SUMMARY
Post-transcriptional adenosine-to-inosine RNA editing mediated by adenosine deaminase acting on RNA1 (ADAR1) promotes cancer progression and therapeutic resistance. However, ADAR1 editase-dependent mechanisms governing leukemia stem cell (LSC) generation have not been elucidated. In blast crisis chronic myeloid leukemia (BC CML), we show that increased JAK2 signaling and BCR-ABL1 amplification activate ADAR1. In a humanized BC CML mouse model, combined JAK2 and BCR-ABL1 inhibition prevents LSC self-renewal commensurate with ADAR1 downregulation. Lentiviral ADAR1 wild-type, but not an editing-defective ADAR1E912 mutant, induces self-renewal gene expression and impairs biogenesis of stem cell regulatory let-7 microRNAs. Combined RNA sequencing, qRT-PCR, CLIP-ADAR1, and pri-let-7 mutagenesis data suggest that ADAR1 promotes LSC generation via let-7 pri-microRNA editing and LIN28B upregulation. A small molecule tool compound antagonizes ADAR1’s effect on LSC self-renewal in stromal co-cultures and restores let-7 biogenesis. Thus, ADAR1 activation represents a unique therapeutic vulnerability in LSC with active JAK2 signaling.
Graphical Abstract
INTRODUCTION
Cumulative evidence suggests that adenosine deaminase acting on RNA (ADAR) editases, such as ADAR1, promote progression and therapeutic resistance of a broad array of human malignancies (Chen et al., 2013; Fumagalli et al., 2015; Han et al., 2015; Jiang et al., 2013; Qi et al., 2014; Qin et al., 2014; Shah et al., 2009; Zipeto et al., 2015). ADAR editases are double stranded (ds) RNA binding proteins that post-transcriptionally deaminate adenosine-to-inosine (A-to-I), most frequently in the context of primate-specific Alu repeat sequences that comprise ten percent of the human genome (Kiran and Baranov, 2010; Picardi et al., 2015). By regulating mRNA and microRNA (miRNA) stability, ADARs exhibit wide-ranging effects on embryonic development and stem cell regulation (Han et al., 2015; Liddicoat et al., 2015; Ota et al., 2013; Solomon et al., 2013; Wang et al., 2000). Genetic ADAR1 deletion, particularly impairment of functional RNA editing, induces embryonic lethality in mice by impairing normal hematopoiesis (Guenzl and Barlow, 2012; Liddicoat et al., 2015; Wang et al., 2000). Conditional ADAR1 deletion in adult mice increases interferon signaling resulting in hematopoietic stem cell (HSC) exhaustion (Essers et al., 2009; Hartner et al., 2009). Cumulative human RNA sequencing (RNA-seq) studies demonstrate that deregulated ADAR expression promotes relapse or progression of lobular breast (Shah et al., 2009), hepatocellular (Chen et al., 2013), and esophageal cancer (Qin et al., 2014) as well as transformation of chronic myeloid leukemia (CML) from chronic phase (CP) to a therapy resistant blast crisis (BC) phase (Jiang et al., 2013).
As the first cancer shown to arise in a clonal HSC population, CP CML is initiated by BCR-ABL1 oncogenic tyrosine kinase expression (Fialkow et al., 1977; Jamieson et al., 2004; Soverini et al., 2015). Progression to BC phase occurs following malignant reprogramming of committed myeloid progenitors into self-renewing progenitor LSCs (Abrahamsson et al., 2009; Goff et al., 2013; Jamieson et al., 2004; Jiang et al., 2013). While BCR-ABL1-targeted tyrosine kinase inhibitor (TKI) therapy (Druker et al., 1996) has greatly reduced morbidity and mortality in CP CML, therapeutic resistance occurs through BCR-ABL1 mutation and/or amplification that leads to additional genetic and epigenetic modifications that promote progression (Abrahamsson et al., 2009; Goff et al., 2013; Jamieson et al., 2004; Quintas-Cardama et al., 2014; Sawyers, 2010). Previously, we showed that increased ADAR1 expression resulted in myeloid progenitor expansion and conversely, lentiviral shRNA knockdown of ADAR1 prevented malignant progenitor self-renewal in a humanized mouse model of BC CML (Jiang et al., 2013). However, 1) the oncogenic drivers of ADAR1 activity, 2) ADAR1’s role in malignant reprogramming of progenitors into self-renewing LSCs, and 3) ADAR1’s role in stem cell regulatory miRNA editing as a post-transcriptional mechanism governing self-renewal were not fully investigated.
Here we demonstrate that increased sensitivity of CML progenitors to JAK2-mediated extrinsic cytokine and intrinsic oncogenic signaling through BCR-ABL1 converge on ADAR1 activation. ADAR1-mediated A-to-I editing drives LSC generation by impairing pri-let-7 biogenesis, resulting in unopposed LIN28B expression, and enhanced self-renewal (Copley et al., 2013; Piskounova et al., 2011). Thus, targeted reversal of ADAR1-mediated self-renewal may enable cancer stem cell (CSC) eradication in a multitude of malignancies that have a propensity for progression in inflammatory microenvironments that activate JAK2 signaling.
RESULTS
JAK2 Signaling Increases ADAR1 Expression in Progenitor LSCs
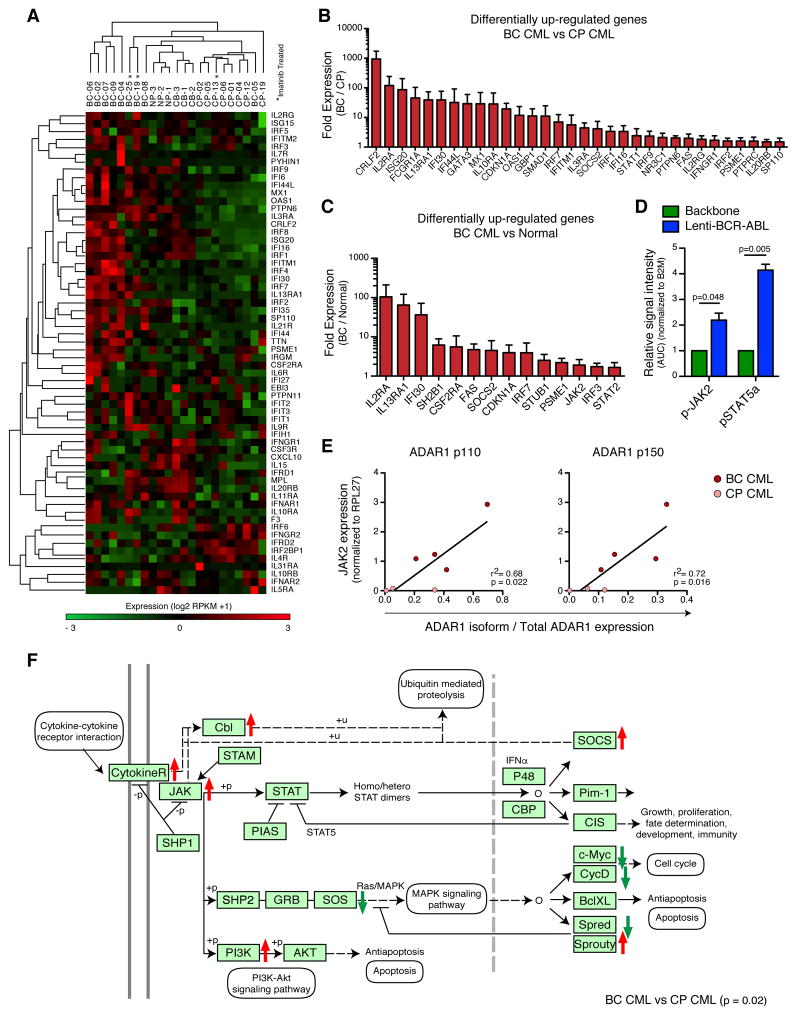
Previous studies suggest that ADAR1 expression is enhanced by inflammatory cytokine signaling that activates STAT binding to the ADAR1 promoter (George et al., 2008; George and Samuel, 2015). Thus, cytokine receptor and downstream target gene expression were analyzed by RNA-seq of fluorescence-activated cell sorting (FACS)-purified normal, CP, and BC progenitors (Table S1). Transcripts of inflammatory cytokine-associated receptor genes, which signal through the JAK-STAT pathway, and their downstream target genes were increased in BC compared with normal and CP progenitors (Figures 1A and S1A–E). Compared with CP, BC progenitors harbored increased levels of IFNγR1 and IL-3Rα, which can signal through JAK2 and activate ADAR1 transcription by binding of STATs to the ADAR1 promoter (Figure 1B) (George and Samuel, 2015). In addition, BC CML harbored increased expression of JAK/STAT signaling pathway genes and JAK2 transcripts compared with normal progenitors (Figures 1C and S1F). Nanoproteomics analysis of protein expression demonstrated elevated p-STAT5a levels following lentiviral BCR-ABL1 amplification (Figures 1D and S1C). Inflammatory cytokine-responsive ADAR1 p150 isoform expression correlated with JAK2 expression in both qRT-PCR and RNA-seq analyses of BC progenitors (Figures 1E and S1G). Moreover, KEGG pathway-based gene set enrichment analysis (GSEA) involving 1495 genes at a FDR of 0.05, revealed that the JAK/STAT signaling pathway was one of the most significantly differentially expressed gene sets (p=0.02, Benjamini adjusted-p value) in BC compared with CP progenitors (Figures 1F and S2A–B). These data highlight the increased sensitivity of BC progenitors to JAK/STAT signaling, which could activate ADAR1-mediated RNA editing in inflammatory microenvironments.
Figure 1. JAK2 Signaling in Progenitor LSCs Increases ADAR1 Expression.
(a) Whole transcriptome RNA sequencing analysis of inflammatory KEGG pathway genes in FACS-sorted hematopoietic progenitor cells (Lin−CD34+CD38+) cells from untreated and imatinib-treated (*) primary CP CML (n=8), BC CML (n=9), normal cord blood (n=3) and normal adult peripheral blood (NP, n=3) samples.
(b) Differentially up-regulated genes in the JAK/STAT/Inflammatory pathway from untreated BC CML (n=6) versus CP CML (n=7) samples.
(c) Differentially up-regulated genes in the JAK/STAT/Inflammatory pathway from untreated BC CML (n=6) versus Normal (n=6) samples.
(d) Area under the curve (AUC) normalized to B2M from nanoproteomic analysis of phospho-JAK2 and phospho-STAT5a levels in CD34+ cord blood cells transduced with lenti-BCR-ABL or backbone control (n=3). Graph shows mean +/− SEM and statistical analysis by paired t-test.
(e) Correlation analysis between mRNA expression levels of JAK2 and ADAR1 p150 isoform expression in primary CP (n=3) and BC (n=4) CML progenitors by quantitative RT-PCR (qRT-PCR) relative to RPL27 expression. Graph depicts best-fit line by Pearson correlation analysis.
(f) Gene set enrichment analysis based on KEGG pathway annotation of JAK/STAT signaling pathway genes in progenitor cells from untreated primary BC compared to CP CML patient samples (p=0.02, Benjamini-Hochberg adjusted). Red and green arrows are indicative of genes that are over-expressed and under-expressed in BC relative to CP, respectively.
See also Figure S1.
JAK2 Signaling Promotes ADAR1-mediated A-to-I Editing
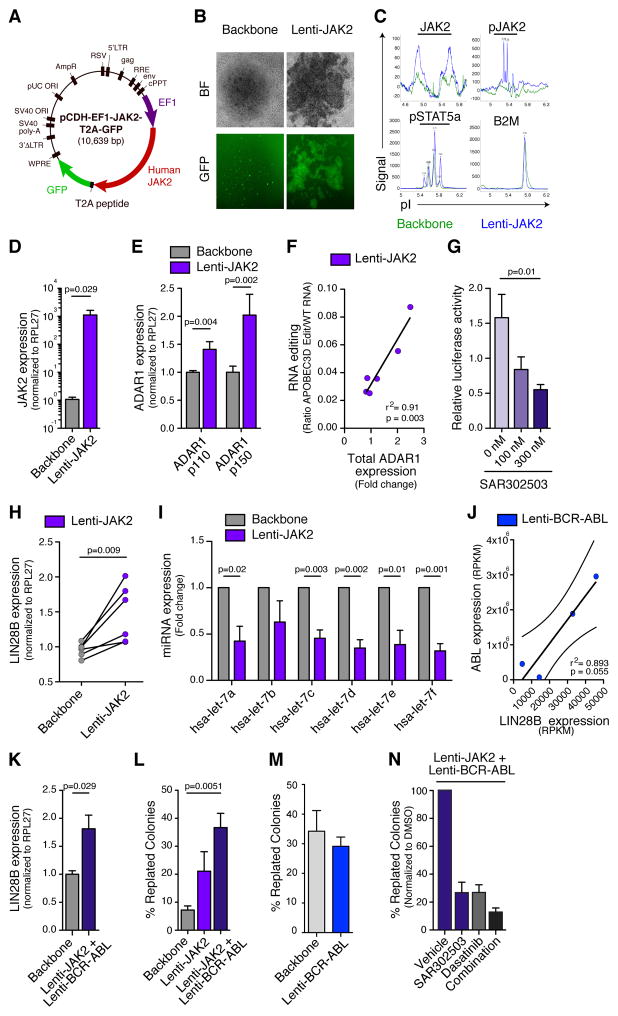
To investigate the direct contribution of JAK2 signaling to activation of ADAR1-mediated A-to-I editing, we developed a lentiviral construct that enabled robust expression of human JAK2-GFP in normal CD34+ progenitors (Figure 2A). Transduction efficiency was confirmed by fluorescence microscopy, nanoproteomics analysis of p-JAK2 and p-STAT5a and JAK2 qRT-PCR (Figures 2B–D and S2C). Lentiviral JAK2 transduction of human CD34+ progenitors enhanced both ADAR1 p150 and p110 transcript expression and ADAR1’s A-to-I editing activity as measured by RNA Editing Site-Specific qPCR (RESSqPCR) (Figures 2E–F) (Crews et al., 2015). While lentiviral ADAR1 transduction of BCR-ABL1+ K562 leukemia cells potentiated A-to-I RNA editing as measured by luciferase reporter activity in both the pCDH lentiviral vector and pDEST26 mammalian expression vector, this activity was further enhanced by addition of JAK2 expression (Figures S2D–F). Selective JAK2 inhibition with a pharmacological inhibitor (SAR302503) (Geron et al., 2008) reversed RNA editing activity in a dose dependent manner in K562 cells (Figure 2G). Taken together, these data suggest that JAK2 signaling enhances ADAR1-mediated RNA editing.
Figure 2. JAK2 Signaling Promotes ADAR1-mediated A-to-I Editing.
(a) Diagram of the human wild-type JAK2 lentiviral vector construct (lenti-JAK2).
(b) Bright-field (BF) and fluorescent (GFP) microscopy showing normal CD34+ cord blood cells transduced with lentiviral vector backbone or human JAK2 vector.
(c) Representative nanoproteomic analysis of total JAK2, phospho-JAK2, phospho-STAT5a and β2 microglobulin (B2M) in 293T control cells transduced with lentiviral human JAK2 (blue) or backbone vector control (green). Peaks represent signal intensity obtained from specific antibodies used.
(d) qRT-PCR analysis of human JAK2 mRNA expression levels relative to RPL27 in normal CD34+ progenitors transduced with lenti-JAK2 or backbone vector control (n=4).
(e) qRT-PCR analysis of human ADAR1 isoform levels in normal CD34+ cord blood cells transduced with lenti-JAK2 or backbone vector (n=7).
(f) Correlation analysis of total ADAR1 mRNA expression, measured by qRT-PCR (normalized to HPRT and backbone control), and A-to-I RNA editing of APOBEC3D, measured by RNA editing site specific qPCR (RESSqPCR), in lenti-JAK2-transduced CD34+ cord blood cells (n=6). Graph depicts best-fit line by Pearson correlation analysis.
(g) Luciferase reporter-based quantification of relative ADAR-mediated A-to-I editing activity in K562 leukemia cells co-transduced with lenti-ADAR1 and lenti-JAK2 following treatment with a JAK2 inhibitor (SAR302503) for 3 hrs at the indicated concentrations. Results represent data from three individual experiments.
(h) qRT-PCR analysis of LIN28B transcript levels normalized to RPL27 in normal CD34+ progenitors transduced with lenti-JAK2 or backbone vector (n=6).
(i) miRNA qRT-PCR analysis of mature let-7 family members in normal CD34+ cells transduced with lenti-JAK2 or backbone vector (n=3).
(j) Correlation analysis between LIN28B and ABL1 RNA expression levels in RNA sequencing-based gene expression data (RPKM) from lenti-BCR-ABL transduced normal CD34+ cells (n=4). Graph depicts best-fit line and 95% confidence intervals by Pearson correlation analysis.
(k) mRNA expression levels of LIN28B in normal CD34+ progenitors co-transduced with lenti-JAK2 and lenti-BCR-ABL or backbone vector (n=4).
(l) Self-renewal capacity as measured by percentage of secondary colonies formed after replating primary colonies from CD34+ cord blood cells transduced with lenti-JAK2 alone or co-transduced with lenti-BCR-ABL (n=3).
(m) Self-renewal capacity as measured by related secondary colonies in CD34+ cord blood cells transduced with lenti-BCR-ABL (n=3).
(n) Self-renewal capacity (normalized to DMSO vehicle) of normal CD34+ cord blood cells co-transduced with lenti-JAK2 and lenti-BCR-ABL following treatment SAR302503, dasatinib or the combination (n=3).
All graphs in Figure 3 show mean +/− SEM and statistical analyses were calculated using Student’s t-test unless otherwise specified.
See also Figures S2–3.
JAK2 and BCR-ABL1 Regulate Let-7 Biogenesis and LSC Self-renewal
While ADAR1 editing targets have not been completely elucidated, emerging data suggest that ADAR1 impairs biogenesis of tumor suppressive miRNAs, thereby contributing to cancer progression (Mariner et al., 2008). Because the JAK2/STAT pathway activated ADAR1, we hypothesized that the LIN28B/let-7 self-renewal axis may also be disrupted by increased JAK2 signaling. Indeed, qRT-PCR assays demonstrated that efficient JAK2 transduction increased LIN28B pluripotency gene expression and inhibited the expression of let-7 family miRNAs (Figures 2H–I).
Amplification of BCR-ABL1 is a hallmark of BC transformation and has been linked to LIN28B upregulation (Viswanathan et al., 2009). We performed RNA-seq analysis of lentiviral BCR-ABL1 transduced normal progenitors. Notably, a BCR-ABL1 transduction efficiency dependent increase in LIN28B expression was observed and was potentiated by co-transduction with JAK2 (Figures 2J–K and S2G). In human LSC-supportive SL/M2 stromal co-cultures, let-7a expression was significantly reduced in BCR-ABL1 transduced progenitors (Figure S2H). However, biogenesis of let-7 family members was more profoundly impaired following co-transduction with JAK2 and BCR-ABL1 (Figure S2I). Treatment of JAK2/BCR-ABL1 transduced normal CD34+ progenitors with the JAK2 inhibitor SAR302503 showed reduced ADAR1 p150 isoform expression, coupled with restored let-7 miRNA biogenesis in a dose-dependent manner (Figures S3A–B). These results confirmed our hypothesis that the JAK2/ADAR1 axis promotes maintenance of self-renewal commensurate with impaired let-7 miRNA biogenesis. Moreover, RNA-seq analysis revealed decreased expression of pri-miRNA processing genes, such as DGCR8 and ILF3, in BC compared with CP progenitors and in normal CD34+ cord blood cells transduced with ADAR1 WT compared with backbone controls (Tables S2 and S3). These results revealed a LSC intrinsic defect in let-7 miRNA maturation. In keeping with disruption of the LIN28B/let-7 self-renewal axis, combined JAK2 and BCR-ABL1 transduction enhanced colony-replating capacity as an in vitro surrogate measure of self-renewal (Figure 2L–M). Conversely, combined inhibition of JAK2 with SAR302503 (Geron et al., 2008) and BCR-ABL1 with dasatinib significantly inhibited their self-renewal potential (Figure 2N). Together these data suggested that JAK2 and BCR-ABL1 enhanced malignant reprogramming of progenitors into LSCs by deregulating LIN28B expression and impairing let-7 miRNA biogenesis.
JAK2 and BCR-ABL1 Inhibition Prevents Self-renewal of ADAR1-expressing LSCs
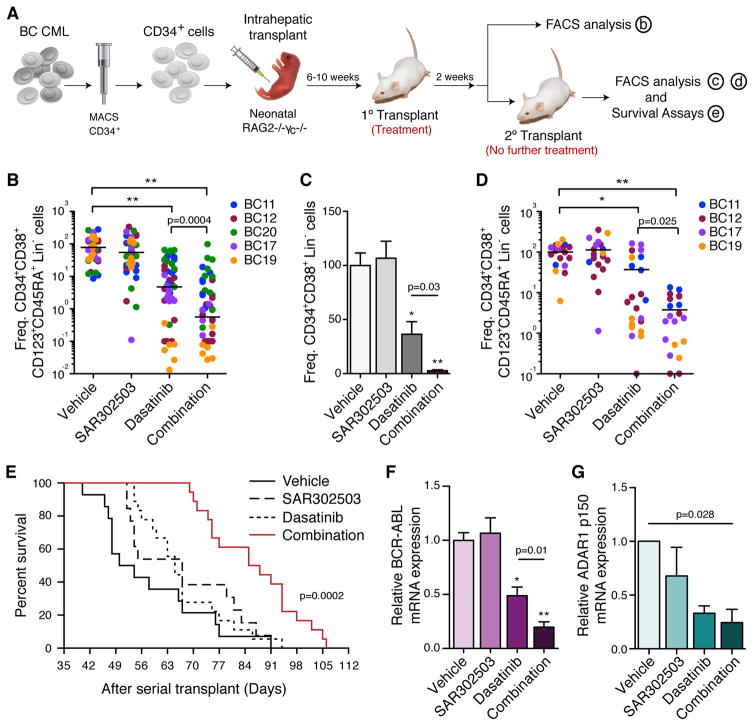
To understand the convergence of JAK2 and ADAR1 pathways on BCR-ABL1+ BC CML LSC maintenance, RAG2−/−γc−/− mice engrafted with human BC CD34+ cells were treated with SAR302503, dasatinib or the combination (Figure 3A). Primary human leukemic engraftment was observed in all hematopoietic tissues and the progenitor cell population was enriched for granulocyte-macrophage progenitors (GMP) that harbored enhanced serial transplantation potential (Abrahamsson et al., 2009; Jamieson et al., 2004) (Figures S4A and B). After two weeks of treatment, FACS analysis of hematopoietic tissues showed that both dasatinib and combination treatment inhibited BC LSC survival compared to vehicle-treated controls (Figures 3B and S4C–E). Selective JAK2 inhibition by SAR302503 in the splenic and bone marrow niches was confirmed by nanoproteomic analysis of p-JAK2, p-CRKL and p-STAT5a levels in BC progenitors isolated from treated mice (Figure S4F). However, only combination treatment significantly reduced self-renewal of BCR-ABL1 expressing BC progenitors, particularly leukemic GMP, in the bone marrow thereby prolonging survival of serially transplanted mice (Figures 3C–F). A significant reduction in ADAR1 p150 transcripts was also observed following combination treatment, suggesting that JAK2 and BCR-ABL1 signaling converge on ADAR1 to enhance LSC propagation (Figure 3G).
Figure 3. JAK2 and BCR-ABL1 Inhibition Prevents Self-renewal of ADAR1-expressing LSCs.
(a) In vivo experimental design of primary and serial transplantation studies (n>400 mice) with CD34+ progenitor cells isolated from BC CML patient samples.
(b) FACS analysis of GMP (CD34+ CD38+ CD123+ CD45RA+ Lin−) engraftment in mouse bone marrow (n=5 primary BC CML patient samples) after treatment with vehicle (n=54), SAR302503 (n=59), dasatinib (n=51) or combination (n=52).
(c) FACS analysis of human progenitor (CD34+ CD38+ Lin−) cell engraftment in mouse bone marrow following serial transplantation of BC CD34+ progenitors from mice treated with vehicle (n=19), SAR302503 (n=19), dasatinib (n=22) or combination (n=22).
(d) FACS analysis of human GMP engraftment in mouse bone marrow following serial transplantation of BC CD34+ progenitor cells from mice treated with vehicle (n=19), SAR302503 (n=19), dasatinib (n=22) or combination (n=22).
(e) Kaplan Meier plot showing percent survival of secondary recipient mice after serial transplantation of an equal amount (20,000–100,000) of BC CD34+ cells isolated from primary transplant recipients treated with vehicle (n=14), SAR302503 (n=13), dasatinib (n=18) or combination (n=18) (p=0.0002 by log-rank test).
(f) qRT-PCR analysis of relative BCR-ABL1 mRNA levels in equal numbers of CD34+ cells isolated from BC CML engrafted mice treated with vehicle (n=10), SAR302503 (n=13), dasatinib (n=11) or combination (n=10) as a measurement of human LSC frequency in bone marrow.
(g) qRT-PCR analysis of ADAR1 p150 isoform levels in equal numbers of FACS purified human CD34+ cells (n=3 individual BC CML patient samples) isolated from bone marrow of mice (3–6 mice per sample per treatment condition) following a 2-day treatment with vehicle, SAR302503, dasatinib or combination.. P value is calculated based on 1-way ANOVA.
All graphs show means +/− SEM; *p<0.05, **p<0.0001 by non-parametric Mann Whitney test unless otherwise specified.
See also Figure S4.
Inhibition of ADAR1-mediated RNA Editing Impairs Self-Renewal
By lentivirally expressing ADAR1 p150 wild-type (ADAR1 WT) or an editing defective mutant vector ADAR1 Mut (ADAR1E912A), we established that an inhibitory tool compound, 8-azadenosine (8-aza) (Veliz et al., 2003) reduced ADAR1’s A-to-I editing activity in K562 leukemic cell line (Figure 4A–B). Treatment with 8-aza showed no effect on BCR-ABL and JAK2 signaling, as demonstrated by qRT-PCR analysis and p-CRKL and p-STAT5a Western blot analysis (Figure S5A–B). To determine if ADAR1 is required for LSC self-renewal, BC CML, normal or lentiviral JAK2 and BCR-ABL1 transduced CD34+ progenitors were plated in SL/M2 stromal cultures and treated with 8-aza, SAR302503, or the combination. After two weeks in stromal co-culture, SAR302503 and 8-aza restored let-7 miRNA biogenesis commensurate with a reduction in ADAR1 expression, RNA editing activity and LIN28B expression in JAK2/BCR-ABL1 transduced progenitors compared with controls (Figures 4C–G). Moreover, replating capacity of primary CML BC progenitors was significantly reduced by combined JAK2 inhibitor and 8-aza treatment in stromal co-cultures (Figures 4H–I). Since normal CD34+ hematopoietic progenitors harbored relatively low JAK2 and ADAR1 levels, their self-renewal capacity was not significantly impaired by 8-aza or combination treatment (Figures 4H–I) indicative of an adequate therapeutic index between normal and malignant progenitors.
Figure 4. Inhibition of ADAR1-mediated RNA Editing Impairs Self-Renewal.
(a) RESS-qPCR analysis of RNA editing ratio of APOBEC3D and qRT-PCR analysis of ADAR1 mRNA expression levels in K562 leukemia cells transduced with lenti-ADAR1 WT, lenti-ADAR1E912A Mutant or pCDH backbone control (n=5 each).
(b) RNA editing ratio of APOBEC3D by RESSqPCR in K562 leukemia cells transduced with ADAR1 WT and ADAR1E912A mutant following 8-aza treatment (n=3). The values are normalized to the editing ratio observed in pCDH control cells.
(c) In vitro experimental design used in the following studies.
(c–i) Cord blood CD34+ cells co-transduced with lenti-JAK2 and lenti-BCR-ABL, normal or BC CD34+ progenitor cells, were treated on SL/M2 stromal cultures with ADAR1 inhibitor 8-azaadenosine (8-Aza, 10–25nM), JAK2 inhibitor (100nM of SAR302503), BCR-ABL inhibitor (dasatinib, 10nM) or combination using same concentrations.
(d–e) miRNA expression levels of let-7 family members (d) and ADAR1 expression (e) in CD34+ cord blood cells co-transduced with lenti-JAK2 and lenti-BCR-ABL following treatment (n=3).
(f) RNA editing ratio of APOBEC3D by RESSqPCR in CD34+ cord blood cells co-transduced with lenti-JAK2 and lenti-BCR-ABL following treatment (n=3).
(g) Expression of LIN28B of CD34+ cord blood cells co-transduced with lenti-JAK2 and lenti-BCR-ABL following treatment (n=3).
(h) Percentage of secondary colonies formed after replating primary colonies from normal bone marrow or BC CD34+ cells treated with different dose of 8-aza (n=3). Graph shows mean +/− SD; p values were calculated using ANOVA and Holm-Sidak method.
(i) Percentage of secondary colonies formed after replating primary colonies from normal bone marrow or BC CML following treatment. Graph shows mean +/− SD; p values were calculated using ANOVA and Holm-Sidak method. See also Figure S5.
ADAR1 Enhances Self-Renewal Gene Expression
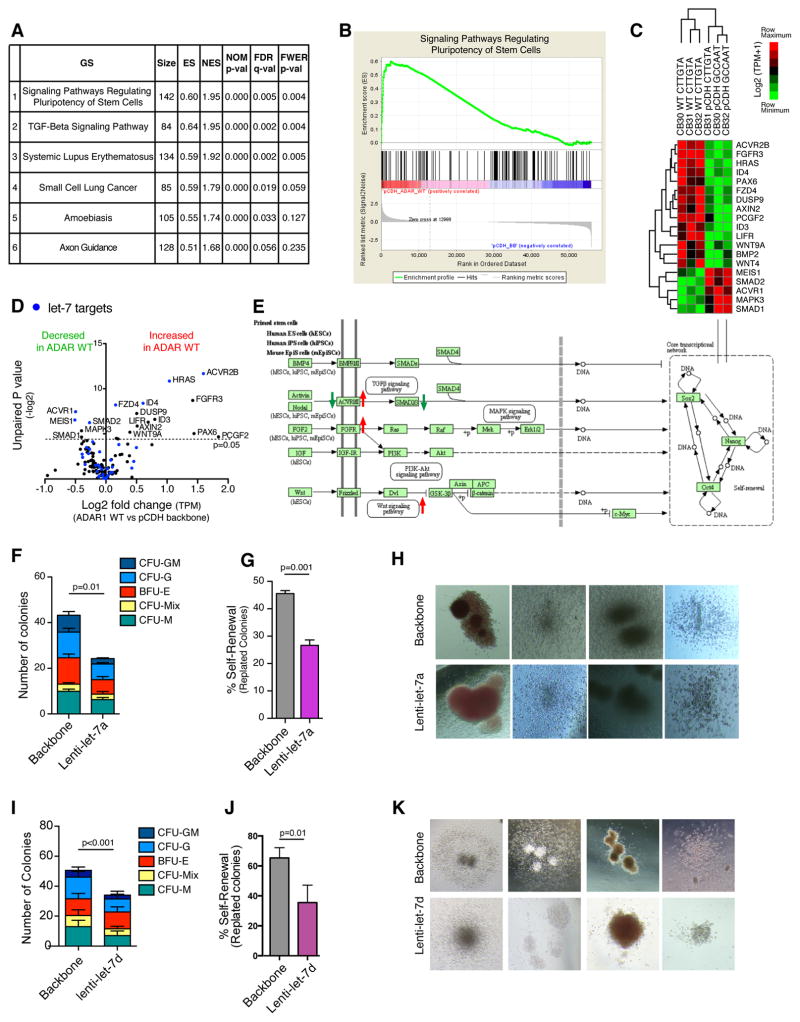
To understand the mechanisms governing ADAR1-mediated LSC self-renewal, we performed RNA-seq analysis of cord blood CD34+ cells lentivirally transduced with lentiviral backbone vector, ADAR1, or an A-to-I editing defective ADAR1E912A mutant vector. A KEGG pathway-based gene set enrichment analysis (GSEA) revealed that ADAR1 overexpression significantly affected genes involved in the regulation of stem cell pluripotency (Figures 5A–B and Table S4). Transcriptomic analysis of ADAR1 compared with backbone-transduced CD34+ cells revealed fourteen upregulated and five downregulated stem cell pluripotency regulatory genes (Figure 5C). Interestingly, the most differentially upregulated genes, including ACVR2B, HRAS, and FGFR3, were let-7 targets, suggesting that ADAR1-mediated impairment of let-7 miRNA biogenesis may also regulate LSC self-renewal capacity (Figure 5D–E). Lentivirally enforced let-7a and let-7d expression reduced progenitor total colony numbers. Colony replating capacity and LIN28B expression decreased thereby underscoring the inhibitory impact of let-7 on self-renewal (Figures 5F–K and S5C–D).
Figure 5. ADAR1 Enhances Self-Renewal Gene Expression.
(a) Gene Set Enrichment Analysis (GSEA) of RNA-seq data revealed the most significantly affected KEGG pathways in normal cord blood CD34+ population transduced with lenti-ADAR1 WT (n=3) or vector controls (n=3).
(b) GSEA plot showing enrichment of genes in “Signaling Pathways Regulating Pluripotency of Stem Cells” in cord blood CD34+ cells transduced with lenti-ADAR1 WT compared with vector controls (n=3).
(c) Heatmap depiction of RNA-seq analysis of cord blood CD34+ population transduced with lenti-pCDH vector controls (n=3) or lenti-ADAR1 WT (n=3) indicates ADAR editing target genes in stem cell signaling pathways regulating pluripoteny of stem cells are significantly differentially expressed (p<0.05, Student’s t-test).
(d) Volcano plot analysis derived from TPM values showing significantly differentially expressed (p<0.05, Student’s t-test) known let-7 target genes (blue) in pluripotency pathway in cord blood CD34+ cells transduced with lenti-pCDH vector controls (n=3) or lenti-ADAR1 WT (n=3).
(e) Gene set enrichment analysis based on KEGG pathway annotation of signaling pathway regulating stem cell pluripotency in progenitor cells from cord blood CD34+ cells transduced with lenti-pCDH vector controls (n=3) or lenti-ADAR1 WT (n=3). Red and green arrows are indicative of genes that are over-expressed and under-expressed in ADAR1 transduced samples compared to pCDH vector controls, respectively.
(f) Number of colonies formed in primary colony-formation assay by normal CD34+ cells transduced with lenti-let-7a or backbone control (n=3). Graph shows average colonies per well and statistical analysis by Student’s t-test.
(g) Percentage of secondary colonies formed after replating primary colonies from CD34+ cord blood cells transduced with lenti-let-7a or backbone control (n=3).
(h) Representative colony pictures of let-7a and backbone transduced CD34+ cord blood cells.
(i) Number of colonies formed in primary colony-formation assay by normal CD34+ cells transduced with lenti-let-7d or backbone control (n=3).
(j) Percentage of secondary colonies formed after replating primary colonies from CD34+ cord blood cells transduced with lenti-let-7d or backbone control (n=3).
(k) Representative colony pictures of let-7d and backbone transduced CD34+ cord blood cells.
All graphs show mean +/− SEM and statistical analysis was calculated using the Student’s t-test.
See also Figure S5.
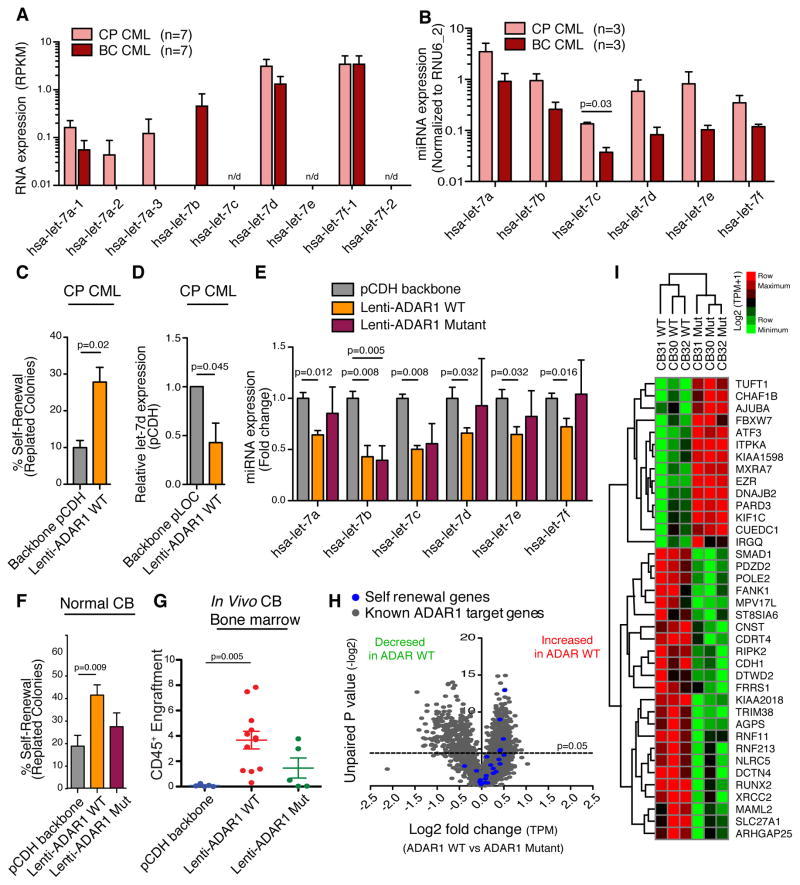
Since JAK2 activates ADAR1-mediated RNA editing and impairs let-7 biogenesis (Figures 2E-F and I), we decided to investigate if let-7 biogenesis in LSC is restricted by cytokine-responsive ADAR1 editing activity. First, we observed a reduction in pri- and pre-let-7a-1, let-7a-2, let7-a-3, pri-let-7d expression using RNA-seq quantification, as well as decreased expression of mature let-7 miRNAs in BC compared to CP progenitors (Figure 6A-B). Lentivirally enforced ADAR1 expression increased replating potential of CP CML coupled with a reduction in mature let-7d expression, suggesting malignant activation of ADAR1 in CP to BC transition is coupled with impaired let-7 biogenesis and enhanced LSC self-renewal (Figures 6C–D). Next, we examined mature let-7 miRNA expression in CD34+ cord blood cells and K562 cells transduced with ADAR1 WT or an ADAR1E912A mutant (ADAR1E912A Mut), which had reduced RNA editing capacity (Figures 6E and S5E–I). Notably, we observed a significant reduction in mature let-7 miRNA levels in cord blood CD34+ cells following transduction with ADAR1 WT but not with ADAR1E912A (Figures 6E). Moreover, only WT ADAR1 significantly increased the self-renewal and survival capacity of normal cord blood CD34+ cells in colony replating and engraftment assays (Figures 6F–G and S5J). In RAG2−/−γc−/− mice transplanted with cord blood CD34+ cells, we also observed significantly enhanced engraftment as measured by increased CD45+ percentage in bone marrow (Figures 6G and S5K). These data suggest that ADAR1 regulates let-7 miRNA biogenesis in an A-to- I editing dependent manner, which has a functional impact on progenitor self-renewal and survival.
Figure 6. ADAR1-Dependent Alterations in Let-7 Biogenesis and Self-Renewal Gene Expression.
(a) RNA sequencing based analysis of let-7 pri-miRNA levels in FACS-sorted hematopoietic progenitor cells (CD34+CD38+ Lin−) cells from untreated primary CP and BC CML (n=7 each) samples. n/d=not detectable.
(b) miRNA PCR analysis of mature let-7 miRNA levels in primary CP and BC progenitors (n=4 each). Values are normalized to RNU6_2 miRNA expression.
(c) Percentage self-renewal (replated colonies) from CP CML CD34+ progenitors transduced with lenti-ADAR1 WT or vector control (n=3).
(d) miRNA PCR analysis of mature let-7d levels in CP CD34+ progenitors transduced with lenti-ADAR1 WT or vector control (n=3).
(e) miRNA PCR analysis of mature let-7 levels in lenti-ADAR1 WT, lenti-ADAR1E912A Mutant or pCDH backbone transduced normal CD34+ cord blood cells (n=5). Values are normalized to RNU6_2 expression.
(f) Percentage of secondary colonies formed after replating primary colonies from CD34+ cord blood cells transduced with lenti-ADAR1 WT, lenti-ADAR1E912A Mutant or backbone vector (n=5).
(g) Percentage of human CD45+ engraftment in bone marrow after transplantation of CD34+ cord blood cells (n=3) transduced with vector control (pCDH), lenti-ADAR1 WT alone, or lenti-ADAR1E912A Mutant into RAG2−/−γc−/− mice (n=5–12 per group).
(h) Volcano plot analysis derived from TPM values showing significantly differentially expressed (p<0.05, Student’s t-test) known ADAR1 target genes (grey) and self-renewal transcripts (blue) in cord blood CD34+ cells transduced with lenti-ADAR1 WT (n=3) compared with lenti-ADAR1E912A Mut (n=3).
(i) Heat map depiction of RNA-seq analysis of cord blood CD34+ population transduced with lenti-ADAR1 WT (n=3) or lenti-ADAR1E912A Mut (n=3) indicates that 38 out of 175 ADAR editing target genes (previously found to be differentially expressed between CP and BC) are significantly differentially expressed (p<0.05, Student’s t-test).
All graphs show mean +/− SEM and statistical analysis was calculated using the Student’s t-test.
Previous reports indicate that ADAR1 depletion induces stress-related apoptosis during fetal hematopoiesis and loss of mouse HSC multi-lineage repopulating potential (Hartner et al., 2009; Wang et al., 2000; Wang et al., 2004). To investigate the contribution of ADAR1-editing to self-renewal, we performed comparative RNA-seq analysis of ADAR1 WT, ADAR1E912A Mut and backbone transduced human CD34+ cells. Volcano plot analysis of RNA-seq data demonstrated distinct differences in expression of known ADAR1 target genes (Roberts et al., 2013) and increased expression of self-renewal transcripts following ADAR1 WT compared to ADAR1E912A Mut transduction (Figures 6H and Table S5). A comparative analysis of A-to-I editing dependent expression profiles with previously identified differentially expressed transcripts in BC compared with CP (Jiang et al., 2013) identified 38 common transcripts. These included genes involved in self-renewal, such as FBXW7 and MAML2, and miRNA regulation, such as SMAD1 (Figure 6I). Taken together, these results indicate that ADAR1-mediated A-to-I editing contributes to LSC self-renewal.
ADAR1 Editase Activity Regulates Let-7 Biogenesis
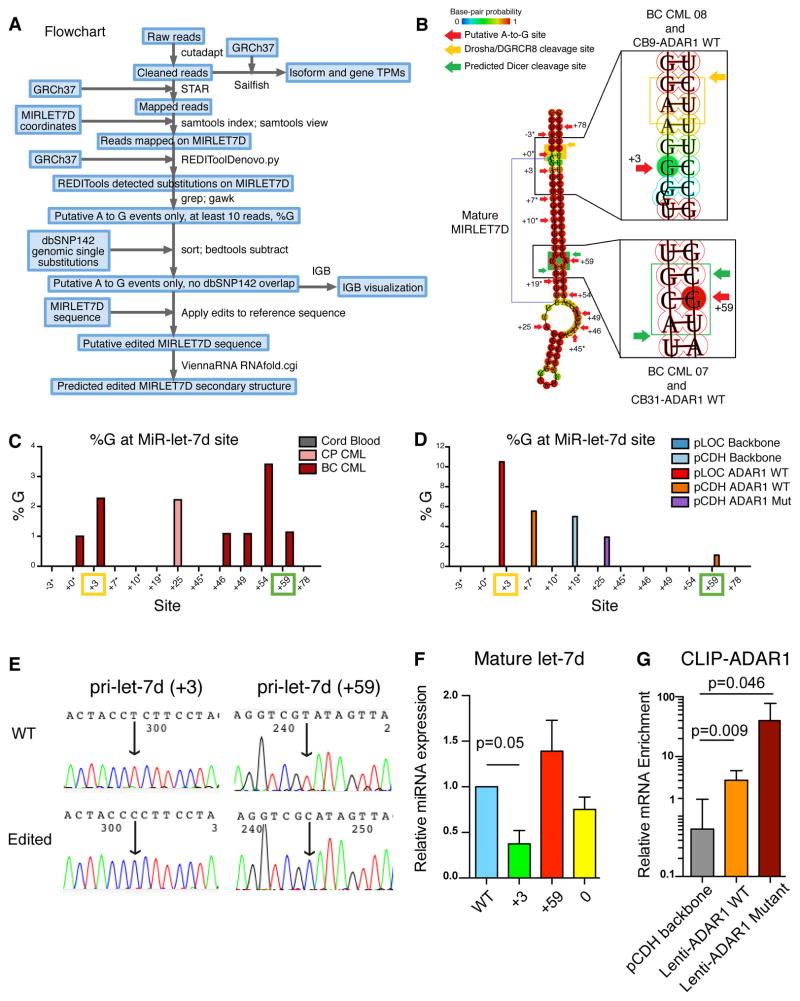
Other studies suggest the ADAR1 mediates miRNA biogenesis by editing polycistronic miRNAs in drosophila (Chawla and Sokol, 2014). Similarly, the primate polycistronic cluster of let-7a, let-7d, and let-7f possesses differential mature miRNA expression potential dependent on ADAR1 RNA editing activity (Figure 6E). Therefore, we set out to investigate the location of A-to-I editing in the let-7 polycistronic cluster. We utilized RNA-seq along with ViennaRNA secondary RNA structure prediction software (Yu et al., 2012) of both primary CML samples and normal progenitors transduced with ADAR1 WT, ADAR1E912A Mut, JAK2, or BCR-ABL1 lentiviral vectors (Figures 7A–B and S6A–B). While a few editing sites were detected in pri-let-7f in CP and BC progenitors (Figure S6C), pri-let-7d was found to be edited at multiple locations (Figures 7C–D and S6D–E). Notably, A-to-G nucleotide changes at the +3 and +59 editing sites were predicted to alter RNA secondary structures at the DROSHA/DGCR8 and DICER cleavage sites, respectively (Figures 7B and S6A–B). Both increased and differential A-to-G editing sites were detected in BC compared with CP and normal progenitors (Figure 7C). The highest editing frequency was observed in close proximity to the predicted DROSHA/DGCR8 cleavage site (+3) in ADAR1 WT transduced samples (Figure 7D). Most importantly, two common editing sites adjacent to DROSHA/DGCR8 and DICER cleavage sites (+3, +59) were detected in BC progenitors and ADAR1 WT transduced cord blood cells, indicative of a pivotal role for ADAR1-mediated editing in BC LSC generation (Figures 7C–D).
Figure 7. ADAR1 Editase Activity Regulates Let-7 Biogenesis.
(a) Flowchart represents the RNA-seq analysis algorithm.
(b) ViennaRNA predicted secondary structure changes in let-7d induced by A-to-I editing occurring near DGCR8/DROSHA (yellow) in BC CML 08 and CB9 ADAR1 WT, and predicted DICER cleavage sites in BC CML 07 and CB31 ADAR1 WT (green). The patient samples with the corresponding A-to-I editing sites are labeled next to +3 and +59 cleavage sites.
(c–d) Depiction of A-to-I editing sites in pri-let-7d, calculated by percentage of guanosine (%G) in (c) primary cord blood, CP CML and BC CML progenitors (n=3–9); or in (d) cord blood CD34+ cells transduced with pLOC Backbone, pCDH Backbone, pLOC ADAR1 WT, pCDH ADAR1 WT or pCDH-ADAR1 Mut (n=3). RNA editing sites are labeled such that the first base of the mature MIRLET7D is +1; sites seen in only one sample are marked by asterisks. The sites located close to DROSHA/DGCR8 and predicted DICER cleavage sites are labeled with yellow and green squares, respectively.
(e) Confirmation of the lentiviral constructs of wild-type (WT) unedited or “pre-edited” pri-let-7d at +3 ad +59 sites. The arrow points to the A-to-G mutations, reverse sequenced as T-to-C changes.
(f) 293T cells were transfected with WT, +3, +59 or 0 pri-let-7d lentiviral constructs and the mature let-7d expression was measured by RT-qPCR. Experiment was performed in triplicates.
(g) Crosslinking RNA Immunoprecipitation (CLIP) in K562 cells stabled transduced with pCDH vector, lenti-ADAR1 WT, and lenti-ADAR1E912A Mutant with an ADAR1 antibody confirms that both ADAR1 WT and ADAR1E912A Mutant are associated with pri-let-7d transcripts. Experiment was performed in triplicates.
All graphs show mean +/− SEM; p values were calculated using Student’s t-test unless otherwise specified.
See also Figures S6–7.
Previous studies revealed that A-to-I editing near the DROSHA or DICER cleavage sites in pri-miRNAs inhibited the cleavage reaction and reduced mature miRNA biogenesis (Nishikura, 2010; Yang et al., 2006). We investigated the effect of A-to-I editing on pri-let-7d biogenesis in 293T cells transfected with edited and unedited pri-let-7d expression plasmids (Figure 7E–F). We examined the unedited wild-type (WT) pri-let-7d plasmid, and “pre-edited” pri-let-7d at +3 and +59 as identified in BC CML progenitors, as well as at 0, which is the DROSHA/DGCR8 cleavage site. Compared to the WT pri-let-7d construct, editing at the +3 site induced a significant reduction in mature let-7d miRNA levels (Figure 7F). Interestingly, the A-to-G changes in +59 or 0 sites did not show any changes in mature miRNA levels, suggesting that the +3 editing site is responsible for the reduced mature let-7d miRNA expression observed in ADAR1-transduced cord blood progenitors. Lastly, cross-linking immunoprecipitation (CLIP) assays were performed in a K562 leukemic cell line stably expressing ADAR1 WT or ADAR1E912A to determine if ADAR1 directly binds to pri-let-7d transcripts (Figure 7G). Using an ADAR1 antibody, we were able to isolate pri-let-7d miRNAs in both ADAR1 WT and ADAR1E912A transduced cells, thereby confirming the capacity of ADAR1 to bind to pri-let-7d transcripts. These findings reveal a pivotal role for ADAR1 editase activity in let-7d biogenesis and BC LSC self-renewal. Further analysis of let-7 clusters demonstrated increased A-to-G editing events within primate-specific Alu repeat sequences with a proclivity for forming dsRNA loops (Figure S7A). Single nucleotide resolution RNA sequencing analysis revealed that pri-let-7d is located in close proximity to multiple Alu repetitive sequences, which may enhance the capacity of ADAR1 to bind and edit pri-let-7d (Figure S7B).
DISCUSSION
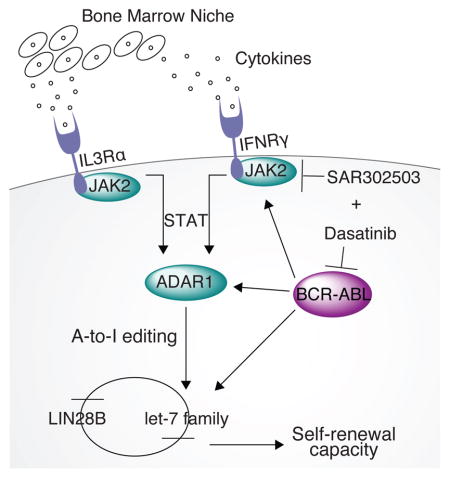
Malignant RNA editing, conferred by ADAR1 activation, has emerged as a dominant driver of cancer relapse and progression (Jiang et al., 2013; Qi et al., 2014; Qin et al., 2014; Shah et al., 2009). Moreover, a recent report describing a genome wide analysis of 6,236 patient samples, representing 17 tumor types in the Cancer Genome Atlas database, revealed non-synonymous A-to-I editing events that were predicted to promote therapeutic resistance (Han et al., 2015). These discoveries have fueled intensive research into the cell type and context specific mechanisms driving ADAR1 activation and the impact on self-renewing CSC generation in malignancies that have a proclivity for therapeutic resistance and progression. In particular, the oncogenic drivers of ADAR1 activation, the A-to-I editing targets, and the non-cell autonomous as well as cell autonomous mechanisms that govern CSC self-renewal had not been elucidated. By employing whole-transcriptome sequencing of normal, CP and BC CML patient progenitor samples and human BC CML progenitor serial transplantation mouse models, our study provides a novel link between increased sensitivity to JAK2-dependent cytokine signaling and ADAR1 editase mediated generation of self-renewing LSCs. Our results show that ADAR1 activation in BC LSCs is triggered by increased JAK2-dependent inflammatory signaling and is further amplified by the presence of BCR-ABL1. Conversely, pharmacologic inhibition of JAK2 and BCR-ABL1 prevented LSC self-renewal commensurate with reduced BCR-ABL1 and ADAR1 p150 expression in humanized BC LSC mouse model. These data highlight a dual mechanism of malignant RNA editing activation in LSCs.
While genetic ablation of ADAR1 in mice leads to embryonic lethality due to severe defects in erythropoiesis (Wang et al., 2000), conditional deletion in the hematopoietic system impairs long-term hematopoietic stem cell (HSC) maintenance, indicative of key roles for ADAR1 in both cell fate specification and self-renewal (Hartner et al., 2009). This suggests that deregulation of editase activity may play a significant role in a variety of blood disorders that have acquired aberrant stem cell self-renewal characteristics. Indeed, in a humanized mouse model of CML, lentiviral shRNA knockdown of ADAR1 inhibited self-renewal of malignant progenitors that promote blast crisis transformation (Jiang et al., 2013). Here we advance this finding by differential gene expression analysis aimed at determining the genes involved in HSC self-renewal. Notably, lentiviral ADAR1 overexpression significantly affected genes involved in the regulation of stem cell pluripotency and self-renewal. Upregulated genes in ADAR1 WT compared to backbone transduced normal CD34+ cells (Figure 5C) included WNT4 and WNT9a signaling proteins. Notably, the WNT pathway is critical for normal HSC homeostasis and self-renewal (Louis et al., 2008; Reya et al., 2003). In addition, we observed upregulation of genes that regulate the TGF-β pathway, including SMAD1 and SMAD2, which transduce extracellular signals to activate transcription of genes that regulate cellular growth, differentiation, and apoptosis (Heldin et al., 1997; Nakao et al., 1997) as well as miRNA expression and maturation (Blahna and Hata, 2012; Davis et al., 2008).
With regard to miRNA maturation mechanisms relevant to LSC function, a seminal study showed that overexpression of LIN28B and a let-7 target gene, HMGA2, coincided with BC transformation of CML (Viswanathan et al., 2009) and that knockdown of LIN28B restored levels of let-7b and let-7g in K562, a BC CML cell line. While LIN28B has been linked to let-7 stem cell regulatory miRNA biogenesis, ADAR1 also contributes to differential expression of miRNAs through direct binding to DROSHA/DGCR8 or DICER proteins (Bahn et al., 2015; Nemlich et al., 2013). Because the LIN28B/Let-7 axis plays a critical role in stem cell maintenance (Copley et al., 2013; Wang et al., 2015), and appears to be deregulated in tumorigenesis (Melton et al., 2010; Piskounova et al., 2011; Viswanathan et al., 2008), we investigated its role in LSC maintenance. Here, we show that ADAR1 enhances LSC self-renewal by impairing let-7 biogenesis. Combined inhibition of ADAR1 and JAK2 restores let-7 expression and inhibits LSC self-renewal. Since ADAR1 mediates differential expression of polycistronic miRNAs transcribed from the lethal-7-Complex (let-7-C) locus by altering DROSHA processing (Chawla and Sokol, 2014), we performed single nucleotide resolution RNA-seq combined with secondary RNA structure prediction (ViennaRNA software) (Yu et al., 2012) analyses and miRNA qRT-PCR. These analyses demonstrated that BC CML and ADAR1 transduced progenitors harbored enhanced editing at the polycistronic let-7 loci and reduced mature let-7 microRNA levels (Melton et al., 2010; Yu et al., 2007). Let-7 loci outside the polycistronic cluster displayed no editing signatures in ADAR1 transduced progenitors. Moreover, RNA editing adjacent to the DROSHA/DGCR8 cleavage site at +3 was associated with reduced let-7d biogenesis in BC LSCs and CD34+ progenitors transduced with ADAR1 but not with ADAR1E912A and empty vectors. Finally, CLIP-ADAR1 assays combined with site-directed mutagenesis mediated introduction of let-7d edits confirmed that ADAR1 directly binds and edits pri-let-7d transcripts thereby reducing the expression of mature let-7d miRNA as measured by qRT-PCR. In BC LSC compared with their CP progenitor counterparts, RNA-seq analysis revealed a reduction in pri-let-7a but not let-7b levels indicative of a LIN28B-independent effect of ADAR1 on let-7 biogenesis. We also detected editing of let-7f (4%), albeit less frequently than for let-7d (10%). In primary LSCs, detection of A-to-I editing of less abundant pri-let-7 transcripts by ADAR1 may require more sensitive methods such as CLIP-sequencing. With regard to clinical relevance, A-to-I RNA editing of transcripts at a level of 10% was recently linked to progression in 17 cancer types (Chen et al., 2013; Han et al., 2015) and will likely represent an important functional biomarker of CSC generation.
The critical advance of this study is that ADAR1 editase activity impairs let-7 family miRNA biogenesis and increases progenitor self-renewal capacity resulting in malignant reprogramming of progenitors into BC LSCs. In addition, we show that enhanced sensitivity to cytokine signaling as a consequence of JAK2-responsive cytokine receptor upregulation and BCR-ABL1 oncogene amplification results in ADAR1 activation. While previous studies have shown that JAK2 signaling is important in the induction of numerous transcriptional mediators, our discovery of a pivotal JAK2-ADAR1-let-7 self-renewal axis provides the first mechanistic link between inflammatory cytokine-driven oncogenic signaling pathways and RNA editing-driven malignant reprogramming of progenitors into LSCs. Perhaps most importantly, targeted reversal of ADAR1 activity may impede the generation of CSCs in a broad array of therapeutically recalcitrant malignancies that evolve in inflammatory microenvironments.
Online Content Methods and additional Supplemental Figures as well as display items are available in the online version of the paper together with references unique to these sections.
EXPERIMENTAL PROCEDURES
Primary normal and CML samples were obtained and RNA-Seq analysis as well as qRT-PCR validation were performed according to published methods (Abrahamsson et al., 2009; Goff et al., 2013; Jiang et al., 2013). MiRNAs were extracted using a RNeasy Micro Kit and qRT-PCR was performed using miRNA human-specific primers normalized to RNU6_2 and SNORD44. Lentiviral human wild-type JAK2, BCR-ABL1, wild-type and mutant ADAR1E912A (overexpression vectors were produced in the pCDH-EF1-T2A-GFP or pLOC lentiviral vector systems. Progenitor transduction, SL/M2 co-culture and colony assays were performed as previously described (Abrahamsson et al., 2009; Goff et al., 2013; Jiang et al., 2013). Immunocompromised RAG2−/−γc−/− mice engrafted with human BC CML progenitors were treated for two weeks with SAR302503, Dasatinib of the combination followed by FACS analysis of human progenitor engraftment in hematopoietic tissues and serial transplantation5. The RNA-sequence data accession numbers are PRJNA319866 and PRJNA214016.
RNA and microRNA extraction and quantitative RT-PCR
Total RNA was isolated from 20,000 to 50,000 FACS-sorted or CD34+ selected (MACS) progenitors cells from normal cord blood, CP CML, BC CML or from xenografted mice and complementary DNA was synthesized according to published methods (Abrahamsson et al., 2009; Goff et al., 2013; Jiang et al., 2013). Then qRT-PCR was performed in duplicate on an iCycler with the use of SYBR GreenER qPCR SuperMix (Invitrogen), 5ng of template mRNA and 0.2μM of each forward and reverse primer (Table S6). Human specific RPL27 primers were used as housekeeping control. MicroRNA extraction was performed using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions. Then 30 ng of cDNA was prepared in a reverse-transcription reaction using miScript RTII kit (Qiagen) and served as a template for the quantification of the expression of mature miRNA of interest. Also, qRT-PCR was performed using miRNA human-specific primers and SYBR Green Kit (Qiagen). MiScript primers, RNU6_2 (Qiagen), were used as housekeeping control. The expression of primary and precursor miRNA transcripts were measured using previous published primers (Patterson et al., 2014).
Human Progenitor Xenotransplantation and Treatment
Immunocompromised RAG2−/−γc−/− mice were bred and maintained in the Moores Cancer Center vivarium according to IACUC approved protocols. Neonatal mice were transplanted intrahepatically with 20,000–100,000 BC CML or human cord blood CD34+ cells according to our published methods (Abrahamsson et al., 2009; Goff et al., 2013; Jiang et al., 2013). Transplanted mice were FACS screened for human engraftment in peripheral blood at 6–10 weeks. Engrafted (>1% human CD45+ cells) mice were treated by oral gavage with SAR302503 (Sanofi-Aventis) twice daily with 60mg/kg (0.5% methylcellulose, 20% tween 80 and H2O), 50 mg/kg dasatinib daily (50% propylene glycol, 50% PBS), combination (SAR302503 plus dasatinib), or drug vehicles for two weeks. Following treatment, mice were euthanized and single cell suspensions of hematopoietic tissues were analyzed by FACS for human engraftment and 20,000–100,000 human CD34+ cells were serially transplanted into neonatal RAG2−/−γc−/− mice. A subgroup of mice was treated for 2 days, and progenitor cells in the bone marrow were analyzed by qRT-PCR.
Supplementary Material
HIGHLIGHTS.
JAK2 signaling activates ADAR1-mediated A-to-I RNA editing
JAK2 and BCR-ABL1 signaling converge on ADAR1 activation through STAT5a
ADAR1-mediated miRNA editing impairs let-7 biogenesis and enhances LSC self-renewal
JAK2 and BCR-ABL1 inhibition reduces ADAR1 expression and prevents LSC self-renewal
eTOC SYNOPSIS.
Zipeto, Court and colleagues show a pivotal role for let-7 microRNA editing in leukemia stem cell self-renewal. Impairment of let-7 is dependent on JAK2 and BCR-ABL-mediated activation of ADAR1 editing. This provides a novel mechanism of malignant reprogramming that can be targeted through combined JAK2 and BCR-ABL or ADAR1 inhibition.
Acknowledgments
This work was supported by CIRM grants (RN2-00910-1; DR1-01430; RS1-00228-1), Cancer Stem Cell Consortium funding from Genome Canada and the Ontario Genomics Institute (OGI-047); NIH NIGMS 5K12GM068524; NIH NCI 2P30CA023100-28; NIH NCI R21CA189705; The Sanford Stem Cell Clinical Center; The San Diego Foundation; The Ratner Family Foundation; The Mizrahi Family Foundation; and the Canadian Institute of Health Research (CSC-105367). This work was also supported in part by the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Cancer Research Program, under Award No. W81XWH-14-1-0121 (C.H.M.J.). Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. The authors wish to thank Dr. Dennis Carson and Dr. Ida Deichaite for scientific advice, Dr. Wenxue Ma (University of California, San Diego) for assistance with humanized mouse model experiments, Dr. Stefan Maas (Lehigh University) for providing the luciferase RNA editing reporter construct, and Canada’s Michael Smith Genome Sciences Centre Library Construction, Sequencing and Bioinformatics teams.
Footnotes
AUTHOR CONTRIBUTIONS
M.A.Z., A.C.C., G.P., N.P.D., Q.J., L.B., L.A.C., C.N.M. and M.P. performed experiments, data analysis and experimental planning. A.S. conducted nanoproteomic experiments. C.H.M.J., H.J.C., N.P.D., C.L.B., A.S., L.A.C., M.A.M. and K.A.F. performed transcriptome profiling, and whole gene expression and pathway analysis. I.G. and R.W. cloned and constructed the lentiviral human wild-type JAK2. D.J.G. and R.W. carried out patient sample FACS sorting, assisted in mouse experiments and data analysis. M.M. provided clinical samples and scientific guidance. Q.J. and C.H.M.J. performed experimental planning, data analysis, and wrote the manuscript, which was reviewed and edited by all authors.
COMPETING FINANCIAL INTERESTS
The authors have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahamsson AE, Geron I, Gotlib J, Dao KH, Barroga CF, Newton IG, Giles FJ, Durocher J, Creusot RS, Karimi M, et al. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3925–3929. doi: 10.1073/pnas.0900189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn JH, Ahn J, Lin X, Zhang Q, Lee JH, Civelek M, Xiao X. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nature communications. 2015;6:6355. doi: 10.1038/ncomms7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blahna MT, Hata A. Smad-mediated regulation of microRNA biosynthesis. FEBS Lett. 2012;586:1906–1912. doi: 10.1016/j.febslet.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G, Sokol NS. ADAR mediates differential expression of polycistronic microRNAs. Nucleic acids research. 2014;42:5245–5255. doi: 10.1093/nar/gku145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, Liu M, Yuan YF, Fu L, Kong KL, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nature medicine. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley MR, Babovic S, Benz C, Knapp DJ, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K, et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nature cell biology. 2013;15:916–925. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- Crews LA, Jiang Q, Zipeto MA, Lazzari E, Court AC, Ali S, Barrett CL, Frazer KA, Jamieson CH. An RNA editing fingerprint of cancer stem cell reprogramming. Journal of translational medicine. 2015;13:52. doi: 10.1186/s12967-014-0370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson LS, Reavie L, Coussens M, Davalos V, Castillo-Martin M, Guijarro MV, Coffre M, Cordon-Cardo C, Aifantis I, Ibrahim S, et al. Limited miR-17–92 overexpression drives hematologic malignancies. Leukemia research. 2015;39:335–341. doi: 10.1016/j.leukres.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nature medicine. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, Trumpp A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- Fialkow PJ, Jacobson RJ, Papayannopoulou T. Chronic myelocytic leukemia: clonal origin in a stem cell common to the granulocyte, erythrocyte, platelet and monocyte/macrophage. The American journal of medicine. 1977;63:125–130. doi: 10.1016/0002-9343(77)90124-3. [DOI] [PubMed] [Google Scholar]
- Fumagalli D, Gacquer D, Rothe F, Lefort A, Libert F, Brown D, Kheddoumi N, Shlien A, Konopka T, Salgado R, et al. Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome. Cell reports. 2015;13:277–289. doi: 10.1016/j.celrep.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Das S, Samuel CE. Organization of the mouse RNA-specific adenosine deaminase Adar1 gene 5′-region and demonstration of STAT1-independent, STAT2-dependent transcriptional activation by interferon. Virology. 2008;380:338–343. doi: 10.1016/j.virol.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Samuel CE. STAT2-dependent induction of RNA adenosine deaminase ADAR1 by type I interferon differs between mouse and human cells in the requirement for STAT1. Virology. 2015;485:363–370. doi: 10.1016/j.virol.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geron I, Abrahamsson AE, Barroga CF, Kavalerchik E, Gotlib J, Hood JD, Durocher J, Mak CC, Noronha G, Soll RM, et al. Selective inhibition of JAK2-driven erythroid differentiation of polycythemia vera progenitors. Cancer cell. 2008;13:321–330. doi: 10.1016/j.ccr.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Goff DJ, Recart AC, Sadarangani A, Chun HJ, Barrett CL, Krajewska M, Leu H, Low-Marchelli J, Ma W, Shih AY, et al. A Pan-BCL2 inhibitor renders bone-marrow-resident human leukemia stem cells sensitive to tyrosine kinase inhibition. Cell stem cell. 2013;12:316–328. doi: 10.1016/j.stem.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenzl PM, Barlow DP. Macro lncRNAs: a new layer of cis-regulatory information in the mammalian genome. RNA biology. 2012;9:731–741. doi: 10.4161/rna.19985. [DOI] [PubMed] [Google Scholar]
- Han J, Zhang J, Chen L, Shen B, Zhou J, Hu B, Du Y, Tate PH, Huang X, Zhang W. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA biology. 2014;11:829–835. doi: 10.4161/rna.29624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner HM, Eterovic AK, Yuan Y, et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nature immunology. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hu YH, Zhang CY, Tian Z, Wang LZ, Li J. Aberrant protein expression and promoter methylation of p16 gene are correlated with malignant transformation of salivary pleomorphic adenoma. Archives of pathology & laboratory medicine. 2011;135:882–889. doi: 10.5858/2010-0181-OARI.1. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. The New England journal of medicine. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Crews LA, Jamieson CH. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1041–1046. doi: 10.1073/pnas.1213021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu W, Sun L, Aifantis KE, Yu B, Fan Y, Feng Q, Cui F, Watari F. Effects of physicochemical properties of nanomaterials on their toxicity. Journal of biomedical materials research Part A. 2014 doi: 10.1002/jbm.a.35384. [DOI] [PubMed] [Google Scholar]
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis I, Heinonen KM, Chagraoui J, Vainio S, Sauvageau G, Perreault C. The signaling protein Wnt4 enhances thymopoiesis and expands multipotent hematopoietic progenitors through beta-catenin-independent signaling. Immunity. 2008;29:57–67. doi: 10.1016/j.immuni.2008.04.023. [DOI] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Molecular cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome research. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K, ten Dijke P. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. The EMBO journal. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemlich Y, Greenberg E, Ortenberg R, Besser MJ, Barshack I, Jacob-Hirsch J, Jacoby E, Eyal E, Rivkin L, Prieto VG, et al. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. The Journal of clinical investigation. 2013;123:2703–2718. doi: 10.1172/JCI62980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annual review of biochemistry. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, Iizasa H, Davuluri RV, Nishikura K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M, Gaeta X, Loo K, Edwards M, Smale S, Cinkornpumin J, Xie Y, Listgarten J, Azghadi S, Douglass SM, et al. let-7 miRNAs can act through notch to regulate human gliogenesis. Stem cell reports. 2014;3:758–773. doi: 10.1016/j.stemcr.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardi E, Manzari C, Mastropasqua F, Aiello I, D’Erchia AM, Pesole G. Profiling RNA editing in human tissues: towards the inosinome Atlas. Scientific reports. 2015;5:14941. doi: 10.1038/srep14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Chan TH, Tenen DG, Chen L. RNA editome imbalance in hepatocellular carcinoma. Cancer research. 2014;74:1301–1306. doi: 10.1158/0008-5472.CAN-13-3485. [DOI] [PubMed] [Google Scholar]
- Qian J, Yao DM, Lin J, Wang YL, Han LX, Xu WR, Wu CY. Methylation of DAPK1 promoter: frequent but not an adverse prognostic factor in myelodysplastic syndrome. International journal of laboratory hematology. 2010;32:74–81. doi: 10.1111/j.1751-553X.2008.01131.x. [DOI] [PubMed] [Google Scholar]
- Qin YR, Qiao JJ, Chan TH, Zhu YH, Li FF, Liu H, Fei J, Li Y, Guan XY, Chen L. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer research. 2014;74:840–851. doi: 10.1158/0008-5472.CAN-13-2545. [DOI] [PubMed] [Google Scholar]
- Quintas-Cardama A, Choi S, Kantarjian H, Jabbour E, Huang X, Cortes J. Predicting outcomes in patients with chronic myeloid leukemia at any time during tyrosine kinase inhibitor therapy. Clinical lymphoma, myeloma & leukemia. 2014;14:327–334. e328. doi: 10.1016/j.clml.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell reports. 2014;9:1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval A, Tanner SM, Byrd JC, Angerman EB, Perko JD, Chen SS, Hackanson B, Grever MR, Lucas DM, Matkovic JJ, et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nature genetics. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers CL. Even better kinase inhibitors for chronic myeloid leukemia. The New England journal of medicine. 2010;362:2314–2315. doi: 10.1056/NEJMe1004430. [DOI] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Solomon O, Oren S, Safran M, Deshet-Unger N, Akiva P, Jacob-Hirsch J, Cesarkas K, Kabesa R, Amariglio N, Unger R, et al. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR) RNA. 2013;19:591–604. doi: 10.1261/rna.038042.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soverini S, de Benedittis C, Mancini M, Martinelli G. Mutations in the BCR-ABL1 Kinase Domain and Elsewhere in Chronic Myeloid Leukemia. Clinical lymphoma, myeloma & leukemia. 2015;15(Suppl):S120–128. doi: 10.1016/j.clml.2015.02.035. [DOI] [PubMed] [Google Scholar]
- Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Molecular cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallawi M, Zebrowski D, Rai R, Roether J, Schubert D, El Fray M, Engel F, Aifantis K, Boccaccini AR. Poly(glycerol sebacate)/poly(butylene succinate-dilinoleate) (PGS/PBS-DLA) fibrous scaffolds for cardiac tissue engineering. Tissue engineering Part C, Methods. 2014 doi: 10.1089/ten.tec.2014.0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veliz EA, Easterwood LM, Beal PA. Substrate analogues for an RNA-editing adenosine deaminase: mechanistic investigation and inhibitor design. Journal of the American Chemical Society. 2003;125:10867–10876. doi: 10.1021/ja029742d. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nature genetics. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LD, Rao TN, Rowe RG, Nguyen PT, Sullivan JL, Pearson DS, Doulatov S, Wu L, Lindsley RC, Zhu H, et al. The role of Lin28b in myeloid and mast cell differentiation and mast cell malignancy. Leukemia. 2015;29:1320–1330. doi: 10.1038/leu.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. The Journal of biological chemistry. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nature structural & molecular biology. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, Xia D, Yang Y, Deng K, Xiao H, et al. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PloS one. 2012;7:e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipeto MA, Jiang Q, Melese E, Jamieson CH. RNA rewriting, recoding, and rewiring in human disease. Trends in molecular medicine. 2015;21:549–559. doi: 10.1016/j.molmed.2015.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.