri-let-7a in HeLa cells (triplicate samples, data shown as mean ± SD). Paired t-test was used for statistical analysis (*** p< 0.001, ** p <0.01, * p<0.05).
(F) RIP assays were performed to measure in vivo association of endogenous Drosha with pri-miRNAs between the control and HP1BP3-depleted or H1.5-depleted HeLa cells (triplicate samples, data shown as mean ± SD). Paired t-test was used for statistical analysis (*** p< 0.001, ** p <0.01, * p<0.05).
To investigate whether HP1BP3 bound dsDNA and pri-miRNA via the same mechanism, we performed in vitro competition experiments by using non-radiolabeled pri-miRNA or dsDNA of the same sequence to compete with binding of radiolabeled pri-miRNA by recombinant HP1BP3. Although dsDNA could not compete off binding of radiolabeled pri-miRNA (Figure 4C), addition of dsDNA upshifted the HP1BP3-pri-miRNA complex (Figure 4C), suggesting that HP1BP3 may bind dsDNA and pri-miRNA simultaneously via different mechanisms.
Following in vivo formaldehyde crosslinking of the RNA/protein (RNP) complexes, we immunoprecipitated HP1BP3-GFP or H1.5-GFP from the BAC transgenic HeLa cells and measured the HP1BP3- or H1.5-associated endogenous pri-miRNAs by Taqman qPCR. This RNA immunoprecipitation (RIP) experiment showed that HP1BP3-GFP associated with all five pri-miRNAs that we examined, whereas H1.5-GFP moderately associated with pri-miR-16, but not four other pri-miRNAs (Figure 4D). Moreover, GFP-tagged HP1BP3 1-250, 1-330 and 1-420 truncated proteins showed increasing affinity for endogenous pri-miRNAs, suggesting that the three GDs of HP1BP3 function cooperatively to bind pri-miRNAs in vivo (Figure 4E). Taken together, these results suggest that HP1BP3 exhibits a specific pri-miRNA binding activity in vitro and in vivo.
HP1BP3 promotes the Drosha/pri-miRNA association in vivo
Furthermore, we compared by RIP the in vivo association of endogenous Drosha and pri-miRNA transcripts between the control and HP1BP3 or H1.5-depleted HeLa cells. Consistently, depletion of HP1BP3, but not depletion of H1.5, resulted in a significant reduction in Drosha binding to all four endogenous pri-miRNAs that we examined (Figure 4F). These results indicate that HP1BP3 promotes the Drosha/pri-miRNA association in vivo. Next, we performed native gel-shift assays to examine whether HP1BP3 directly enhance the binding of Drosha-DGCR8 complex to pri-miRNA in vitro (Figure S5D). Although HP1BP3 did not enhance pri-miRNA binding, it resulted in a supershift of the Drosha-DGCR8/pri-miRNA complex (Figure S5D), suggesting that HP1BP3 and Drosha-DGCR8 could co-occupy the same pri-miRNA to form a higher order complex. In our reconstitution system, HP1BP3 also slightly inhibited the pri-miRNA processing activity of recombinant Drosha-DGCR8 complex (Figure S5E). However, it should be noted that the lack of chromatin component and/or use of truncated Drosha/DGCR8 proteins could explain the inability to fully recapitulate the physiological activity of HP1BP3 in vitro.
HP1BP3 promotes chromatin retention of nascent pri-miRNA transcripts
One plausible explanation was that knockdown of HP1BP3 could cause the premature release of pri-miRNA transcripts from the chromatin, resulting in diminished Drosha/pri-miRNA association. In fact, Steitz and colleagues previously showed that pri-miRNA transcripts retained on chromatin, e.g. by inhibition of 3′ polyadenylation, were more efficiently processed by the Microprocessor (Pawlicki and Steitz, 2008). Inspired by this study, we used the same strategy to test our hypothesis that HP1BP3 promote chromatin retention of nascent pri-miRNA transcripts by comparing ectopic miRNA expression from transfected Pri-lin-4 or Pri-lin-4ΔpA constructs with or without the “AAUAAA” cleavage and polyadenylation (CPA) signal (Figure 5A).
Figure 5. HP1BP3 facilitate pri-miRNA processing by enhancing its chromatin retention.
(A) A schematic diagram of the Pri-lin-4 and Pri-lin-4ΔpA constructs (Pawlicki et al., 2008).
(B, C) Semi-quantitative (B, by RT-PCR and agarose gel electrophoresis) and quantitative (C, by Taqman qPCR) analysis of the pri-lin-4 and mature lin-4 expression from transfected Pri-lin-4 and Pri-lin-4ΔpA constructs between the control and HP1BP3-depleted HeLa cells (triplicate experiments, data shown as mean ± SD). Paired t-test was used for statistical analysis (*** p< 0.001).
(D) A schematic diagram for the chromatin and nucleoplasm fractionation procedure.
(E, F) Semi-quantitative RT-PCR (top) and Taqman qPCR (bottom) were performed to measure the abundance of chromatin-associated (E) and nucleoplasmic (F) pri-lin-4 transcripts expressed from transfected Pri-lin-4 and Pri-lin-4ΔpA constructs between the control and HP1BP3-depleted HeLa cells (triplicate samples, data present as mean ± SD). Paired t-test was used for statistical analysis (*** p< 0.001, ** p <0.01, * p<0.05).
(G, H) Taqman qPCR were performed to measure of the relative abundance of endogenous pri-miRNA and histone H4 transcripts in the chromatin (G) and nucleoplasm (H) fractions between the control and HP1BP3-depleted HeLa cells (quadruplet samples, data present as mean ± SD). Notably, primers for amplifying the introns of nascent pri-miR-25 and pri-miR-140 transcripts were used. 18S rRNA was used as a loading control. Paired t-test was used for statistical analysis (*** p< 0.001, ** p <0.01, * p<0.05).
We first found that HP1BP3 associated with the transfected Pri-lin-4 plasmid and was required for lin-4 expression from Pri-lin-4 in HeLa cells (Figure S6A and S6B). Next, we compared the expression of pri-lin-4 and mature lin-4 from the Pri-lin-4 and Pri-lin-4ΔpA constructs, respectively, in the control and HP1BP3-depleted HeLa cells (Figure 5B and 5C). Depletion of HP1BP3 did not affect pri-lin-4 expression, but reduced lin-4 production from Pri-lin-4. In contrast, neither pri-lin-4 nor lin-4 expression from Pri-lin-4ΔpA was affected by HP1BP3 depletion. These results are consistent with our hypothesis that HP1BP3 promotes chromatin retention of pri-lin-4 transcript to enhance co-transcriptional pri-lin-4 processing. For Pri-lin-4ΔpA, artificial retention of pri-lin-4 transcript at the transcription site, due to deletion of CPA, allows for efficient co-transcriptional pri-lin-4 processing in the absence of HP1BP3.
To directly measure chromatin retention of pri-miRNA, we isolated the chromatin and nucleoplasm fractions from the control and HP1BP3-depleted HeLa cells (Gagnon et al., 2014; Morlando et al., 2008; Pawlicki and Steitz, 2008), and quantified the relative abundance of pri-lin-4 in either fraction by Taqman qPCR (Figure 5D–5F). For the Pri-lin-4 construct, knockdown of HP1BP3 resulted in a 2 to 4-fold reduction of pri-lin-4 from the chromatin as well as a >2-fold increase in pri-lin-4 in the nucleoplasm. Since the total amount of pri-lin-4 was unchanged (Figure 5C), depletion of HP1BP3 likely resulted in the premature release of pri-lin-4 transcript from the chromatin into the nucleoplasm. For the Pri-lin-4ΔpA construct, depletion of HP1BP3 did not change the abundance of pri-lin-4 in either the chromatin or nucleoplasm fraction. These results confirm that HP1BP3 helps retain pri-lin-4 transcript at the transcription site to enhance co-transcriptional pri-lin-4 processing.
Furthermore, we showed that depletion of HP1BP3 resulted in a significant reduction of seven endogenous pri-miRNAs from the chromatin and their concomitant increase in the nucleoplasm (Figure 5G and 5H). We also verified that nascent pri-miRNA transcripts were prematurely released from the chromatin by using PCR primers spanning the introns of pri-miR-25 and pri-miR140 (Figure 5G and 5H). By contrast, the abundance of histone H4 transcript remained the same in the chromatin and nucleoplasm fractions in HP1BP3-depleted cells (Figure 5G and 5H). Knockdown of H1.2 or H1.5 did not significantly affect the levels of pri-miRNAs in either the chromatin or nucleoplasm fractions (Figure S6C–S6E). Taken together, these series of experiments strongly suggest that HP1BP3 promotes co-transcriptional miRNA processing through chromatin retention of nascent pri-miRNA transcripts.
DISCUSSION
Chromatin retention factors for co-transcriptional miRNA processing
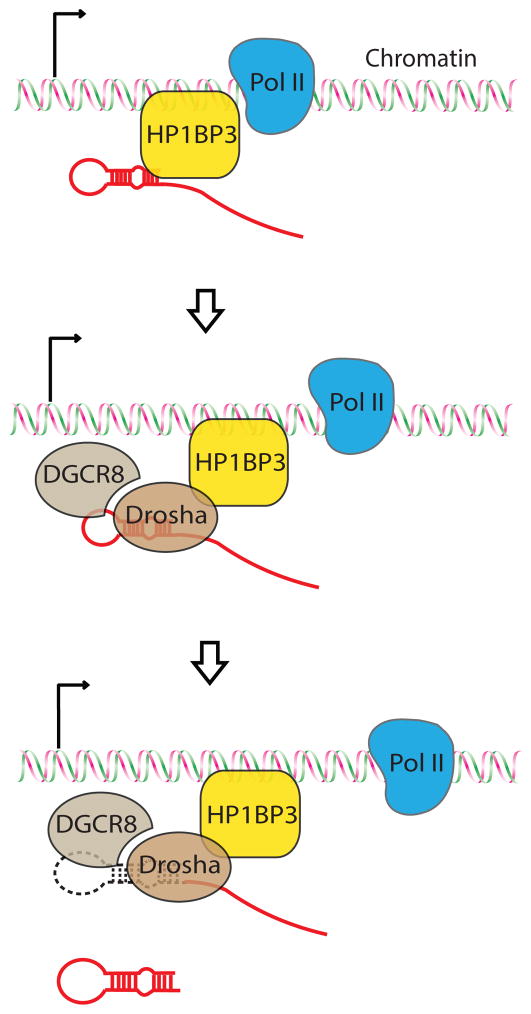
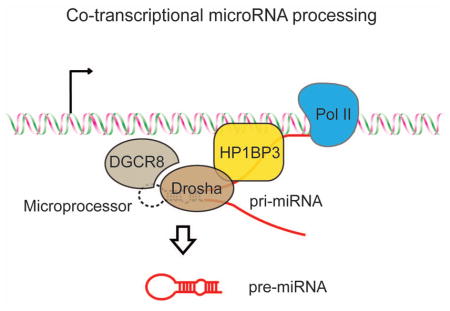
Based on our findings, we propose a working model for co-transcriptional miRNA processing in mammalian cells (Figure 6): First, the pri-miRNA transcript is transcribed by RNA polymerase II (Pol II) from the miRNA gene. Second, HP1BP3 binds chromatin DNA and nascent pri-miRNA transcript to help retain pri-miRNA on chromatin. Third, retention of pri-miRNA at the site of transcription provides more opportunity for the Drosha-DGCR8 complex to bind pri-miRNA on chromatin. Since HP1BP3 associates with both Drosha/DGCR8 and pri-miRNA, HP1BP3 may actively facilitate the Microprocessor to bind or process pri-miRNA co-transcriptionally in vivo. In the absence of HP1BP3, however, pri-miRNA transcripts are prematurely released from the chromatin and co-transcriptional pri-miRNA processing is compromised. In the nucleoplasm, the released pri-miRNA transcripts may become vulnerable to nuclease-mediated decay, or localize in regions, such as the SC35 bodies, that prevent efficient binding or processing by the Microprocessor (Pawlicki and Steitz, 2009). Furthermore, we found that Drosophila H1 exhibited a similar pri-miRNA binding activity and promoted miRNA biogenesis in S2 cells (Figure S1). Thus, it will be interesting for future studies to investigate whether histone H1 and related proteins play a conserved role in co-transcriptional miRNA processing in flies, worms and other organisms.
Figure 6. A working model for co-transcriptional pri-miRNA processing.
A schematic diagram showing that HP1BP3 promotes co-transcriptional pri-miRNA processing through chromatin retention of nascent pri-miRNA transcript.
Regulation of co-transcriptional pri-miRNA processing
Recent studies have identified a number of proteins that regulate pri-miRNA processing. For example, transcription activators, such as SMADs and p53, promote the processing of a specific subset of pri-miRNAs through interaction with DDX5/p68 helicase that associates with the Microprocessor (Davis et al., 2008; Suzuki et al., 2009). RNA-binding proteins, such as LIN28, hnRNPA1 and KSRP, recognize the terminal loop of a subset of pri-miRNAs and inhibit or enhance their processing by the Microprocessor (Guil and Caceres, 2007; Newman et al., 2008; Trabucchi et al., 2009). In most of these studies, it is unclear whether these regulatory events occur on the chromatin or in the nucleoplasm. Because our studies suggest that most pri-miRNA transcripts undergo co-transcriptional miRNA processing, a logical prediction would be that many of these regulators should actually regulate co-transcriptional miRNA processing on chromatin. It is entirely plausible for transcription activators (e.g. SMADs and p53) to bind specific chromatin elements and recruit the Microprocessor to adjacent pri-miRNA transcripts to facilitate co-transcriptional miRNA processing. LIN28 has recently been shown to bind nascent pri-let-7 transcript co-transcriptionally and inhibits Drosha-mediated pri-let-7 processing in C. elegans and human ES cells (Van Wynsberghe et al., 2011). Moreover, FUS/TLS (fused in sarcoma/translocated in liposaroma) protein, an RNA binding protein that is linked to Amyotrophic Lateral Sclerosis (ALS), stimulates co-transcriptional miRNA processing by facilitating Drosha recruitment to specific miRNA loci (Morlando et al., 2012). It is possible that some of these RNA binding proteins may also possess DNA binding activity or interacts with chromatin factors to promote co-transcriptional miRNA processing. Therefore, mounting evidence suggest an emerging theme that the chromatin regulation of co-transcriptional miRNA processing is a widespread and conserved mechanism in eukaryotes.
Essential roles of HP1BP3 in miRNA processing during development
HP1BP3 is ubiquitously expressed in all somatic tissues and is highly enriched in the brain (Garfinkel et al., 2015b). Knockout of HP1BP3 in mice causes early post-natal lethality (~60% pups die within 24 hours after birth) (Garfinkel et al., 2015a; Garfinkel et al., 2015b). Homozygous Hp1bp3−/− mice that survive to weaning are fertile and have a normal life span, but are significantly smaller than their littermates since birth (Garfinkel et al., 2015a; Garfinkel et al., 2015b). These mutant mice show proportional reduction in body weight, body length and organ weight as well as severe impairment in bone development. However, wild-type and Hp1bp3−/− mouse embryonic fibroblast (MEF) exhibit a similar sensitivity of chromatin to micrococcal nuclease digestion (Garfinkel et al., 2015b). Moreover, the lack of HP1BP3 only impacts a limited set of gene expression (Garfinkel et al., 2015b). These observations suggest that the phenotypes of Hp1bp3−/− mice are probably not due to a global defect in chromatin organization. Rather, that HP1BP3 promotes global miRNA processing provides a likely explanation for the lethality and growth defects of Hp1bp3−/− mice.
Conclusions
The H1 family of linker histones is often assumed to be non-specific DNA binding proteins with prosaic and well-understood functions. However, it is still a mystery why there is a major expansion from single H1 in flies to eleven H1 variants plus HP1BP3 in mammals. Because of their proximity to chromatin, histone H1 and related proteins are well positioned to play important roles in various transcriptional and co-transcriptional processes. The possibility of unanticipated functions of various linker histones is underscored by our surprising discovery that HP1BP3 possesses a specific pri-miRNA binding activity and promotes co-transcriptional miRNA processing in human cells. The current work significantly expands the functional repertoire of the H1 family of proteins, and suggests a mechanism that chromatin factors retain nascent transcripts at the site of transcription to enhance co-transcriptional RNA processing.
MATERIALS AND METHODS
Northern blotting
Northern blotting was performed to measure expression of pre- and mature miRNAs in the control and H1.5 or HP1BP3-depleted HeLa cells as previously described (Liang et al., 2013). In brief, 30 μg total RNA was resolved by 12% Urea-PAGE, transferred to GT membrane (Bio-Rad), and crosslinked by ultra violet (UV) light. The sequences of antisense RNA probes for detection of miRNAs and 5S RNA are listed in Table S7. The RNA probes were 5′ radiolabeled with γ-32P ATP by T4 polynucleotide kinase (NEB). Hybridization was carried in Ultrasensitive Hybridization solution (Ambion) at 40°C overnight. The membrane was washed 3 times at 40°C for 15 minutes with 2 ×SSC, 0.5% SDS followed by autoradiography.
Small RNA sequencing
Sequence libraries were filtered for adapter contamination using cutadapt (v1.8.3) (Martin, 2011) software tool. This software cut the adapter sequence from the sequencing reads and filtered the reads whose length is greater than or equal to 15bp for further analysis. Filtered reads were aligned to the human reference genome (hg19) using Bowtie (v.2.2.5) (Langmead et al., 2009). Reads that mapped with ≤ 2 mismatches to the reference sequence were retained for further analysis. For siBP3 replicates one and two we obtained 4,863,552 reads and 4,369,227 reads respectively, while for control replicates one and two we obtained 3,908,404 reads and 1,907,245 reads respectively. We used the 523 re-annotated miRNA list (Fromm et al., 2015), 624 tRNA regions (GenCode) and 1,769 rRNA regions (UCSC genome browser) to count the mapped reads using featurecounts (v1.4.6) module from subread package (Liao et al., 2014). These counts were normalized to library size and performed differential expression analysis using edgeR bioconductor package (v.3.8.6) (Robinson et al., 2010). Limma (v.3.22.7) (Ritchie et al., 2015) package was used to calculate differential expression change.
Chromatin immunoprecipitation (ChIP) and RNA immunoprecipitation (RIP)
ChIP and RIP were performed as described previously (Sakurai et al., 2010). Cells were crosslinked with 1% formaldehyde and then nuclear fractions were isolated. The nuclear lysates were incubated with anti-Drosha antibody (2 μg; Abcam) or rabbit IgG (2 μg; Millipore) overnight and antibody-protein-DNA/RNA complexes were recovered using protein G plus/protein A agarose beads (Millipore). After reverse crosslinking and proteinase K treatment, immunoprecipitated DNA/RNA were purified by phenol-chloroform extraction and ethanol precipitation. For RIP, the samples were treated with DNase I to remove genomic DNA and then reverse transcribed to generate cDNA. Real time RT-PCR was performed using Taqman pri-miRNA kit in RIP or designed primers for miRNA gene locus (listed in table S6).
Chromatin and nucleoplasm fractionation
Chromatin fractionation was performed as previously described (Gagnon et al., 2014). HeLa cells were lysed in hypotonic buffer (HLB) (10mM Tris pH 7.5, 10mM NaCl, 3mM MgCl2, 0.3% (vol/vol) NP-40, 1% protease inhibitor cocktail). Cell nuclei were isolated by centrifugation at 1000g for 4min and washed three times using 1mL of HLB. The nuclei were suspended in Modified Wuarin-Schibler buffer (MWS) (10mM Tris pH 7.0, 4mM EDTA, 0.3M NaCl, 1M urea, and 1% (vol/vol) NP-40) and spun at 1000g for 4min at 4 °C. While the supernatant was collected as the nucleoplasm fraction, the pellet was washed three times with 1mL MWS buffer and then collected as the chromatin fraction. RNA from nucleoplasm was precipitated by ethanol and extracted by Trizol. RNA from chromatin fraction was directly isolated by Trizol extraction. Taqman qPCR was performed to measure the relative abundance of chromatin bound and nucleoplasmic pri-miRNAs.
Supplementary Material
Acknowledgments
We thank Drs. J.A. Steitz and N.V. Kim for generously sharing reagents, Drs. Y. Liu, J. Mendell and N. Conrad for comments on the manuscript; K. Wu, S. Liu and R. Ji for technical assistance. H.L. received Sara and Frank McKnight Graduate Student Fellowship. R.K. is John L. Roach Scholar in Biomedical Research. Y.N. is Southwestern Medical Foundation Scholar in Biomedical Research. Q. L. is W.A. “Tex” Moncrief Jr. Scholar in Biomedical Research. This study is supported by grants from the Welch foundation [Q.L.(I-1608), Y.N.(I-1851)], the National Institute of Health [Q.L.(GM111367), D.R.C.(GM118103)], the Cancer Prevention Research Institute of Texas (CPRIT) [Y.N.(R1221)], and a JSPS KAKENHI grant [Q.L.(16H04740)] and the World Premium Initiative (WPI) program from Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT).
Footnotes
ACCESSION NUMBERS
The accession number for the sequencing data reported in this paper is GSE77856
AUTHOR CONTRIBUTIONS
Q.L., H.L. and C.L. conceived the project. H.L. and C.L., assisted by X.K., Z.W. and G.P.Y., performed most of the experiments. R.K. and R.K.K. conducted ChIP-Seq, RNA-Seq and bioinformatics analyses. D.R.C. and M.M. helped ChIP and RIP studies. L.N.K. and N.V.G performed sequence alignment and phylogenetic tree analysis. Y.N., B-C.J. and K.S.Y. helped in vitro reconstitution studies. Q.L. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ballarino M, Pagano F, Girardi E, Morlando M, Cacchiarelli D, Marchioni M, Proudfoot NJ, Bozzoni I. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol Cell Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annual review of genetics. 2015;49:213–242. doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell reports. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel BP, Arad S, Le PT, Bustin M, Rosen CJ, Gabet Y, Orly J. Proportionate Dwarfism in Mice Lacking Heterochromatin Protein 1 Binding Protein 3 (HP1BP3) Is Associated With Alterations in the Endocrine IGF-1 Pathway. Endocrinology. 2015a;156:4558–4570. doi: 10.1210/en.2015-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel BP, Melamed-Book N, Anuka E, Bustin M, Orly J. HP1BP3 is a novel histone H1 related protein with essential roles in viability and growth. Nucleic Acids Res. 2015b;43:2074–2090. doi: 10.1093/nar/gkv089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Gromak N, Dienstbier M, Macias S, Plass M, Eyras E, Caceres JF, Proudfoot NJ. Drosha regulates gene expression independently of RNA cleavage function. Cell reports. 2013;5:1499–1510. doi: 10.1016/j.celrep.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashihara K, Uchiyama S, Shimamoto S, Kobayashi S, Tomschik M, Wakamatsu H, No D, Sugahara H, Hori N, Noda M, et al. The middle region of an HP1-binding protein, HP1-BP74, associates with linker DNA at the entry/exit site of nucleosomal DNA. J Biol Chem. 2010;285:6498–6507. doi: 10.1074/jbc.M109.092833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hergeth SP, Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015;16:1439–1453. doi: 10.15252/embr.201540749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT, 2nd, Nelson S, Rosbash M. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Ma C, Poser I, Fischer S, Hyman AA, Buchholz F. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc Natl Acad Sci U S A. 2005;102:2396–2401. doi: 10.1073/pnas.0409861102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Zhou J, Hua S, Ma L, Liu Y, Pendleton E, Cheng C, Gerstein M, White KP. A comprehensive nuclear receptor network for breast cancer cells. Cell reports. 2013;3:538–551. doi: 10.1016/j.celrep.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Knuckles P, Vogt MA, Lugert S, Milo M, Chong MM, Hautbergue GM, Wilson SA, Littman DR, Taylor V. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nature neuroscience. 2012;15:962–969. doi: 10.1038/nn.3139. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Xiong K, Szulwach KE, Zhang Y, Wang Z, Peng J, Fu M, Jin P, Suzuki HI, Liu Q. Sjogren syndrome antigen B (SSB)/La promotes global microRNA expression by binding microRNA precursors through stem-loop recognition. J Biol Chem. 2013;288:723–736. doi: 10.1074/jbc.M112.401323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, Shen J, Liu J, Sun X, Zhao G, Chang Y, Xu L, Li X, Zhao Y, Zheng H, et al. Genome-wide identification and functional annotation of Plasmodium falciparum long noncoding RNAs from RNA-seq data. Parasitology research. 2014;113:1269–1281. doi: 10.1007/s00436-014-3765-4. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011;2011:17. [Google Scholar]
- Millan-Arino L, Islam AB, Izquierdo-Bouldstridge A, Mayor R, Terme JM, Luque N, Sancho M, Lopez-Bigas N, Jordan A. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014;42:4474–4493. doi: 10.1093/nar/gku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Dini Modigliani S, Torrelli G, Rosa A, Di Carlo V, Caffarelli E, Bozzoni I. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31:4502–4510. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Gomes T, Grosso AR, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ. Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell. 2015;161:526–540. doi: 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA. Subnuclear compartmentalization of transiently expressed polyadenylated pri-microRNAs: processing at transcription sites or accumulation in SC35 foci. Cell Cycle. 2009;8:345–356. doi: 10.4161/cc.8.3.7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Bai H, Konno T, Ideta A, Aoyagi Y, Godkin JD, Imakawa K. Function of a transcription factor CDX2 beyond its trophectoderm lineage specification. Endocrinology. 2010;151:5873–5881. doi: 10.1210/en.2010-0458. [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Yu Y, Reed R. Primary microRNA processing is functionally coupled to RNAP II transcription in vitro. Scientific reports. 2015;5:11992. doi: 10.1038/srep11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.