Abstract
CRISPR-Cas9 has recently emerged as a promising system for multiplexed genome editing as well as epigenome and transcriptome perturbation. Due to its specificity, ease of use and highly modular programmable nature, it has been widely adopted for a variety of applications such as genome editing, transcriptional inhibition and activation, genetic screening, DNA localization imaging, and many more. In this review, we will discuss non-editing applications of CRISPR-Cas9 for transcriptome perturbation, metabolic engineering, and synthetic biology.
Graphical abstract
Introduction
Since the early days of genetic engineering there has been a need for control of gene expression. Naturally occurring transcription factors (TFs) have traditionally been used to achieve this goal (reviewed in [1]). However, their limited DNA binding sequence space required installing specific sequences within the transcription regulatory elements of the target genes. This can be technically difficult and may have unintended consequences on gene expression. Zinc fingers (ZFs) and transcription activator-like effectors (TALEs) were developed to overcome the fixed binding sequence requirements of native TFs. However, both ZFs and TALEs have significant limitations. ZFs have complicated design criteria and large highly repetitive TALE genes are difficult to synthesize and clone (reviewed in [2,3]). These challenges have recently been overcome using CRISPR-Cas9 based TFs. In this article we overview the biochemical properties of CRISPR-Cas9 based TFs that enable such flexibility and describe their applications to synthetic gene circuit design and multiplexed perturbation of native gene networks.
Transcriptional regulation with CRISPR-Cas9
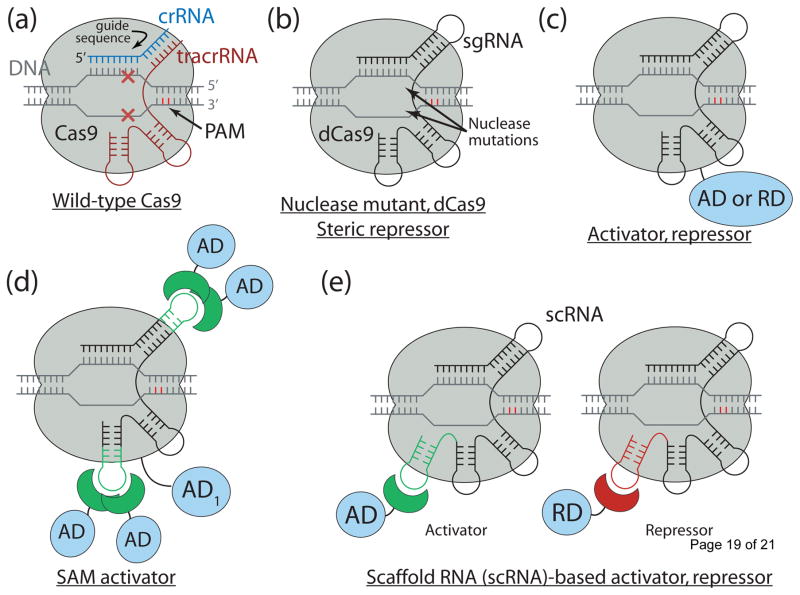
Cas9 is a key protein of bacterial Type II CRISPR adaptive immune system (reviewed in [4]). In its native context, Cas9 is an RNA-guided endonuclease that is responsible for targeted degradation of the invading foreign DNA – plasmids and phages. Cas9 is directed to its DNA targets by forming a ribonucleoprotein complex with two small non-coding RNAs: CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) (Fig 1(a)). By elegant engineering, crRNA and tracrRNA can be joined end-to-end and transcribed as a single guide RNA (sgRNA) that too efficiently directs Cas9 protein to DNA targets encoded within the guide sequence of sgRNA [5]. The optimal DNA target of the complex is determined by a Watson-Crick base pairing of a short ~20-nt sequence within sgRNA (within the crRNA in wild-type), termed the guide sequence, adjacent to a few nucleotide long conserved motif recognized directly by Cas9 protein (protospacer adjacent motif, PAM) [5,6]. Despite this, a few mismatches between guide sequence and target DNA can be tolerated [5,7–9], more so within the 5′ proximal position of the guide sequence. Cas9 nuclease can be converted into deactivated Cas9 (dCas9), an RNA-programmable DNA-binding protein, by mutating two key residues within its nuclease domains (Fig. 1(b)) [5,6].
Fig. 1.
Overview of Cas9 nuclease and dCas9-based transcription factors. (a) Wild-type Cas9 endonuclease guided by crRNA:tracrRNA to a specific site in DNA creates a double-stranded DNA break. (b) dCas9, nuclease deactivated mutant of Cas9, is an RNA programmable DNA binding protein. It can act as a steric repressor of transcription in pro- and eukaryotes. sgRNA is an artificial chimeric molecule consisting of crRNA and tracrRNA molecules connected with a short loop. (c) dCas9 fusion with various transcription effectors can be used to repress or activate transcription. (d) Effector domains can be recruited by sgRNA in addition to dCas9 for enhanced activity. (e) sgRNA can be modified with specific protein binding hairpins to concurrently recruit repressor or activator domains in the same cell.
In the simplest case, dCas9 can repress transcription by sterically interfering with transcription initiation or elongation by being targeted to the gene of interest with a properly chosen sgRNA [5–8,10–14]. The repression strength is strongly dependent on the position with respect to the target promoter as well as the nature of promoter itself [7,8,10,11]. In prokaryotes, repression of up to 1000-fold was achieved when targeting dCas9 to either DNA strand within a promoter or to the non-template DNA strand downstream [7,8,10,15–18]. However, in eukaryotic cells such steric repression is weaker: only up to 2- and 20-fold repression was observed with natural promoters in mammalian and yeast cells correspondingly [7,12,13]. As a notable exception, synthetic promoters specifically constructed for direct repression by dCas9 can be repressed up to 100-fold in mammalian cells [14]. Potentially stronger downregulation of natural promoters in eukaryotic cells can be achieved by fusing dCas9 with transcriptional repressor domains (RD in Fig. 1(c)) [12,19–21]. Up to 50-fold repression was achieved using a fusion with a transcriptional repressor Mxi1 in yeast [12]. In mammalian cells, dCas9 fusion with histone demethylase LSD1 can also be used to repress transcription by distal enhancers [22]. The most widely used dCas9-KRAB fusion is strong and highly specific in both yeast and mammalian cells [23–26].
Transcriptional activation can also be achieved by engineering dCas9, sgRNA, or both to recruit transcription effectors to the DNA (Fig. 1(c)–(e)). In E. coli, an activator has been constructed by fusing the ω subunit of native RNA polymerase to dCas9 in a ω subunit deficient host [8] (Fig. 1(c)). This activator was moderately active with weak promoters (up to 23-fold activation), but had progressively less effect with the increasing basal strength of the target promoter. The simplest eukaryotic activators can be engineered by fusing dCas9 with VP64 (and its variations), EDLL, TAD, p65, and p300core effector domains [12,21,27–32]. Activation levels using a single sgRNA per target promoter are generally weak. However, activation can be further increased by appropriately tiling the target promoter with multiple sgRNAs [29–31]. This observation led to the construction of activators that contain multiple effector domains per dCas9 molecule. First, three different activation domains (VP64, p65, and Rta) were fused to form a so-called VPR activator [33]. This activator was 1–2 orders of magnitude more efficient than a single VP64 fusion. Second, dCas9 was fused with SunTag, a peptide containing an array of 10 or more tandem repeats [19,34]. Each repeat then recruited a VP64 transcription activator fused to an engineered antibody fragment scFv. One potential drawback of this system was an aggregation prone nature of scFv. Nevertheless, about 50-fold transcription activation was achieved using this activator guided by a single sgRNA in human K562 cells [34]. In addition to dCas9 fusions, sgRNA can also be engineered to recruit effectors [35,36]. Konermann et al. created so-called Synergistic Activation Mediator (SAM) by inserting two bacteriophage MS2 RNA hairpins into non-essential regions of sgRNA (Fig. 1(d)). Each MS2 hairpin binds two molecules of MS2 coat protein MCP, each fused with a pair of transcriptional activators: p65 and human heat shock factor HSF1. Guiding dCas9-VP64 fusion protein by this chimeric sgRNA results in activation within 2- to >104-fold range and is generally better than activation of the same promoter with dCas9-VP64 guided by 8 tiled standard sgRNAs. An elegant corollary of sgRNA scaffolding is that it enables concurrent independent use of repressor, activator, or other effector domains in the same cell using a single dCas9 ortholog. Zalatan et al. used three mutually orthogonal RNA hairpin:RNA binding protein pairs (MS2:MCP, com:Com, PP7:PCP) to simultaneously recruit VP64 and KRAB effectors (Fig 1(e)) [24]. In principle, a repertoire of independent TFs can be further expanded using engineered dCas9 variants with altered specificity, natural Cas9 orthologs, as well as other CRISPR systems [37–41]. Recently, the efficiency of most modern eukaryotic dCas9-based activators and their combinations was compared directly by Chavez et al. within human, mouse, and fruit fly cells [42]. SAM, VPR, and SunTag activators were the most efficient, with SAM typically having up to 5-fold advantage over the other activators in human cell lines.
Beneath the apparent simplicity of dCas9-mediated transcriptional regulation, the rules that determine kinetics and specificity of DNA binding as well as the strength of transcriptional regulation are nuanced depending on context. The main categories of context are the degree of similarity between the target DNA site and the guide sequence of sgRNA, the nucleotide composition of the sgRNA guide sequence, the chromatin state of the target DNA, the host organism, and the specific ortholog of dCas9 used. In terms of DNA targeting specificity, complementarity between DNA and sgRNA is generally less important at the 5′ end of the guide sequence, even tolerating DNA or RNA bulges in the bound conformation [43,44]. Both Cas9 and dCas9 appear to tolerate similar numbers of mismatches between the sgRNA guide and the target DNA [8,19] and have similar binding kinetics [45]. However, DNA cleavage by Cas9 has an effectively higher sensitivity to mismatches, presumably since it requires an additional conformational change to induce cleavage [46,47]. This property can be leveraged to achieve transcriptional regulation while avoiding DNA cleavage using nucleolytically active Cas9 guided by sgRNA with mismatches or truncations within the guide sequence [8,48]. Mismatch sensitivity is further affected by the concentration of dCas9/sgRNA complex relative to the target DNA [49], which itself depends on the nature and relative amounts of sgRNAs expressed [50]. In eukaryotes, efficiency of regulation is diminished by guanine enrichment and adenine depletion of the guide sequence since it decreases sgRNA stability [51]. Chromatin state also affects dCas9 DNA binding: it is much weaker within condensed chromatin regions, therefore very good match between sgRNA and target DNA sequence is required [9,46,52]. Further studies of dCas9 and specificity-improved Cas9 proteins [53,54] in the aforementioned contexts will inform further development of computational algorithms [11,43,51,52,55] that incorporate these effects to precisely select the types and amounts of sgRNAs that are most appropriate to specific applications.
CRISPR-Cas9 regulatable synthetic promoters and gene circuits
The ability of dCas9 to regulate practically arbitrary transcriptional regulatory elements enables their interconnection into de novo networks, also known as synthetic gene circuits. These collections of interacting genes can be programmed with sensing, storage, or actuation functions combined to perform desired tasks [56,1]. Synthetic circuit design is facilitated by having a large panel of modular promoters with defined characteristics that can be predictably interfaced with each other. Such synthetic dCas9-regulatable promoters have been created by strategically installing dCas9/sgRNA binding sites within a parent promoter. By appropriately choosing these binding sites and their corresponding sgRNA guide sequences a panel of distinct promoter/transcription factor pairs can be produced.
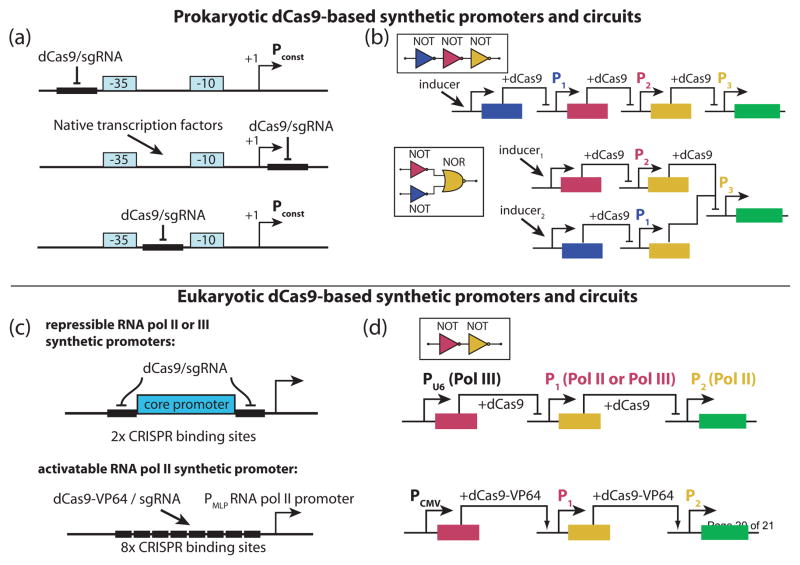
In prokaryotes, custom promoters have been constructed by introducing dCas9/sgRNA binding sites in the direct vicinity of the RNA polymerase binding site of parent natural promoters (Fig. 2(a)). Four orthogonal sgRNA/promoter pairs were computationally designed and experimentally validated within two synthetic circuits, double and triple repression cascades (Fig. 2(a,b), top). These dCas9-based repressors were then interfaced with native inducible promoters PBAD, PLlacO-1, and PLuxI, resulting in predictable regulation with both synthetic and native TFs (Fig. 2(a), middle). Nielsen et al. constructed and validated five orthogonal promoter/repressor pairs (Fig. 2(a), bottom) [10]. These promoters were used to construct logical NOT and NOR gates and their combinations (Fig. 2(b), bottom); the output of a synthetic NOT[NOR(A,B)] logic circuit was connected to native E. coli network via an sgRNA directing repression of an endogenous transcription factor gene. In a separate study, a panel of orthogonal T7 RNA polymerase driven dCas9-regulatable hybrid promoters was designed, tested, and used to control a reconstituted metabolic pathway [49].
Fig. 2.
dCas9-based synthetic promoters and circuits. (a) Orthogonal modular dCas9-repressible promoters have been designed for building synthetic gene circuits in Escherichia coli. (b) Proof of principle logic circuits were built using the promoters shown in (a) [10,11]. (c) dCas9-repressible (top) and dCas9-activatable (bottom) eukaryotic modular promoters have been designed for building synthetic circuits in eukaryotes. (d) Proof of principle regulatory circuits were constructed using the promoters shown in (c) [14,57].
Eukaryotic synthetic gene circuits require additional considerations for efficient linking between input and output of their components. Both RNA polymerase II and III promoters have been used to transcribe active sgRNAs. RNA polymerase III promoters can directly produce active nuclear localized sgRNA, but are significantly limited in their scope of regulation. RNA polymerase II promoters do not suffer from this drawback, but their transcripts are typically exported out of the nucleus so special methods need to be used to generate active sgRNA within the nucleus. This can be done by a) using non-standard RNA polymerase III terminators (U1 3′ Box and MASC [35]), b) releasing active sgRNA using site-specific RNA nucleases or self-cleaving ribozymes [57,58], or c) releasing it from an intron [14,57]. Sterically repressible promoters with high fold repression (~100-fold) have been constructed by flanking a minimal RNA polymerase II or RNA polymerase III promoter with multiple dCas9 binding sites [14,48] (Fig 2(c), top). A modular dCas9-VP64 activatable synthetic promoter has been engineered by inserting multiple dCas9 binding sites upstream of a weak RNA polymerase II promoter [57] (Fig 2(c), bottom). Combining these custom promoters and the approaches for production of active sgRNAs enabled construction of two-stage repressor and activator circuits [14,48,57] (Fig 2(d)), as well as a programmable kill switch, by combining transcription regulation and nucleolytic activity of wild-type Cas9 [48]. To demonstrate a medically relevant application, a dCas9-based logic circuit was wired to native gene network to sense and kill cancer cells [59].
Common to both pro- and eukaryotic synthetic circuit design are two fundamental considerations: a) the crosstalk within a circuit, as well as between the circuit and the host, and b) the dynamics of interactions between the circuit components. First, naïve choice of sgRNA guide sequences can result in undesired crosstalk that leads to interference with the circuit or host function [8]. This problem can be addressed by computationally designing sgRNA guide sequences and the corresponding target DNA sites in order to minimize undesired interactions [11]. Second, while dCas9/sgRNA complex binds its cognate target DNA very tightly, the dissociation kinetics is slow (half-life of ~6hrs or more in vitro [45,60]). The DNA dissociation rate of dCas9/sgRNA complex can likely be increased while maintaining sufficiently tight DNA binding by introducing a small number of strategically designed mismatches or truncations within the sgRNA guide sequence [61]. Alternatively, by tagging dCas9 for degradation [32,62] the effective half-life of DNA-bound dCas9/sgRNA may be decreased. Further advances in addressing these considerations will enable construction of complex circuits with precise dynamics expanding the scope of applications.
Applications
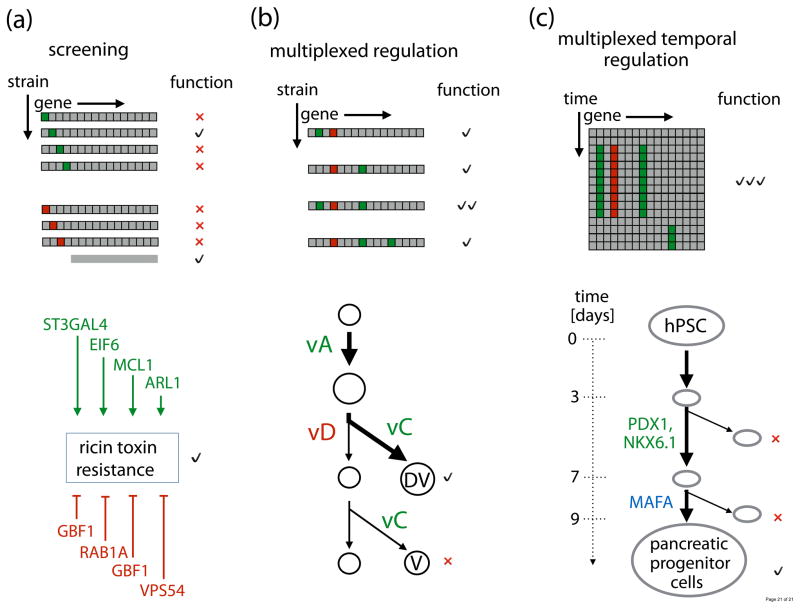
dCas9-based transcriptional regulation is a useful tool for basic science and biotechnology applications, especially those employing screening and network engineering (Fig. 3). Forward genomic screens enable identification of genes involved in cellular functions of interest by creating several strain variants with single gene expression perturbations and observing which variants produce phenotypes of interest (Fig. 3(a), top). Gene knockdowns using dCas9 can be more effective than traditional RNAi based approaches, which are susceptible to incomplete knockdown and off-target effects (reviewed in [63,64]). This was explicitly demonstrated by comparing shRNA and dCas9-KRAB/sgRNA in screens for genes responsible for resistance to the ricin toxin in human K562 cells [19] (Fig. 3(a), bottom). More importantly, not only does dCas9-based screening extend the scope of RNAi regulation to cells that do not use it (for example, prokaryotes), it can also easily be adapted for gain of function screening. Recently, Peters et al. screened the microbe B. subtilis with dCas9 to identify the essential gene target of newly discovered antibiotic MAC-0170636 as well as find genetic determinants of cell morphology and growth [17]. A gain of function screen with dCas9-VP64 identified genes that modulate resistance to inhibition of BRAF, a human proto-oncogene [36]. Combining dCas9-based activation and repression screening can lead to new regulatory network information; this approach enabled characterization of complementary pathways in glycosphingolipid biosynthesis related to cholera and diphtheria toxin resistance [19]. These studies foreshadow the potential of dCas9 for next-generation functional genomic screens.
Fig. 3.
Applications of transcriptional regulation with dCas9. (a) Gain of function (green) and loss of function (red) monogenic screening. Below is an example of ricin toxin resistance screen [19]. (b) Combinatorial screening combining multiple transcriptional perturbations. Below is an example of this approach to maximize deoxyviolacein (DV) production relative to violacein (V) by upregulating vA and vC (green) and downregulating vD genes (red) [24]. (c) Reversible perturbations to multiple genes. Below is an example of using a conditionally degradable dCas9-based activator to differentiate human pluripotent stem cells to pancreatic progenitor cells. Upregulation of PDX1 and NKX6.1 (green) during days 3–7 followed by activation of MAFA (blue) during days 7–9 leads to differentiation [32].
The ability to guide dCas9 with multiple sgRNAs concurrently allows straightforward extension of screening results to testing strain variants with multiple transcriptional perturbations (Fig. 3(b), top). This is particularly useful for adjusting groups of enzymes in metabolic pathways to optimize production of desired metabolites. While broad genetic screens using nucleolytically active Cas9 have been widely used to find genes that increase metabolite yield [65], there are many cases in which graded dCas9-based modulation of multiple enzyme concentrations is required for optimal metabolite production. This has been demonstrated in E. coli and C. glutamicum, where multiplexed promoter repression resulted in increased yields of naringenin [66], 4-hydroxybutyrate [67], as well as lysine and glutamate [16]. Such an approach has also been successfully utilized in E. coli [49] and S. cerevisiae [24] to increase the flux in the violacein metabolic pathway (Fig. 3(b), bottom). Here, the authors employed concurrent up- and downregulation of enzymes at several metabolic flux branch points enabling quick construction of subtle metabolic pathway variants. While dCas9-based metabolic engineering has thus far been limited to unicellular organisms or cells in tissue culture, first steps are being taken for engineering complex multicellular organisms used in agriculture [21]. Besides improving production of valuable compounds, we envisage that dCas9 will be used to engineer metabolic networks for bioremediation [68], biosensing [69] and endowing cells with other useful properties (e.g. stress resistance [70]).
Controlling cell fate holds great promise for regenerative medicine and oncology. Regulation of cell pluripotency and tissue morphogenesis is achieved by transcriptional regulation of unique combinations of gene targets at precise times during development. The multiplexibility and reversibility of the dCas9/sgRNA system is thus well suited to study these programs (Fig. 3(c), top). Both VP64 activator and KRAB repressor versions of dCas9 have been used to reprogram fibroblasts to myocytes [26], guide human pluripotent stem cell differentiation [23,32], or revert fibroblasts to induced pluripotent stem cells [32]. To quickly relieve dCas9-mediated activation, Balboa et al. fused an inducible destabilization domain (DHFR from E. coli) to dCas9. This enabled the execution of a transient overexpression of differentiation genes PDX1 and NKX6.1 that achieved a differentiation of human pluripotent stem cells into pancreatic progenitor cells [32] (Fig. 3(c), bottom). To detect and retard bladder cancer cell growth Liu et al. integrated oncogenic hTERT and bladder-specific hUP II gene upregulation with dCas9/sgRNA-induced apoptosis [59]. These studies recapitulate that dCas9-mediated interactions with transcriptional programs enable researchers to both explore the coordination of genetic networks that underlie cellular functions and to leverage this knowledge for regulation of these functions. Harnessing this ability in a diversity of environmental circumstances is germane to applications spanning all of the life sciences.
Conclusion
In this review we have described how dCas9 has been developed to efficiently regulate transcription in a variety of contexts, including generation of novel gene regulatory circuits, screens for discovering functions of genes, and perturbations enabling efficient production of compounds and cell types. As the technology matures and new applications emerge we foresee an increasing importance of CRISPR-based tools for basic science, biotechnology, and medicine.
Highlights.
CRISPR-Cas9 is a powerful tool to modulate transcription in wide range of cell types
An expanding set of CRISPR-based transcription effectors is available
Gene networks can be efficiently probed and modified for biotechnology applications
Acknowledgments
The authors thank Pavel Chubukov, Omar Din, Robert Cooper, and Philip Bittihn for discussion and comments on the manuscript. This work was supported by NIH Grant R01 GM069811 and the San Diego Center for Systems Biology, NIH Grant P50 GM085764.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Brophy JAN, Voigt CA. Principles of genetic circuit design. Nat Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strauss A, Lahaye T. Zinc fingers, TAL effectors, or Cas9-based DNA binding proteins: What’s best for targeting desired genome loci? Mol Plant. 2013;6:1384–1387. doi: 10.1093/mp/sst075. [DOI] [PubMed] [Google Scholar]
- 3.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 5**.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA– Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. Seminal paper that characterized the fundamental in vitro biochemistry of S. pyogenes Cas9 and also introduced the dCas9 mutant and the sgRNA chimeric molecule. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–83. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–37. doi: 10.1093/nar/gkt520. Constructed and characterized dCas9-based transcription repressors and activators in E.coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 10*.Nielsen AAK, Voigt CA. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol Syst Biol. 2014;10:763. doi: 10.15252/msb.20145735. Constructed and characterized orthogonal dCas9-based promoters in E. coli and used them to build simple synthetic circuits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11*.Didovyk A, Borek B, Hasty J, Tsimring L. Orthogonal Modular Gene Repression in Escherichia coli Using Engineered CRISPR/Cas9. ACS Synth Biol. 2016;5:81–88. doi: 10.1021/acssynbio.5b00147. Designed and experimentally validated a computational algorithm for automated building of multiplexed dCas9-based prokaryotic synthetic gene circuits. Implemented a strategy to integrate orthogonal dCas9-based regulation into native promoters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farzadfard F, Perli SD, Lu TK. Tunable and Multi-Functional Eukaryotic Transcription Factors Based on CRISPR / Cas. ACS Synth Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Kiani S, Beal J, Ebrahimkhani MR, Huh J, Hall RN, Xie Z, Li Y, Weiss R. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat Methods. 2014;11:723–726. doi: 10.1038/nmeth.2969. Created synthetic modular dCas9-repressible eukaryotic promoters and used them to construct simple gene circuits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong Y, Charusanti P, Zhang L, Weber T, Lee SY. CRISPR-Cas9 Based Engineering of Actinomycetal Genomes. ACS Synth Biol. 2015;4:1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 16.Cleto S, Jensen JK, Wendisch VF, Lu TK. Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi) ACS Synth Biol. 2016 doi: 10.1021/acssynbio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters JM, PhD, Larson MH, Hawkins JS, Lu CHS, Koo B, Shiver AL, Whitehead EH, Weissman JS, Brown ED, et al. A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. Cell. 2016 doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Zhang Z-T, Seo S-O, Lynn P, Lu T, Jin Y-S, Blaschek HP. Gene transcription repression in Clostridium beijerinckii using CRISPR-dCas9. Biotechnol Bioeng. 2016 doi: 10.1002/bit.26020. [DOI] [PubMed] [Google Scholar]
- 19**.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. Introduced and characterized application of dCas9, dCas9-KRAB repressor, and SunTag activator for gain and loss of function screens in eukaryotic cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J. 2015;13:578–589. doi: 10.1111/pbi.12284. [DOI] [PubMed] [Google Scholar]
- 22.Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearns NA, Genga RMJ, Enuameh MS, Garber M, Wolfe SA, Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. Designed dCas9/sgRNA scaffolds for multiplexed regulation and applied them for tuning of metabolic pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakore PI, Ippolito AMD, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakraborty S, Ji H, Kabadi AM, Gersbach CA, Christoforou N, Leong KW. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Reports. 2014;3:940–947. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA – guided activation of endogenous human genes. Nat Methods. 2013;10:977–981. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–71. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Balboa D, Weltner J, Eurola S, Trokovic R, Wartiovaara K, Otonkoski T. Conditionally Stabilized dCas9 Activator for Controlling Gene Expression in Human Cell Reprogramming and Differentiation. Stem Cell Reports. 2015;5:448–459. doi: 10.1016/j.stemcr.2015.08.001. Introduced inducible degradation of dCas9 and applied it to control differentiation of human induced pluripotent stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer REP, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. Highly Efficient Cas9- Mediated Transcriptional Programming. Nat Methods. 2015;12:2–6. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12:664–670. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–8. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.System CC, Zetsche B, Gootenberg JS, Abudayyeh OO, Regev A, Koonin EV, Zhang F, Pam T, Zetsche B, Gootenberg JS, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:1–13. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A. 2015;112:3002–7. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–21. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh J-RJ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–5. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braff JL, Yaung SJ, Esvelt KM, Church GM. Characterization of Cas9–Guide RNA Orthologs. Cold Spring Harb Protoc. 2016;2016 doi: 10.1101/pdb.top086793. pdb.top086793. [DOI] [PubMed] [Google Scholar]
- 42**.Chavez A, Tuttle M, Pruitt BW, Ewen-campen B, Chari R, Ter-ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, et al. Comparison of Cas9 activators in multiple species. Nat Methods. 2016 doi: 10.1038/nmeth.3871. Experimentally compared all modern eukaryotic dCas9-based activators and their combinations within human, mouse, and fruit fly cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, Wile BM, Vertino PM, Stewart FJ, Bao G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016:34. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 46.Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, Bosanac L, Zhang ET, El Beheiry M, Masson J-B, Dahan M, et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- 47.Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR–Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiani S, Chavez A, Tuttle M, Hall RN, Chari R, Ter-Ovanesyan D, Qian J, Pruitt BW, Beal J, Vora S, et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cress BF, Jones JA, Kim DC, Leitz QD, Englaender JA, Collins SM, Linhardt RJ, Koffas MAG. Rapid generation of CRISPR/dCas9-regulated, orthogonally repressible hybrid T7-lac promoters for modular, tuneable control of metabolic pathway fluxes in Escherichia coli. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mekler V, Minakhin L, Semenova E, Kuznedelov K, Severinov K. Kinetics of the CRISPR-Cas9 effector complex assembly and the role of 3???-terminal segment of guide RNA. Nucleic Acids Res. 2016;44:2837–2845. doi: 10.1093/nar/gkw138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreno-Mateos MA, Vejnar CE, Beaudoin J-D, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh R, Kuscu C, Quinlan a, Qi Y, Adli M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 2015;43:1–8. doi: 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Keith Joung J. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 56.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 57*.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and Programmable Regulation of Gene Networks with an Integrated RNA and CRISPR/Cas Toolkit in Human Cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. Created synthetic modular dCas9-activatable eukaryotic promoters and used them to construct simple gene circuits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2014;56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Zeng Y, Liu L, Zhuang C, Fu X, Huang W, Cai Z. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat Commun. 2014;5:5393. doi: 10.1038/ncomms6393. [DOI] [PubMed] [Google Scholar]
- 60.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna Ja. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–7. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Josephs EA, Kocak DD, Fitzgibbon CJ, McMenemy J, Gersbach CA, Marszalek PE. Structure and specificity of the RNA-guided endonuclease Cas9 during DNA interrogation, target binding and cleavage. Nucleic Acids Res. 2015;43:8924–41. doi: 10.1093/nar/gkv892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khakhar A, Bolten NJ, Nemhauser J, Klavins E. Cell-cell communication in yeast using auxin biosynthesis and auxin responsive CRISPR transcription factors. ACS Synth Biol. 2016;5:279–286. doi: 10.1021/acssynbio.5b00064. [DOI] [PubMed] [Google Scholar]
- 63.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR–Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agrotis A, Ketteler R. A new age in functional genomics using CRISPR/Cas9 in arrayed library screening. Front Genet. 2015;6:1–15. doi: 10.3389/fgene.2015.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jakočiūnas T, Bonde I, Herrgård M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–222. doi: 10.1016/j.ymben.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 66.Lv L, Ren Y-L, Chen J-C, Wu Q, Chen G-Q. Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: Controllable P(3HB-co-4HB) biosynthesis. Metab Eng. 2015;29:160–168. doi: 10.1016/j.ymben.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 67.Cress BF, Toparlak ÖD, Guleria S, Lebovich M, Stieglitz JT, Englaender JA, Jones JA, Linhardt RJ, Koffas MAG. CRISPathBrick: Modular Combinatorial Assembly of Type II-A CRISPR Arrays for dCas9-Mediated Multiplex Transcriptional Repression in E. coli. ACS Synth Biol. 2015;4:987–1000. doi: 10.1021/acssynbio.5b00012. [DOI] [PubMed] [Google Scholar]
- 68.Díaz E. Bacterial degradation of aromatic pollutants: A paradigm of metabolic versatility. Int Microbiol. 2004;7:173–180. [PubMed] [Google Scholar]
- 69.Checa SK, Zurbriggen MD, Soncini FC. Bacterial signaling systems as platforms for rational design of new generations of biosensors. Curr Opin Biotechnol. 2012;23:766–772. doi: 10.1016/j.copbio.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 70.Jia H, Fan Y, Feng X, Li C. Enhancing stress-resistance for efficient microbial biotransformations by synthetic biology. Front Bioeng Biotechnol. 2014;2:44. doi: 10.3389/fbioe.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]