Abstract
The constitutive androstane receptor (CAR; NR1I3) is a member of the nuclear receptor superfamily that functions as a xenosensor, serving to regulate xenobiotic detoxification, lipid homeostasis and energy metabolism. CAR activation is also a key contributor to the development of chemical hepatocarcinogenesis in mice. The underlying pathways affected by CAR in these processes are complex and not fully elucidated. microRNAs (miRNAs) have emerged as critical modulators of gene expression and appear to impact many cellular pathways, including those involved in chemical detoxification and liver tumor development. In this study, we used deep sequencing approaches with an Illumina HiSeq platform to differentially profile microRNA expression patterns in livers from wild type C57BL/6J mice following CAR activation with the mouse CAR-specific ligand activator, 1,4-bis-[2-(3,5,-dichloropyridyloxy)] benzene (TCPOBOP). Bioinformatic analyses and pathway evaluations were performed leading to the identification of 51 miRNAs whose expression levels were significantly altered by TCPOBOP treatment, including mmu-miR-802-5p and miR-485-3p. Ingenuity Pathway Analysis of the differentially expressed microRNAs revealed altered effector pathways, including those involved in liver cell growth and proliferation. A functional network among CAR targeted genes and the affected microRNAs was constructed to illustrate how CAR modulation of microRNA expression may potentially mediate its biological role in mouse hepatocyte proliferation.
Keywords: microRNA, liver cancer, constitutive androstane receptor, TCPOBOP, mouse
Introduction
The CAR receptor (NR1I3) is a xenosensor that regulates biotransformation function and many physiological processes, including lipid metabolism, glucose metabolism, hormonal regulation, cell growth and apoptosis [24,26,58,61]. CAR is unusual among nuclear receptors in that it is constitutively active in the absence of any ligand and primarily expressed in liver hepatocytes. The receptor’s activity is modulated through chemical interactions which serve to release the receptor from a cytoplasmic tethering complex, allowing its translocation to the nucleus. A wide range of chemicals can alter CAR activity, including direct receptor ligand activators, inverse receptor agonists as well as indirect CAR activators [23,27,44]. CAR modulators vary in structure and origin and certain CAR ligands function in a species-specific manner. CITCO (6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehydeO-(3,4-dichlorobenzyl)oxime) is a direct human-specific CAR ligand [31], whereas TCPOBOP (1,4-bis[2-(3,5-dichloropyridyloxy)]benzene) is a direct mouse-specific CAR ligand [42,57]. In contrast, phenobarbital (PB) is an indirect activator of human, mouse and rat CAR [37,53]. Indirect CAR activators function through a signaling network, triggering the dephosphorylation of CAR and its subsequent nuclear translocation [36,67]. In the nucleus, CAR heterodimerizes with RXR and interacts with DNA targets to regulate gene expression networks that modulate downstream cellular biology [15,38].
Activation or alteration of CAR signaling may contribute to the development of liver hyperplasia and hepatomegaly [19,46], although species differences in these responses among rodents and humans have been reported [46,63]. Biological implications of CAR function also likely impact pathophysiological processes including cholestatic liver disease [25,68], hyperbilirubinemia [18], and obesity and type II diabetes [10,14]. Thus, given its central role as a xenosensor, CAR signaling pathways impinge upon a variety of disease scenarios.
MicroRNAs (miRNAs) are small noncoding RNAs that are increasingly recognized for their important regulatory roles as regulators of gene expression and modulators of many critical processes, including cell proliferation, apoptosis, organ development, metabolism, inflammation, and cancer [21,35,47]. In these respects, detection of certain miRNAs in tissues or in body fluids has been advanced as diagnostic markers for various disease states [21,32,69]. Given their reported roles as regulators of xenobiotic detoxification, liver development, and liver cancer, the biological functions of hepatic miRNAs have been actively investigated [6,17]. For example, miR-122, a liver-specific miRNA, was suggested as a diagnostic marker for hepatocellular carcinoma (HCC) [30]. Its expression is down-regulated in HCC [8,56], and the restoration of miR-122 expression decreased tumorigenesis, angiogenesis and intrahepatic metastasis in mouse HCC models [56].
A limited number of studies have implicated CAR as a regulator of miRNA expression. For example, Shizu and co-workers reported that PB-induced CAR activation down-regulated the expression of hepatic miR-122 [49]. Takwi et al reported an apparent inverse correlation of miR-137 and CAR expression in parental and doxorubicin-resistant neuroblastoma cells [55]. However, the potential interrelationships of CAR activation with miRNA regulation in liver cancer progression remains poorly understood. In the current study, we used deep sequencing methodologies to differentially profile the miRNA populations in liver tissue from mice comparatively treated with the specific mouse CAR ligand, TCPOBOP. Pathway analysis and integrative analysis of CAR-targeted miRNAs and mRNAs was performed to further inform the involvement of CAR signaling in hepatocarcinogenesis.
Methods and Materials
Materials
Sequencing reagents were obtained from Illumina (San Diego, CA). TCPOBOP (>99%) was synthesized by the Environmental Health Laboratory in the Department of Environmental and Occupational Health Safety at the University of Washington (Seattle, WA).
Animal maintenance and chemical treatments
All animal care and experimental procedures complied with protocols approved by the Institutional Animal Care and Use Committee at The Pennsylvania State University. Wild type C57BL/6 male mice were purchased from Charles River and permitted to acclimate at least one week prior to treatment. The mice were maintained under a standard 12 h light, 12 h dark cycle at a constant temperature (23 ± 1 C) with 45-65% humidity. Water and standard chow were provided ad libitum. A total of 6–8-week-old male C57BL/6 mice were treated with a single dose of 2 mg/kg of TCPOBOP (n=3) or vehicle (DMSO) (n=3), via single IP injection. TCPOBOP is a potent and persistent mouse CAR agonist. After 72h, liver tissues were immediately harvested from the mice following euthanasia via CO2 asphyxiation.
RNA extraction and miRNA sequencing
Total RNA was extracted from the collected liver tissue using Trizol (Qiagen, Germnatown, MD), and the mirVana miRNA Isolation Kit (Invitrogen, Carlsbad, CA) according to the manufacturers’ protocol. Total RNA was submitted to the Penn State Genomics Core Facility (Penn State University, PA) for sequencing. Briefly, RNA quality was assessed using NanoDrop 1000 (ThermoScientific, Waltham, MA), Qubit 2.0 (ThermoFisher, Waltham, MA), and Agilent Bioanalyzer 2100 (Agilent Technologies Inc., Santa Clara, CA) instrumentation. RNA (260/280>2.0 and 260/230 ratios of 1.5–2.0 as determined by the Nanodrop, and RIN > 9.0 as determined by the Bioanalyzer) was used for further analysis. Libraries were prepared using the Illumina TruSeq Small RNA Sample Preparation Kit (Illumina, San Diego, CA) according to the manufacturer's protocol. A total of six miRNA libraries (three vehicle-treated samples and three TCPOBOP-treated samples) were sequenced on the Illumina HiSeq 2500 platform (Illumina) in Rapid Run mode using 50 nt single read sequencing. The raw sequence data were processed with the miRDeep2 bioinformatics module and differentially expressed microRNAs were assessed using the Bioconductor statistics package, as detailed below.
miRNA sequencing data analysis
Raw reads from miRNA sequencing were processed using miRDeep2 [13]. Briefly, 3’ adapter sequences were removed from raw sequencing reads. Reads shorter than 14 nucleotides were discarded, then the mouse reference genome GRCm38 (NCBI_Assembly:GCA_000001635.2) was used for alignment. Only reads that mapped perfectly to the genome were used for miRNA detection. Expression values for the detected miRNAs were exported using miRDeep2.
Expression data of the known miRNAs exported from miRDeep2 were further analyzed in the R Bioconductor DESeq2 package [29]. After quality checking with scatter plots and Principle Component Analysis (PCA), a negative binomial-based generalized linear model and Wald statistics were used according to the DESeq2 package to test for the differential expression in the miRNA sequencing datasets. The list of differentially expressed miRNAs exported from DESeq2 was further filtered with exclusion of those with p-values >0.05. A heatmap of differentially expressed miRNAs was produced using the pheatmap function from the R package.
RT-qPCR
For data validation, stem-loop qPCR was performed using the TaqMan® MicroRNA Assay system (Applied Biosystems, Grand Island, NY). The selective assays included hsa-miR-34a (Assay ID#000426), hsa-miR-122-5p (Assay ID#002245), mmu-miR-182 (Assay ID#002599), , hsa-miR-200b (Assay ID#002251), mmu-miR-203-5p (Assay ID#002580) , mmu-miR-329 (Assay ID#000192), hsa-miR-222b (Assay ID#002276), and mmu-miR-802-3p (Assay ID#463561_mat), qPCR reactions were performed on a CFX96 Real Time PCR system (Bio-Rad, Hercules, CA). Each sample was run in triplicate. miRNA expression was normalized with the reference miRNA U6 snRNA (Assay ID#001973). The ΔΔCt method was used to calculate fold change in gene expression as previously described [39].
Pathway analysis and miRNA:mRNA integrative analyses by IPA
The Ingenuity Pathway analysis (IPA, Ingenuity Systems, Redwood City, CA; www.ingenuity.com) was used to identify functional groups and molecular networks from the differentially expressed (DE) miRNA datasets generated by Bioconductor.
Transcriptome datasets generated from TCPOBOP treated and vehicle treated C57BL/6 mice by microarray were downloaded from GEO (GSE13688). The lists of differentially expressed mRNAs and miRNAs were imported separately into IPA, followed by integrated analyses using the microRNA Target Filter tool within IPA. This tool prioritizes experimentally validated and predicted mRNA targets for each miRNA, and generates the pairing results of miRNAs and mRNAs, with opposite or same expression patterns. The paired miRNAs and mRNAs were further pooled in a sub list for pathway enrichment by IPA, and then a putative network was built among these miRNAs and mRNAs using the “liver cancer” functional category.
Results
Deep sequencing of miRNAs from TCPOBOP treated and vehicle-treated mice and data analysis using miRDeep2
To investigate whether CAR activation by TCPOBOP would alter miRNA expression profiles, we performed high-throughput short read deep sequencing of small RNAs using the Illumina HiSeq platform. The flow of the experimental scheme is shown in Figure 1. Total RNA was submitted for sequencing, since the Illumina TrueSeq library kit provides a 3’ adaptor fishing strategy to separate the small RNAs from long sequence mRNAs. The library was then sequenced from the 3’ adaptor end, reading the small RNAs. The raw reads were analyzed using miRDeep2 [13], a probabilistic algorithm designed to detect miRNAs from deep sequencing data based on the miRNA biogenesis model. Since the reads generated from the sequencer include a mixed pool of all the small RNAs, including mRNA degradation products and stRNAs, we performed 5’ adaptor decontamination, 3’ adaptor removal, and then filtered out reads smaller than 14 nt. The clean reads were used for alignment against the mouse reference genome. A list of 626 annotated mature miRNAs was generated and the sequences of unannotated small RNAs were then separated to another pool for identification of novel miRNA candidates.
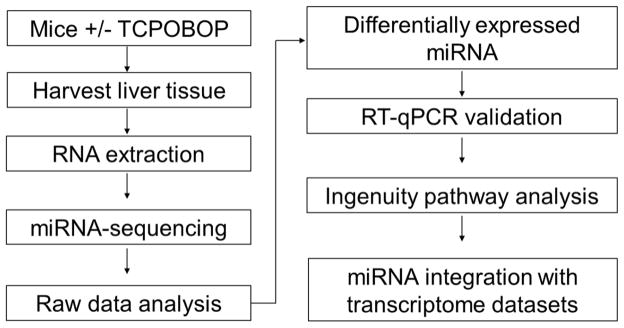
Figure 1.
Scheme of the experimental workflow. Mice were treated with TCPOBOP, and liver tissues were harvested, followed by total RNA extraction. RNA was sequenced on the Illumina HiSeq 2500 platform. Sequencing data were processed using miRDeep2 [29], and differentially expressed miRNAs were determined using DESeq2 package of Bioconductor R. Selected sequencing data were validated using RT-qPCR. The differentially expressed miRNAs were exported into IPA for pathway analysis and integrative analysis with a transcriptome dataset (GSE13688) [45].
Determination of differentially expressed (DE) known miRNAs in TCPOBOP treated and vehicle-treated mice
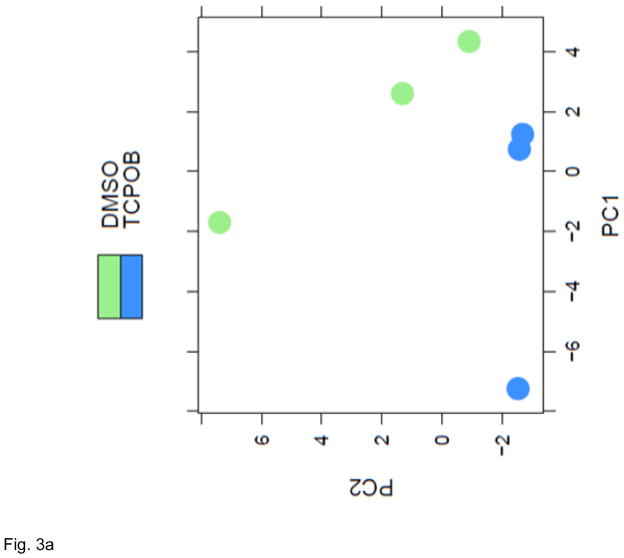
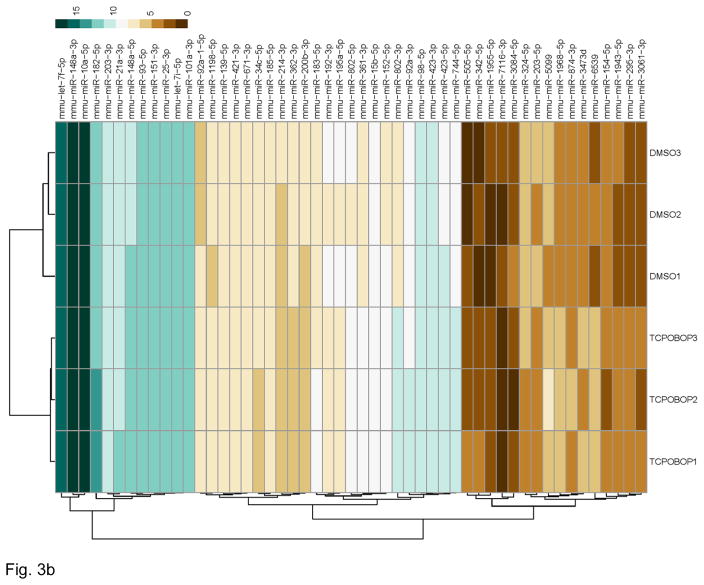
The annotated miRNA list was further subjected to quality check and differential expression analysis using the R Bioconductor DESeq2 package [29]. The quality of the dataset was assessed using its scatter plot function and through Principle Component Analysis (PCA). The scatterplot (Figure 2) of the datasets produced with the R hexbin package showed that the miRNA reads were very similar for all the samples, and that low counts were common. The plot shows the expected result that samples from the same treatment are more similar to each other, since the data are close to the diagonal, i.e., the data points are closely scattered around the line y=x. When the data scatter close to that diagonal, the samples are positively correlated and each treatment group exhibited this feature. From these analyses, the data were judged to exhibit high quality. The PCA analyses demonstrated that the respective treatment groups, TCPOBOP (blue) and vehicle (green), exhibited clear separation (Figure 3A). Following these quality checks, a negative binomial-based generalized linear regression was applied to the data using the R Bioconductor DESeq2 package. Differentially expressed miRNA profiles were obtained by comparing the data generated from TCPOBOP and vehicle control treated animals. A total of 51 differentially expressed miRNA genes were identified. A heat map of the 51 significantly differentially expressed miRNA genes is shown in Figure 3B. There were 36 miRNAs induced by the TCPOBOP treatment. In addition to induction, TCPOBOP treatment also resulted in the repression of 15 miRNAs. An extracted list of the top 10 differentially up-regulated and down-regulated miRNAs is presented in Table 1. A more comprehensive list of the raw data and their associated statistical features, i.e., all of the miRNAs that were significantly up- or down-regulated by TCPOBOP, using a p<.05 statistic, is presented in Supplemental Table 1.
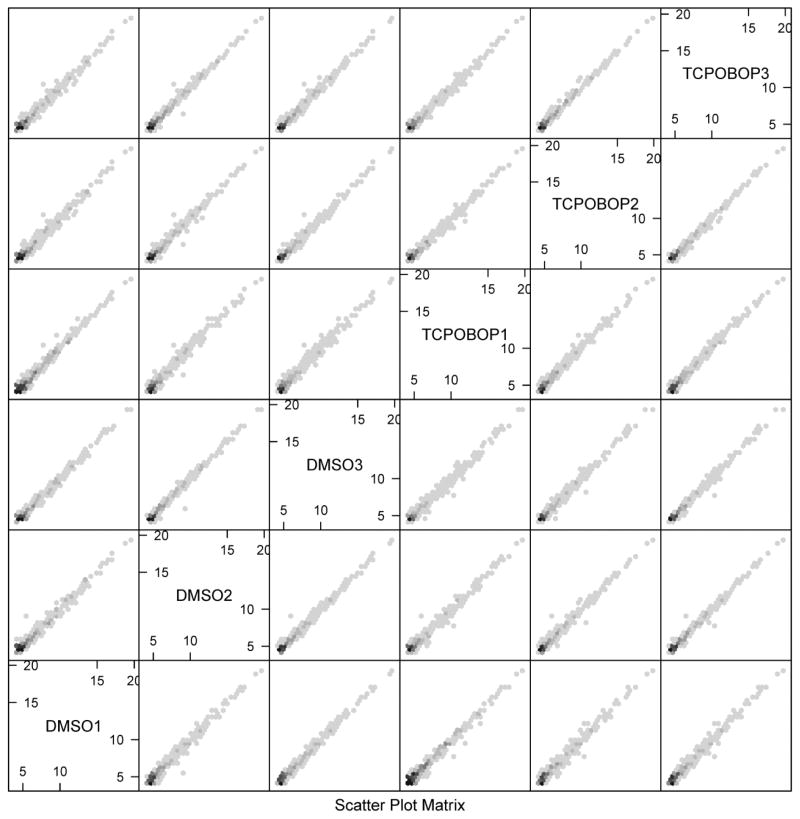
Figure 2.
Scatterplot matrix of the miRNA-seq data. As a quality check, a scatterplot matrix of the miRNA expression datasets exported from miRDeep2 was produced with the R hexbin package. Examining the pairwise correlation between all treatment groups, the scatterplot matrix shows all the pairwise scatter plots of the log2-transformed miRNA expression values from the 3 vehicle (DMSO) and 3 TCPOBOP treatment groups. Overall highly positive correlations were observed between all treatment groups, indicating that the raw sequence data were of high quality.
Figure 3.
CAR activation by TCPOBOP altered miRNA profiling. (A) PCA plot of normalized expression values of miRNAs. PCA analysis was performed using R Bioconductor DESeq2. A clear separation between the DMSO and TCPOBOP treatment samples was observed. (B) Hierarchical clustering analysis of differentially expressed miRNAs. A heatmap was produced using the pheatmap function from the pheatmap package in R. Differentially expressed miRNAs shown at the rows of the heatmap were clustered using Euclidean distance and complete linkage. The treatment groups shown in the columns were clustered using correlation distance and complete linkage. The color scale at the top-right corner illustrates the expression value of a particular miRNA on a log2 scale.
Table 1.
Selected TCPOBOP differentially expressed miRNAs.
| miRNA | Ratio | P-value |
|---|---|---|
| Up-regulated | ||
| mmu-miR-802-3p | 6.63 | 1.97E-52 |
| mmu-miR-6539 | 5.76 | 1.31E-12 |
| mmu-miR-1968-5p | 2.84 | 1.48E-06 |
| mmu-miR-1943-5p | 2.08 | 3.88E-03 |
| mmu-miR-802-5p | 2.05 | 3.17E-10 |
| mmu-miR-295-3p | 1.95 | 1.94E-02 |
| mmu-miR-342-5p | 1.92 | 3.66E-02 |
| mmu-miR-1955-5p | 1.87 | 4.60E-02 |
| mmu-miR-505-5p | 1.85 | 5.00E-02 |
| mmu-miR-182-5p | 1.83 | 5.04E-04 |
| Down-regulated | ||
| mmu-miR-122-5p | −1.01 | 0.86 |
| mmu-miR-3084-5p | −1.89 | 4.19E-02 |
| mmu-miR-214-3p | −1.87 | 6.62E-03 |
| mmu-miR-7116-3p | −1.74 | 3.94E-02 |
| mmu-miR-154-5p | −1.69 | 5.35E-02 |
| mmu-miR-203-5p | −1.65 | 2.01E-02 |
| mmu-miR-324-5p | −1.62 | 1.81E-02 |
| mmu-miR-34c-5p | −1.50 | 2.46E-02 |
| mmu-miR-139-5p | −1.39 | 5.49E-03 |
| mmu-miR-362-3p | −1.37 | 2.85E-02 |
Validation of expression of DE miRNA by Taqman assay
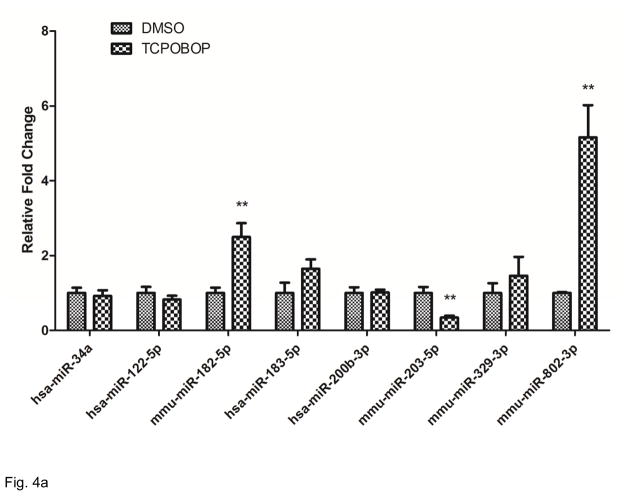
Based on results presented in Table 1 and Figure 4B, we used RT-PCR Taqman assays to selectively and quantitatively assess several of miRNAs of interest, chosen either for their dysregulated features or for their potential biological significance in the downstream pathway analysis. The results are presented in Figure 4A. Consistent with the miRNA deep sequencing analysis, mmu-miR-802-3p was detected as highly TCPOBOP up-regulated and mmu-miR-203-5p and mmu-miR-203-5P were significantly down-regulated. Although not statistically significant in our study, expression levels of miR-122-5p trended lower in TCPOBOP samples. Since this miRNA was reported previously as a repressed CAR target [49], we included miR-122-5p in the differentially expressed miRNA list for subsequent pathway analyses, resulting in a list of 52 miRNAs identified as TCPOBOP/CAR-regulated. Several other mRNA targets were selectively quantified as potentially implicated in our subsequent pathway analyses, but differential expression of these mRNAs did not reach statistically significant levels.
Figure 4.
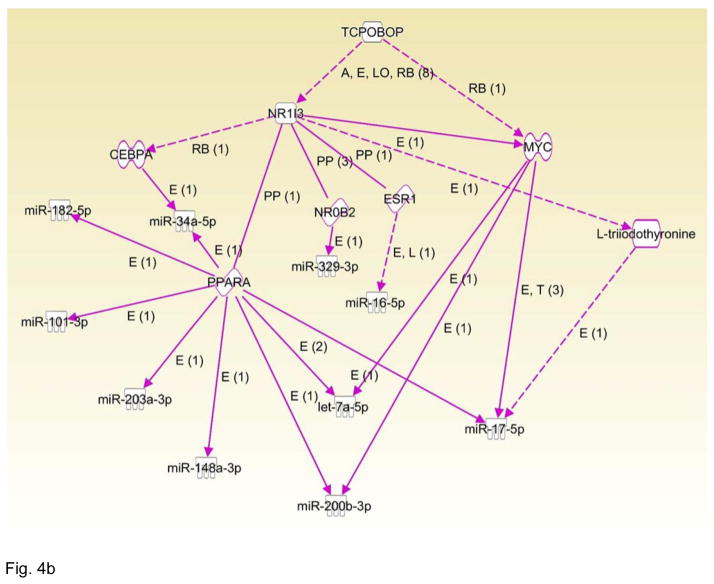
CAR targeted miRNAs. (A) Selected miRNAs were validated by RT-qPCR. Expression of levels of miR-182-5p, miR-802-3p and miR-203-5p showed results consistent with the derived RNA-seq data; **p<0.05. Though the trends of the expression levels of miR-122-5p and mi-183-5p agreed with the differential expression patterns ascertained by the RNA-seq analyses, these changes were not statistically significant RT-qPCR at the p<0.05 level. (B) CAR targeted miRNA network built by IPA. The network indicates that CAR regulates several miRNAs through MYC, ESR1, NO0B2, PPARa, and CEBPA.
CAR targeted analysis and global pathway analysis of DE miRNAs by IPA
To further investigate the differentially expressed miRNAs, we used IPA analysis to identify reported CAR target miRNAs in the dataset. Differentially expressed miRNAs were imported into IPA, and a network was built among CAR and the differentially expressed miRNAs. As shown in Figure 4B, several differentially expressed miRNAs were reported as indirectly regulated by CAR, through the C/EBPα, PPARα, NR0B2, ESR1, and MYC pathways. However, based on the data mining by IPA, none of the miRNAs were identified as directly regulated by CAR, likely due to the limited number of published studies available in this area.
In a strategy to further illuminate the functional pathways of the differentially expressed miRNAs, we used IPA to perform disease prediction and functional pathway enrichment of the 52 differentially expressed miRNAs (p≤0.05). Selected disease and function enrichments are shown in Table 2. For example, 22 miRNAs were significantly enriched in the digestive system cancer category, 13 miRNAs in the liver cancer category, 8 in hepatocellular carcinoma, and 10 in hematologic cancer. Other than diseases, molecular and cellular functional categories were also enriched, for example, 16 miRNAs were significantly enriched in the ‘proliferation of cells’ category. In addition, the ‘inflammation of organs’ category was significantly enriched (10 miRNAs; Table 2).
Table 2.
Selected disease and function enrichment.
| Diseases or functions |
No. of miRNAs |
P-value | miRNAs |
|---|---|---|---|
| Digestive system cancer | 22 | 4.58E-02 | let-7f-5p, miR-10a-5p, miR-128-3p, miR-139-5p, miR-148a-3p, miR-151-3p, miR-16-5p, miR-17-5p, miR-183-5p, miR-200b-3p, miR-203a-3p, miR-214-3p, miR-324-5p, miR-329-3p, miR-34c-5p, miR-361-3p, miR-421-3p, miR-423-5p, miR-671-3p, miR-874-3p, miR-92a-3p, miR-122-5p |
| Gastrointestinal tract cancer | 20 | 4.81E-04 | let-7f-5p, miR-10a-5p, miR-128-3p, miR-139-5p, miR-148a-3p, miR-151-3p, miR-16-5p, miR-17-5p, miR-183-5p, miR-200b-3p, miR-203a-3p, miR-214-3p, miR-324-5p, miR-329-3p, miR-34c-5p, miR-361-3p, miR-421-3p, miR-423-5p, miR-874-3p, miR-92a-3p |
| Liver cancer | 13 | 9.26E-02 | let-7f-5p, miR-10a-5p, miR-148a-3p, miR-151-3p, miR-16-5p, miR-17-5p, miR-183-5p, miR-200b-3p, miR-203a-3p, miR-34c-5p, miR-671-3p, miR-92a-3p, miR-122-5p |
| Hepatocellular carcinoma | 8 | 4.12E-06 | let-7f-5p, miR-148a-3p, miR-16-5p, , miR-122-5p, miR-17-5p, miR-183-5p, miR-34c-5p, miR-92a-3p |
| Hematologic cancer | 10 | 2.09E-03 | let-7f-5p, miR-101-3p, miR-10a-5p, miR-148a-3p, miR-16-5p, miR-17-5p, miR-342-5p, miR-34c-5p, miR-361-3p, miR-92a-3p |
| Proliferation of cells | 16 | 2.76E-03 | let-7f-5p, miR-101-3p, miR-10a-5p, miR-128-3p, miR-139-5p, miR-148a-3p, miR-16-5p, miR-17-5p, miR-182-5p, miR-183-5p, miR-185-5p, miR-200b-3p, miR-203a-3p, miR-34c-5p, miR-92a-3p, miR-122-5p |
| Cell apoptosis | 10 | 3.08E-02 | let-7f-5p,miR-122-5p,miR-16-5p,miR-17-5p,miR-183-5p,miR-185-5p,miR-200b-3p,miR-203a-3p,miR-214-3p,miR-34c-5p |
| Inflammation of organ | 10 | 4.03E-05 | let-7f-5p, miR-16-5p, miR-17-5p, miR-185-5p, miR-200b-3p, miR-203a-3p, miR-324-5p, miR-423-3p, miR-423-5p, miR-92a-3p |
Integrative analysis of miRNA and mRNA expression profiles by IPA
A particular focus of our interests was on liver cancer, as activation of CAR has been reported as a key driver of mouse liver tumor promotion [19,65], and independently, alteration of miRNA expression profiles have been linked to liver cancer development [6,30,56,62]. To further investigate how the alteration of miRNA profiles may affect biological functions in the liver upon TCPOBOP treatment, we investigated the association of miRNA and their target mRNAs, using a published TCPOBOP- treated mouse transcriptome dataset (GSE13688) [45]. This dataset was generated using microarray analyses from liver tissues of the C56Bl/6J mice treated with TCPOBOP, highly comparable to that used in our own study. We downloaded the dataset from the GEO database (http://www.ncbi.nlm.nih.gov/geo/), extracted 60 differentially expressed, TCPOBOP-regulated mRNAs (fold-change ≥ 1.4, p≤0.05), and performed integrative analysis using the differentially expressed miRNAs from our dataset and the differentially expressed mRNAs from the microarray dataset.
The differentially expressed lists of 52 miRNAs and 60 mRNAs were imported to IPA independently, and the miRNA target filter analysis was applied to compare the two datasets. Comprehensive miRNA target information is built into the IPA target filtering tool, using a combination of multiple target prediction tools such as TargetScan, miRecord and TarBase [59]. Combining our own derived differentially expressed miRNA list with IPA’s comprehensive list of miRNA inferences resulted in the generation of an IPA correlation analysis encompassing 92 miRNA and mRNA pairs, with different confidence levels, including those experimentally-observed (based on TarBase, and miRecords data mining tools), and those exhibiting designations of high prediction and moderate prediction (based on target prediction from TargetScan tools, using the seed sequences of the mature miRNAs). The resulting IPA list of miRNA and mRNA pairing results is shown in Table 3. Twenty two of the 28 miRNAs generated from the IPA analysis also were identified as significantly differentially expressed in our deep sequencing dataset (Supplemental Table 1). Certain of the IPA identified miRNAs were predicted to pair with multiple mRNAs. For example, miR-361-3p paired to ppara, ogt, mlxipl, cyp2a6, cebpa, abcc3, and abcc1; and, miR-185-5p paired to serping1, scarb1,ppara, nr2f6, insig1, cpt1a, and abcc1. Some miRNAs paired only with a single mRNA, for example, miR-671-3p and insig1, and some mRNAs paired with multiple miRNAs. For example, ppara paired with miR-361-3p, miR-185-5p, miR-128-3p, let-7a-5p, miR-10a-5p, miR-16-5p, miR-101-3p, miR-17-5p, miR-291a-3p, and miR-154-5p. insig1 paired with miR-185-5p, miR-182-5p, miR-203a-3p, miR-92a-3p, miR-183-5p, and miR-671-3p. In addition, adipor2 associated with miR-6967-5p, let-7a-5p, miR-17-5p, miR-329-3p, miR-423-3p, miR-200b-3p, and miR-34a-5p (Table 3).
Table 3.
miRNA and mRNA pairing.
| miRNA | No. of target mRNAs | mRNA |
|---|---|---|
| miR-361-3p | 7 | ppara, ogt, mlxipl, cyp2a6, cebpa, abcc3, abcc1 |
| miR-185-5p | 7 | serping1, scarb1,ppara, nr2f6, insig1, cpt1a, abcc1 |
| miR-128-3p | 6 | ppara, nr2f6, nr1d2, igf1, cyp39a1, igf1 |
| miR-6967-5p | 6 | igf1, cyp8b1, cyp2b6, cebpa, adipor2, abcc3 |
| miR-182-5p | 5 | ogt, nr1d2, insig1, cyp8b1, cebpa |
| let-7a-5p | 5 | ppara, igf1, c2,adipor2, abcc2 |
| miR-10a-5p | 5 | uap1, stat3, ppara, insig2, abcc3 |
| miR-342-5p | 4 | nr2f6, fasn, cyp8b1, cpt1a |
| miR-874-3p | 4 | stat3, mlxipl, cpt1a, c4a/c4b |
| miR-16-5p | 4 | ppara, ogt, igf1, fasn |
| miR-148a-3p | 4 | lipa, igf1, cpt1a, abcc3 |
| miR-101-3p | 4 | ppara, ogt, nr1d2, cebpa |
| miR-17-5p | 4 | stat3, ppara, mbtps1, adipor2 |
| miR-203a-3p | 4 | nr1d2, insig1, cyp8b1, cyp1a2 |
| miR-92a-3p | 3 | insig1, fgb, cebpa |
| miR-214-3p | 3 | scarb1, igf1, cyp8b1 |
| miR-291a-3p | 2 | ppara, insig2 |
| miR-183-5p | 2 | ppp1r3c, insig1 |
| miR-423-5p | 2 | c2, apoa5 |
| miR-744-5p | 2 | cyp2b6, cyp21a2 |
| miR-329-3p | 2 | apoa5, adipor2 |
| miR-324-5p | 2 | fgb, cpt1a |
| miR-671-3p | 1 | insig1 |
| miR-421-3p | 1 | nr1d2 |
| miR-423-3p | 1 | adipor2 |
| miR-200b-3p | 1 | adipor2 |
| miR-34a-5p | 1 | adipor2 |
| miR-154-5p | 1 | ppara |
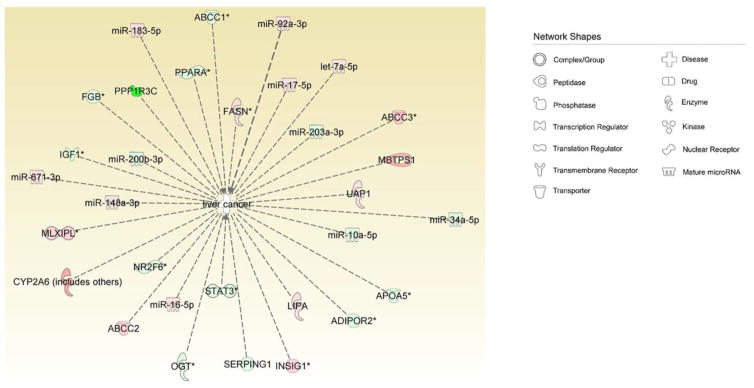
To further comprehend and visualize the biological effects of the mRNA and miRNA pairing, we pooled the paired miRNAs and mRNAs, imported these into IPA, and performed pathway analysis. Consistently, the digestive system cancer and liver cancer categories were significantly enriched from the miRNA and mRNA pool, as shown in Table 4 and Figure 5. In addition, cell proliferation, cell death and inflammation of organs were all enriched from the mRNA and miRNA pool (Table 4). As demonstrated in Figure 5, 11 miRNAs and 20 mRNAs from the pairing miRNAs and mRNA pools associated with liver cancer.
Table 4.
Selected diseases and function enrichment of pooled miRNAs and mRNAs.
| Diseases or functions |
P-value | Molecules |
|---|---|---|
| Digestive system cancer | 1.76E-05 | ABCC1,ABCC2,ABCC3,ADIPOR2,APOA5,CEBPA,CYP1A2,CYP21 A2,CYP2A6 (includes others),FASN,FGB,IGF1,INSIG1,INSIG2,let-7a-5p,LIPA,MBTPS1,miR-10a-5p,miR-128-3p,miR-148a-3p,miR-16-5p,miR-17-5p,miR-183-5p,miR-200b-3p,miR-203a-3p,miR-214-3p,miR-324-5p,miR-329-3p,miR-34a-5p,miR-361-3p,miR-421-3p,miR-423-5p,miR-671-3p,miR-874-3p,miR-92a-3p,MLXIPL,NR1D2,NR2F6,OGT,PPARA,PPP1R3C,SCARB1,SERPING1,STAT3,UAP1 |
| Liver cancer | 6.53E-05 | ABCC1, ABCC2, ABCC3, ADIPOR2, APOA5, CYP2A6 (includes others), FASN, FGB, IGF1, INSIG1, LIPA, MBTPS1, MLXIPL, NR2F6, OGT, PPARA, PPP1R3C, SERPING1, STAT3, UAP1, let-7a-5p, miR-10a-5p, miR-148a-3p, miR-16-5p, miR-17-5p, miR-183-5p, miR-200b-3p, miR-203a-3p, miR-34a-5p, miR-671-3p, miR-92a-3p |
| Proliferation of cells | 8.77E-07 | ABCC1,ABCC2,ABCC3,ADIPOR2,ATP,CEBPA,CYP2A6 (includes others),CYP2B6,FASN,IGF1,INSIG1,let-7a-5p,LIPA,miR-101-3p,miR-10a-5p,miR-128-3p,miR-148a-3p,miR-16-5p,miR-17-5p,miR-182-5p,miR-183-5p,miR-185-5p,miR-200b-3p,miR-203a-3p,miR-34a-5p,miR-92a-3p,MLXIPL,NR2F6,OGT,PPARA,SCARB1,STAT3 |
| Cell death | 1.36E-03 | ABCC1,ABCC3,ATP,C4A/C4B,CEBPA,CYP2B6,FASN,IGF1,let-7a-5p,LIPA,MBTPS1,miR-16-5p,miR-17-5p,miR-183-5p,miR-185-5p,miR-200b-3p,miR-203a-3p,miR-214-3p,miR-34a-5p,miR-92a-3p,OGT,PPARA,SCARB1,STAT3 |
| Inflammation of organ | 2.75E-09 | ABCC2,ADIPOR2,ATP,C4A/C4B,CEBPA,IGF1,INSIG1,let-7a-5p,LIPA,miR-16-5p,miR-17-5p,miR-185-5p,miR-200b-3p,miR-203a-3p,miR-324-5p,miR-423-3p,miR-423-5p,miR-92a-3p,PPARA,STAT3 |
Figure 5.
CAR targeted miRNA and mRNA in liver cancer. Paired CAR targeted miRNAs and mRNAs were pooled for IPA pathway analysis. The “liver cancer” category was significantly enriched from multiple miRNAs and mRNAs. Network shapes are shown in the figure inset. Red labels represent up-regulated miRNAs or mRNAs. Green labels represent down-regulated miRNAs or mRNAs.
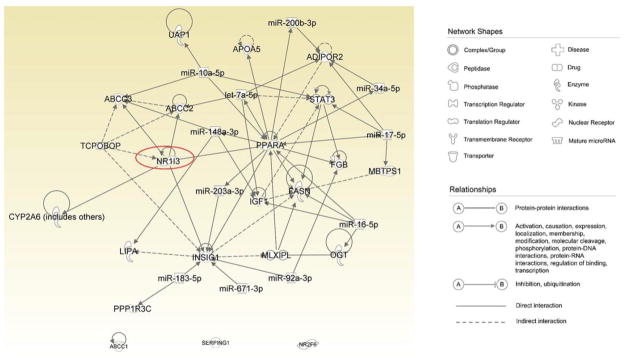
Further, a network of these affected mRNA and miRNAs was assembled using IPA, as a means for predicting interactions of the miRNAs and mRNA that may coalesce in the development of liver cancer. Figure 6 illustrates the predictive CAR-mediated miRNA:mRNA pathways activated by TCPOBOP in the liver cancer scheme. These analyses suggest that activation of CAR by TCPOBOP may contribute to liver cancer development through alteration of miRNA:mRNA interactions.
Figure 6.
Putative miRNA:mRNA interaction upon activation of CAR by TCPOBOP in liver cancer. Paired miRNAs and mRNAs enriched in the “liver cancer” category were imported inIPA to assess the interactions among these parameters. Connections were established by IPA based on data mining or prediction according to miRNA seed sequences. Network shapes and relationships are shown in the figure inset. Red colors represent miRNAs while green colors represent mRNAs.
Discussion
In this study, the effects of CAR activation by TCPOBOP on miRNA profiling was investigated along with the predictive effects on miRNA-targeted mRNA profiling. 51 miRNAs were significantly altered by TCPOBOP-treatment. To facilitate prediction of the biological disruption caused by the miRNA dysregulation, we performed functional analysis on the altered miRNAs. Further, miRNA target genes were predicted based on data mining using IPA.
As shown in Figure 3B, Table 1 and Supplementary Table 1, a list of differentially expressed mouse liver miRNAs was identified as either up- or down-regulated upon CAR activation by TCPOBOP. Previous reports have indicated miR-122-5p as a direct CAR target miRNA [49]. Although exhibiting a similar down-regulated trend to the previous report, miR-122-5p differential expression levels did not reach statistical significant in our study, perhaps due to the relatively high abundance of this miRNA in the liver combined with the variation among individual mice. Another reported CAR target miRNA is miR-137, which also regulates expression of CAR in neuroblastoma cells [55], but was not significantly altered in our system. We used a data mining approach by IPA to predict how CAR activation by TCPOBOP may regulate the newly identified miRNAs in our dataset. As shown in Figure 4B, most of the miRNAs appear indirectly regulated by CAR, via CAR-targeted genes. For example, miR-34a-5p was regulated by cebpa and pparα, which are regulated by CAR. These interactions are first based on the previous reports that C/EBPα and HNF4α synergistically cooperate with CAR to transactivate CYP2B6 in human hepatocytes [4], and that CAR regulates transcription and transactivation of PPARα [26]. Pulikkan et al reported that miR-34a is regulated by C/EBPα during granulopoiesis in AML [43]. miR-34a is also a reported PPARα target in mouse liver, according to the miRNA microarray experiments performed by Shah et al [48]. In addition, several other miRNAs identified in our dataset, including miR-182-5p, miR-101-3p, miR-203a-3p, mir-148a-3p, mir-200b-3p, let-7a-5p, mir-17-5p, and mir-34a-5p, were also reported in Shah’s study, and further confirmed the crosstalk of PPARα and CAR on regulating miRNA profiling [48]. Furthermore, miR-329-3p was regulated by NR0B2 (SHP) [51], reported to interact with CAR and inhibit transcriptional activity of CAR [1,40,41]. CAR also regulates mir-17-5p [21,48,50], miR-200b-3p [3] and let-7a-5p [34] via myc, which was induced by CAR in the mouse liver upon TCPOBOP activation [5]. Additionally, CAR reportedly regulates miR-16-5p via esr1 [64]. Lastly, CAR signaling and ER signaling exhibit crosstalk as CAR was reported to be activated by estrogens [33], and Grip-1 was shown to be a coactivator of both CAR and ER [33].
To further understand how these miRNAs may function in the liver, we performed pathway enrichment analysis using IPA. As shown in Table 2, a panel of miRNAs is involved in “cell proliferation” and “liver cancer” categories, which is consistent with previous reports demonstrating a likely association of CAR activation with cellular proliferation and development of hepatocellular carcinoma [19,46,65]. The potential roles of miRNAs in the disruption of these biological processes are not well elucidated. Our results suggest that CAR regulation in these biological processes may involve alteration of miRNA profiling. Functionally, these enriched miRNAs have been reported as involved in cell proliferation in other tissues or cell lines. For example, miR-122 decreases growth of SK-HEP-1 cells [56], and growth of HepG2 cells in cell culture [9]. In addition, inhibition of mmu-miR-182 decreases expansion of mouse T-helper cell in culture [52]. Inhibition of human miR-183 (the homologue of mouse miR-183), increases proliferation of A549 cells in cell culture following impairment by carbon dioxide [60]. Further, mouse mmu-miR-34c (also known as mmu-miR-34a), was reported to decrease proliferation of mouse ovarian surface epithelial cells [7], and growth of M23 cells [66].
With respect to liver cancer category, decrease of human miR-122 (homologue of mouse miR-122) was associated with liver cancer in humans [16,56]. Down-regulation of mouse miR-122 is also associated with liver cancer in mouse [11], consistent with our dataset (Tables 1 and 2). Up-regulation of hsa-miR-148a (homologue of mouse miR-148a) is associated with HCC in human [20], also consistent with the up-regulation of mouse miR-148a in our dataset (Table 2; Suppl Table 1). Further, up-regulation of rat rno-miR-671 (homologue of mouse miR-671) in serum was associated with experimental hepatocarcinogenesis in rat, which is in line with the up-regulation of miR-671 in our data (Suppl Table 1). Moreover, human let-7f (hsa-let-7f, homologue of mmu-let-7f), and rat let-7f (rno-let-7f, homologue of mmu-let-7f), were associated with hepatocellular carcinoma in humans [20], and experimental hepatocarcinogenesis in rat, respectively [54]. These reports are in line with our observation that mmu-let-7f was up-regulated 1.21-fold upon CAR activation (Suppl Table 1). In sum, the differentially miRNAs identified as enriched in the “liver cancer” and “cell proliferation” categories in our analyses are well supported by previous studies.
To further understand how these altered miRNAs disrupt biological process and lead to liver cancer, we performed miRNA and mRNA integrative analysis, by combining two datasets: using miRNA sequencing data from our study and published transcriptomics data generated from TCPOBOP treatment. The analysis yielded 92 miRNA and mRNA pairs, with multiple miRNAs sharing the same mRNA targets, or vice versa. Of all the pairs, only two pairs are experimentally observed according to TarBase and miRecords data mining tools, miR-295-3p and insig2 [28], and miR-93-5p and stat3 [12]. The remaining pairs are highly or moderate predicted based on the seed sequences of the miRNAs, using the TargetScan prediction tool. However, through manual data mining, we identified the PPARα activation microRNA / microarray datasets published by Shah et al [48]. According to that study, miR-101-3p, let 7a-5p (also termed let-7f-5p) and mir-17-5p (also termed miR-93-5p) are PPARα target miRNAs, consistent with our network prediction. In addition, several other CAR targeted miRNAs in our dataset, such as miR-182-5p, miR-203a-3p, mir-148a-3p, mir-200b-3p and mir-34a-5p, were reported as PPARα target miRNAs [48]; however, IPA failed to extract this information. Furthermore, Pulikkan et al reported that cebpα regulated miR-34a-5p during granulopoiesis in acute myeloid leukemia [43], which also was not predicted by IPA. On the other hand, IPA predicted several other cebpa targeted miRNAs based on their seed sequences, including miR-361-3p, miR-6967-5p, miR-182-5p, miR-101-3p, and miR-92a-3p. In summary, IPA provides powerful data mining functions and prediction tools by integrating several databases, but based on our experience, manual literature research is still recommended to obtain more comprehensive information.
Since a principle aim of our investigation was to gather more insight with respect to how CAR regulated miRNAs function in liver cancer, we performed another enrichment of the pooled miRNAs and mRNAs, and used those enriched in the ‘liver cancer’ category to build an integrative network using IPA (Figure 5). Some of the interactions were predicted by TargetScan based on the seed sequences of the miRNAs, while others were experimentally supported. For example, ppara and mmu-miR-203, let-7a-5p (mmu-let-7c), mir-200b-3p, mir-17-5p, and mir-148a-3p were reported by Shah and colleagues [48]. CAR was reported to bind the PPREs (peroxisome proliferator response elements) upon heterodimerizing with RXR [26]. Our findings further suggested cross-talk of CAR signaling and PPARα signaling on the regulation of miRNAs in liver cancer. In addition, CAR also appears to interact with other miRNAs via abcc2 (encoding ATP-binding cassette, sub-family C, member 2), abcc1 (encoding ATP-binding cassette, sub-family C, member 3), and insig1 (encoding Insulin-Induced Gene 1), which together with PPARα, are all involved in lipid metabolism. These associations suggest that dysregulation of lipid metabolism is associated with liver cancer, in line with previous studies (reviewed in [2] and [22]). However, validation of these data mining-based conclusions will require follow-up with further experimentation. In future studies, CAR knockout mice could be used to affirm the miRNA expression profiles upon TCPOBOP treatment. Similarly, the miRNA:mRNA pair data will need to be experimentally verified by using loss of function or gain of function studies in animal models or in primary hepatocyte cultures.
Conclusions
In summary, we examined differential miRNA signaling in mouse liver as regulated by TCPOBOP, a selective CAR ligand activator. We conducted pathway analyses on the CAR-targeted miRNAs in relation to liver cancer and using the IPA miRNA:mRNA integrative tool, offer a predictive framework as to how these miRNAs may interact with mRNAs. These approaches enabled focus onto a subset of miRNA:mRNA pairs representing potential new diagnostic markers for liver cancer.
Supplementary Material
Illumina HiSeq 2500-derived, TCPOBOP differentially expressed miRNA sequencing counts and statistics normalized to library size. Individual count data from 3 DMSO control mice and 3 TCPOBOP treated mice are shown.
Highlights.
microRNA expression is altered in mouse liver by TCPOBOP activation of CAR
51 miRNAs were significantly altered by TCPOBOP-treatment
CAR activation may contribute to liver cancer by altering miRNA:mRNA interactions
Acknowledgments
This study was supported by a grant from the NIH General Medical Sciences Institute, GM066411 (CJO), and by DuPont/Haskell Laboratories (RH, XH). The authors also acknowledge the expert assistance of Dr. Craig Praul and the PSU Huck Life Sciences Institutes Genomics Core Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bae Y, Kemper JK, Kemper B. Repression of CAR-mediated transactivation of CYP2B genes by the orphan nuclear receptor, short heterodimer partner (SHP) 1. DNA Cell Biol. 2004;23:81. doi: 10.1089/104454904322759894. [DOI] [PubMed] [Google Scholar]
- 2.Baenke F, Peck B, Miess H, Schulze A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech. 2013;6:1353. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai JX, Yan B, Zhao ZN, Xiao X, Qin WW, Zhang R, Jia LT, Meng YL, Jin BQ, Fan DM, Wang T, Yang AG. Tamoxifen represses miR-200 microRNAs and promotes epithelial-to-mesenchymal transition by up-regulating c-Myc in endometrial carcinoma cell lines. Endocrinology. 2013;154:635. doi: 10.1210/en.2012-1607. [DOI] [PubMed] [Google Scholar]
- 4.Benet M, Lahoz A, Guzman C, Castell JV, Jover R. CCAAT/enhancer-binding protein alpha (C/EBPalpha) and hepatocyte nuclear factor 4alpha (HNF4alpha) synergistically cooperate with constitutive androstane receptor to transactivate the human cytochrome P450 2B6 (CYP2B6) gene: application to the development of a metabolically competent human hepatic cell model. J Biol Chem. 2010;285:28457. doi: 10.1074/jbc.M110.118364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco-Bose WE, Murphy MJ, Ehninger A, Offner S, Dubey C, Huang W, Moore DD, Trumpp A. C-Myc and its target FoxM1 are critical downstream effectors of constitutive androstane receptor (CAR) mediated direct liver hyperplasia. Hepatology. 2008;48:1302. doi: 10.1002/hep.22475. [DOI] [PubMed] [Google Scholar]
- 6.Chen XM. MicroRNA signatures in liver diseases. World J Gastroenterol. 2009;15:1665. doi: 10.3748/wjg.15.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 8.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diao S, Zhang JF, Wang H, He ML, Lin MC, Chen Y, Kung HF. Proteomic identification of microRNA-122a target proteins in hepatocellular carcinoma. Proteomics. 2010;10:3723. doi: 10.1002/pmic.201000050. [DOI] [PubMed] [Google Scholar]
- 10.Dong B, Saha PK, Huang W, Chen W, Abu-Elheiga LA, Wakil SJ, Stevens RD, Ilkayeva O, Newgard CB, Chan L, Moore DD. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci U S A. 2009;106:18831. doi: 10.1073/pnas.0909731106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Z, Zhang C, Wu R, Hu W. Tumor suppressor p53 meets microRNAs. J Mol Cell Biol. 2011;3:44. doi: 10.1093/jmcb/mjq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foshay KM, Gallicano GI. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev Biol. 2009;326:431. doi: 10.1016/j.ydbio.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, He J, Zhai Y, Wada T, Xie W. The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J Biol Chem. 2009;284:25984. doi: 10.1074/jbc.M109.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Xie W. PXR and CAR at the crossroad of drug metabolism and energy metabolism. Drug Metab Dispos. 2010;38:2091. doi: 10.1124/dmd.110.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garofalo M, Croce CM. microRNAs: Master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 17.Gooderham NJ, Koufaris C. Using microRNA profiles to predict and evaluate hepatic carcinogenic potential. Toxicol Lett. 2014;228:127. doi: 10.1016/j.toxlet.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Huang W, Zhang J, Chua SS, Qatanani M, Han Y, Granata R, Moore DD. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc Natl Acad Sci U S A. 2003;100:4156. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, Moore DD. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol Endocrinol. 2005;19:1646. doi: 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- 20.Huang YS, Dai Y, Yu XF, Bao SY, Yin YB, Tang M, Hu CX. Microarray analysis of microRNA expression in hepatocellular carcinoma and non-tumorous tissues without viral hepatitis. J Gastroenterol Hepatol. 2008;23:87. doi: 10.1111/j.1440-1746.2007.05223.x. [DOI] [PubMed] [Google Scholar]
- 21.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–R44. [PubMed] [Google Scholar]
- 22.Jiang JT, Xu N, Zhang XY, Wu CP. Lipids changes in liver cancer. J Zhejiang Univ Sci B. 2007;8:398. doi: 10.1631/jzus.2007.B0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jyrkkarinne J, Windshugel B, Makinen J, Ylisirnio M, Perakyla M, Poso A, Sippl W, Honkakoski P. Amino acids important for ligand specificity of the human constitutive androstane receptor. J Biol Chem. 2005;280:5960. doi: 10.1074/jbc.M411241200. [DOI] [PubMed] [Google Scholar]
- 24.Kachaylo EM, Pustylnyak VO, Lyakhovich VV, Gulyaeva LF. Constitutive androstane receptor (CAR) is a xenosensor and target for therapy. Biochemistry (Mosc ) 2011;76:1087. doi: 10.1134/S0006297911100026. [DOI] [PubMed] [Google Scholar]
- 25.Kakizaki S, Takizawa D, Tojima H, Yamazaki Y, Mori M. Xenobiotic-sensing nuclear receptors CAR and PXR as drug targets in cholestatic liver disease 10. Curr Drug Targets. 2009;10:1156. doi: 10.2174/138945009789735174. [DOI] [PubMed] [Google Scholar]
- 26.Kassam A, Winrow CJ, Fernandez-Rachubinski F, Capone JP, Rachubinski RA. The peroxisome proliferator response element of the gene encoding the peroxisomal beta-oxidation enzyme enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase is a target for constitutive androstane receptor beta/9-cis-retinoic acid receptor-mediated transactivation. J Biol Chem. 2000;275:4345. doi: 10.1074/jbc.275.6.4345. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Wang H. Activation of xenobiotic receptors: driving into the nucleus. Expert Opin Drug Metab Toxicol. 2010;6:409. doi: 10.1517/17425251003598886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo J, Chen M, Huang H, Yuan T, Zhang M, Zhang K, Deng S. Circulating microRNA-122a as a diagnostic marker for hepatocellular carcinoma. Onco Targets Ther. 2013;6:577. doi: 10.2147/OTT.S44215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, Moore JT. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278:17277. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- 32.Merkel O, Hamacher F, Laimer D, Sifft E, Trajanoski Z, Scheideler M, Egger G, Hassler MR, Thallinger C, Schmatz A, Turner SD, Greil R, Kenner L. Identification of differential and functionally active miRNAs in both anaplastic lymphoma kinase (ALK)+ and ALK- anaplastic large-cell lymphoma. Proc Natl Acad Sci U S A. 2010;107:16228. doi: 10.1073/pnas.1009719107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min G, Kim H, Bae Y, Petz L, Kemper JK. Inhibitory cross-talk between estrogen receptor (ER) and constitutively activated androstane receptor (CAR). CAR inhibits ER-mediated signaling pathway by squelching p160 coactivators. J Biol Chem. 2002;277:34626. doi: 10.1074/jbc.M205239200. [DOI] [PubMed] [Google Scholar]
- 34.Mongroo PS, Noubissi FK, Cuatrecasas M, Kalabis J, King CE, Johnstone CN, Bowser MJ, Castells A, Spiegelman VS, Rustgi AK. IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Res. 2011;71:2172. doi: 10.1158/0008-5472.CAN-10-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morishita A, Masaki T. miRNA in hepatocellular carcinoma. Hepatol Res. 2015;45:128. doi: 10.1111/hepr.12386. [DOI] [PubMed] [Google Scholar]
- 36.Mutoh S, Osabe M, Inoue K, Moore R, Pedersen L, Perera L, Rebolloso Y, Sueyoshi T, Negishi M. Dephosphorylation of threonine 38 is required for nuclear translocation and activation of human xenobiotic receptor CAR (NR1I3) J Biol Chem. 2009;284:34785. doi: 10.1074/jbc.M109.048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Omiecinski CJ, Coslo DM, Chen T, Laurenzana EM, Peffer RC. Multi-species analyses of direct activators of the constitutive androstane receptor. Toxicol Sci. 2011;123:550. doi: 10.1093/toxsci/kfr191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omiecinski CJ, Vanden Heuvel JP, Perdew GH, Peters JM. Xenobiotic metabolism, disposition, and regulation by receptors: from biochemical phenomenon to predictors of major toxicities. Toxicol Sci. 2011;120(Suppl 1):S49–S75. doi: 10.1093/toxsci/kfq338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Page JL, Johnson MC, Olsavsky KM, Strom SC, Zarbl H, Omiecinski CJ. Gene expression profiling of extracellular matrix as an effector of human hepatocyte phenotype in primary cell culture. Toxicol Sci. 2007;97:384. doi: 10.1093/toxsci/kfm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmieri G, Perego M, Alkabes M, Palmieri R. ACTH-adrenal system and exogenous steroids. Ric Clin Lab. 1979;9:253. doi: 10.1007/BF02932177. [DOI] [PubMed] [Google Scholar]
- 41.Park YY, Kim HJ, Kim JY, Kim MY, Song KH, Cheol PK, Yu KY, Shong M, Kim KH, Choi HS. Differential role of the loop region between helices H6 and H7 within the orphan nuclear receptors small heterodimer partner and DAX-1. Mol Endocrinol. 2004;18:1082. doi: 10.1210/me.2003-0339. [DOI] [PubMed] [Google Scholar]
- 42.Poland A, Mak I, Glover E, Boatman RJ, Ebetino FH, Kende AS. 1,4-Bis[2-(3,5-dichloropyridyloxy)]benzene, a potent phenobarbital-like inducer of microsomal monooxygenase activity. Mol Pharmacol. 1980;18:571. [PubMed] [Google Scholar]
- 43.Pulikkan JA, Peramangalam PS, Dengler V, Ho PA, Preudhomme C, Meshinchi S, Christopeit M, Nibourel O, Muller-Tidow C, Bohlander SK, Tenen DG, Behre G. C/EBPalpha regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood. 2010;116:5638. doi: 10.1182/blood-2010-04-281600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pustylnyak VO, Gulyaeva LF, Lyakhovich VV. Induction of cytochrome P4502B: role of regulatory elements and nuclear receptors. Biochemistry (Mosc ) 2007;72:608. doi: 10.1134/s000629790706003x. [DOI] [PubMed] [Google Scholar]
- 45.Rezen T, Tamasi V, Lovgren-Sandblom A, Bjorkhem I, Meyer UA, Rozman D. Effect of CAR activation on selected metabolic pathways in normal and hyperlipidemic mouse livers. BMC Genomics. 2009;10:384. doi: 10.1186/1471-2164-10-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross J, Plummer SM, Rode A, Scheer N, Bower CC, Vogel O, Henderson CJ, Wolf CR, Elcombe CR. Human constitutive androstane receptor (CAR) and pregnane X receptor (PXR) support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogens phenobarbital and chlordane in vivo 3. Toxicol Sci. 2010;116:452. doi: 10.1093/toxsci/kfq118. [DOI] [PubMed] [Google Scholar]
- 47.Ross SA, Davis CD. The emerging role of microRNAs and nutrition in modulating health and disease. Annu Rev Nutr. 2014;34:305. doi: 10.1146/annurev-nutr-071813-105729. [DOI] [PubMed] [Google Scholar]
- 48.Shah YM, Morimura K, Yang Q, Tanabe T, Takagi M, Gonzalez FJ. Peroxisome proliferator-activated receptor alpha regulates a microRNA-mediated signaling cascade responsible for hepatocellular proliferation. Mol Cell Biol. 2007;27:4238. doi: 10.1128/MCB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shizu R, Shindo S, Yoshida T, Numazawa S. MicroRNA-122 down-regulation is involved in phenobarbital-mediated activation of the constitutive androstane receptor. PLoS One. 2012;7:e41291. doi: 10.1371/journal.pone.0041291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song G, Wang L. Transcriptional mechanism for the paired miR-433 and miR-127 genes by nuclear receptors SHP and ERRgamma. Nucleic Acids Res. 2008;36:5727. doi: 10.1093/nar/gkn567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F, Heinz GA, Fang Z, Li N, Bissels U, Hatam F, Jahn A, Hammoud B, Matz M, Schulze FM, Baumgrass R, Bosio A, Mollenkopf HJ, Grun J, Thiel A, Chen W, Hofer T, Loddenkemper C, Lohning M, Chang HD, Rajewsky N, Radbruch A, Mashreghi MF. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 53.Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274:6043. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 54.Sukata T, Sumida K, Kushida M, Ogata K, Miyata K, Yabushita S, Uwagawa S. Circulating microRNAs, possible indicators of progress of rat hepatocarcinogenesis from early stages. Toxicol Lett. 2011;200:46. doi: 10.1016/j.toxlet.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Takwi AA, Wang YM, Wu J, Michaelis M, Cinatl J, Chen T. miR-137 regulates the constitutive androstane receptor and modulates doxorubicin sensitivity in parental and doxorubicin-resistant neuroblastoma cells. Oncogene. 2014;33:3717. doi: 10.1038/onc.2013.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, Hsu MT, Hsiao M, Huang HD, Tsou AP. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 57.Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20:2951. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol. 2002;61:1. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- 59.Viollet C, Davis DA, Reczko M, Ziegelbauer JM, Pezzella F, Ragoussis J, Yarchoan R. Next-Generation Sequencing Analysis Reveals Differential Expression Profiles of MiRNA-mRNA Target Pairs in KSHV-Infected Cells. PLoS One. 2015;10:e0126439. doi: 10.1371/journal.pone.0126439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vohwinkel CU, Lecuona E, Sun H, Sommer N, Vadasz I, Chandel NS, Sznajder JI. Elevated CO(2) levels cause mitochondrial dysfunction and impair cell proliferation. J Biol Chem. 2011;286:37067. doi: 10.1074/jbc.M111.290056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- 62.Wong CM, Kai AK, Tsang FH, Ng IO. Regulation of hepatocarcinogenesis by microRNAs. Front Biosci (Elite Ed) 2013;5:49. doi: 10.2741/e595. [DOI] [PubMed] [Google Scholar]
- 63.Yamada T, Okuda Y, Kushida M, Sumida K, Takeuchi H, Nagahori H, Fukuda T, Lake BG, Cohen SM, Kawamura S. Human hepatocytes support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogen sodium phenobarbital in an in vivo study using a chimeric mouse with humanized liver 1. Toxicol Sci. 2014;142:137. doi: 10.1093/toxsci/kfu173. [DOI] [PubMed] [Google Scholar]
- 64.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K, Minami Y, O'Malley BW, Kato S. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 65.Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64:7197. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- 66.Yan D, Zhou X, Chen X, Hu DN, Dong XD, Wang J, Lu F, Tu L, Qu J. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Invest Ophthalmol Vis Sci. 2009;50:1559. doi: 10.1167/iovs.08-2681. [DOI] [PubMed] [Google Scholar]
- 67.Yoshinari K, Kobayashi K, Moore R, Kawamoto T, Negishi M. Identification of the nuclear receptor CAR:HSP90 complex in mouse liver and recruitment of protein phosphatase 2A in response to phenobarbital. FEBS Lett. 2003;548:17. doi: 10.1016/s0014-5793(03)00720-8. [DOI] [PubMed] [Google Scholar]
- 68.Yoshinari K, Ohno H, Benoki S, Yamazoe Y. Constitutive androstane receptor transactivates the hepatic expression of mouse Dhcr24 and human DHCR24 encoding a cholesterogenic enzyme 24-dehydrocholesterol reductase 3. Toxicol Lett. 2012;208:185. doi: 10.1016/j.toxlet.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 69.Zhi F, Cao X, Xie X, Wang B, Dong W, Gu W, Ling Y, Wang R, Yang Y, Liu Y. Identification of circulating microRNAs as potential biomarkers for detecting acute myeloid leukemia. PLoS One. 2013;8:e56718. doi: 10.1371/journal.pone.0056718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Illumina HiSeq 2500-derived, TCPOBOP differentially expressed miRNA sequencing counts and statistics normalized to library size. Individual count data from 3 DMSO control mice and 3 TCPOBOP treated mice are shown.