Abstract
Cells sense mechanical and other biophysical properties of their environment, altering their morphology, migration and differentiation in response. In turn, cells also influence microenvironmental structure and mechanics by secreting, digesting and remodeling matrix components. This dynamic mechanobiological relationship features centrally in development, tissue homeostasis, and disease progression. To explore and control these processes, a wide range of approaches for engineering the cellular microenvironment have been developed. It is only in more recent years that the field has begun to create complementary strategies to control mechanobiological signaling pathways from within the cell. These “inside-out” approaches have been greatly accelerated by incorporating tools and concepts from synthetic biology, a field that seeks to build novel systems from living components.
Complex phenotypes often arise from the activation of signals at specific times and places within tissue (or nanoscale regions of cells), and so comprehensive control of mechanobiological behaviors requires precise modulation of these signals over a wide range of spatial and temporal scales. For instance, whereas stress-induced activation of Src occurs over a few hundred milliseconds[1], mechanically-driven stem cell differentiation responds to stimuli presented over the course of several hours or days. Furthermore, because signaling events are frequently associated with accumulation of a molecular effector to some critical, local concentration[2–4] the duration of a signaling event is important in driving phenotype[5]. Similarly, gradients in biophysical cues and signaling molecules are important for facilitating cell polarity and directing migration [6,7]. Finally, mechanobiological signaling pathways and cell phenotype are typically strongly dependent on the dimensionality of their growth environment. A complete synthetic mechanobiology toolbox therefore requires approaches that can permeate tissue scaffolds or even take advantage of three-dimensional biomaterials, in addition to controlled temporal and spatial cues.
In this review, we explore inside-out control of mechanobiological signaling and phenotype (summarized in Table 1), with emphasis on spatial specificity and temporal dynamics. First, we will discuss studies that direct cell behavior by changing the expression of a target protein. Second, we will explore strategies that control behavior by changing the activity of a target protein. In the former category, we focus on inducible/repressible gene expression systems in which mechanotransductive signals are placed under the control of soluble inputs. In the latter category, we emphasize small-molecule induction of protein complexation. We then consider technologies in which nominally mechanotransductive signaling systems are re-engineered to be induced by non-mechanical inputs such as light and magnetic fields.
Table 1.
Major mechanobiological pathways manipulated by synthetic biology tools.
| Mechanobiological process |
Key signaling molecules | Engineering approach |
|---|---|---|
| Actomyosin contractility | RhoA, ROCK, non-muscle myosin activation |
Genetic[16–18], Chemical[23], Optical [38,47] |
|
| ||
| Non-muscle cells: MLCK, pMLC |
Genetic[16,18] | |
|
| ||
| Muscle cells: Ca2+-CaM, MLCK, caldesmon, pMLC |
Optical[42,44–46] | |
|
| ||
| Actin polymerization | Tiam1, Rac1, WAVE, PAK1, Arp2/3 |
Genetic[17], Chemical[23,29– 31,33,34,36,37], Optical[38,39,41,43,48], Magnetic[53] |
|
| ||
| Cdc42, N-WASP, Arp2/3, Intersectin |
Chemical [23,26], Optical[48], Magnetic [53] |
|
|
| ||
|
Focal adhesion
assembly |
Src, FAK, p130Cas, Paxillin | Chemical [24,32] |
|
| ||
| Microtubule assembly | RCC1, RanGTP, Microtubule associated proteins (MAPs) |
Magnetic[54,56] |
Controlled Induction of Gene Expression
Gene transcription represents an early point of control in regulating protein abundance and therefore activity. A range of conditional promoter systems have been deployed in mammalian cells, most of which place the transcription of specific genes under the control of light [8,9] or small molecules that can be added to the culture medium, such as antibiotics[10,11], steroid hormones[12,13], or metabolites[14,15]. These systems are typically reversible, such that removal of the stimulus restores expression to basal levels. While these systems allow control of expression rates, they neither directly control protein activity levels nor evade native cell regulatory mechanisms.
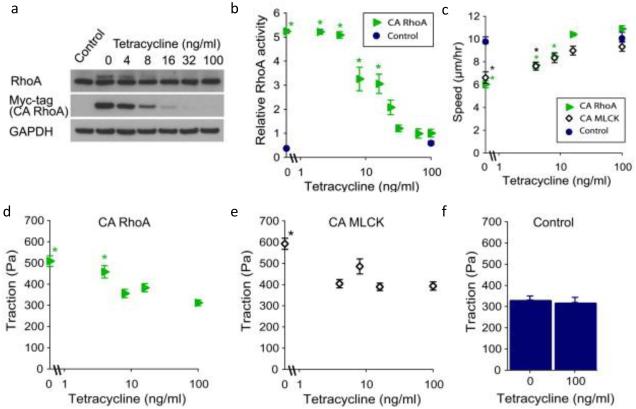
To apply these strategies to mechanobiological signaling while circumventing endogenous feedback regulation, our laboratory has placed constitutively active (CA) mutants of key mechanotransductive genes under the control of conditional promoters. In an early effort, we used lentiviral delivery to create stable human glioma cell lines that express CA RhoA or CA myosin light chain kinase (MLCK) under the control of a tetracycline-repressible promoter. By varying the concentration of tetracycline in the medium, we achieved stably graded expression levels of these proteins. Moreover, because both RhoA and MLCK promote activation of the actin cytoskeletal motor non-muscle myosin II, we were able to apply this strategy to control a variety of mechanobiological phenotypes in a graded and stable way (Figure 1), including random migration speed, cortical stiffness and traction force generation[16].
Figure 1. Graded control of cell mechanpbiology using conditional gene expression.
Delivery of a constitutively active (CA) RhoA under the control of a tetracycline-repressible promoter allowed for concentration-dependent expression (a) and activity (b) of RhoA. This graded expression of CA RhoA or CA MLCK produced graded cell migration speeds (c) or traction force generation (d,e) compared to control (f). Reproduced with permission from Mackay et al. [16]
Provided mutually orthogonal promoters are selected, this strategy can be multiplexed to independently and simultaneously control several target proteins. This approach could allow for engineering more complex mechanobiological behavior, or permit one to map the “phase space” that describes how multiple proteins interact to control cell phenotype. For example, we used dual lentiviral transduction to simultaneously express CA RhoA and CA Rac1 under the control of doxycycline-and cumate-inducible promoters, respectively[17]. These GTPases are canonically regarded to regulate opposing aspects of cell motility and mutually antagonize one another at several levels, making it challenging to independently manipulate them. By using this orthogonal promoter strategy, we circumvented this crosstalk and mapped the range of phenotypes observed in the otherwise inaccessible state of high-RhoA activation and high-Rac1 activation.
Inducible/repressible promoter strategies offer a number of important advantages, including highly stable expression and the ability to uniformly control gene expression in an entire population of cells, and in an easily scalable way. These features can be leveraged to study and control the biomechanical role of target proteins in mice grafted with genetically engineered cells.[18] However, there are also a number of limitations, perhaps the most important of which is the slow dynamics of the expression system and the protein of interest. While in some scenarios, cells respond phenotypically within six hours[16,19], some systems may take as long as ten days to reach a steady-state response[19,20]. This is compounded by system-to-system variations in the kinetics of transcriptional activation, protein folding and post-translational modifications, protein transport, and protein degradation, all of which may be key to the final phenotype.
Additionally, this strategy has inherently limited spatial resolution. Once the gene has been transcribed, there is no control over subcellular protein localization. However, several approaches for spatial control of gene expression at the cell population level have been proposed. For instance, inducers and repressors can be restricted to certain areas of a cell population through microfluidic control[20], by occlusion of membrane pores[19], or by sequestration of the agent within the material scaffold[21,22]. Several factors influence the extent of control over spatial activation of gene expression and thus pattern fidelity. Cell migration and slow delivery, induction, and expression kinetics may disrupt intended patterns. Shorter lag times between introduction of the inducer/repressor and protein expression allows for more faithful pattern formation [19].
Controlled Activation of Protein Activity
While modulating gene expression can produce graded and reversible changes in cell mechanobiology, the response time of this system is limited by transcription and translation rates as well as by protein and mRNA degradation rates. As a result, these approaches are most relevant for modulating processes that occur on time scales of hours to days or at a steady-state of protein activity. Direct control of protein activity, on the other hand, can influence signaling and thus cell behavior on the scale of seconds to minutes. Importantly, these rapid kinetics may also better replicate time scales associated with soluble ligand-induced signaling in cells (e.g. growth factors). Several techniques for selective activation of target proteins have been devised and are beginning to be applied to mechanobiology, including chemical, optical, and magnetic actuation.
Chemically Induced Protein Activity
One commonly employed chemical actuation system is to bring two proteins of interest into proximity through chemically induced dimerization (CID), such as the rapamycin-induced dimerization of FK506 binding protein (FKPB) and FKBP rapamycin-binding domain (FRB). In this approach, FKBP and FRB are each genetically fused to two target proteins. Subsequent addition of the small molecule rapamycin to the culture then induces the association and binding of the two proteins, triggering downstream signaling events. This approach also allows for control of protein localization by fusing one of these domains to an organelle-targeting domain. For example, by fusing FKBP to a membrane-localization sequence, FRB-RhoA and FRB-Rac chimeras have been recruited to the cell membrane upon addition of rapamycin, leading respectively to cell contraction and membrane ruffling[23].
Further refinements to this approach have enabled increasingly precise control of signaling. For instance, Karginov et al. selectively activated certain branches of the Src signaling cascade by inserting FKBP into the catalytic domain of Src, thereby deactivating its kinase activity. Addition of rapamycin induced binding of this construct with FRB, which allosterically rescued kinase activity[24,25]. This protein was co-expressed with either FAK or p130Cas constructs containing FRB instead of the Src-binding domain, allowing for specific induction of FAK-mediated or p130Cas-mediated effects of Src activation. Kapp et al. used computational methods to design versions of Cdc42 and its activator intersectin that acted orthogonally to the cell’s native signaling machinery and dimerized upon rapamycin addition[26]. Cells expressing both synthetic signaling molecules or both wild-type Cdc42 and intersectin demonstrated increased lamellipodium formation, while cells expressing one wild-type and one synthetic protein did not form lamellipodia.
While rapamycin-induced dimerization allows for the rapid and highly specific localization and activation of proteins, these effects are largely irreversible[27]. Various reversible CID systems have been reported[28,29]. Alternatively, a second, orthogonal dimerizing agent can be used to relocate a rapamycin-induced FRB-FKBP complex from the plasma membrane to mitochondrial membranes to produce a pulse of localized protein activation[30,31].
Whereas manipulation of gene expression produces changes in protein levels on a cell-wide scale, chemically-induced dimerization offers the opportunity to manipulate protein activity through the directed delivery of the inducer (e.g. rapamycin). Variants of these dimerization inducers can be conjugated with photocleavable domains that greatly reduce inducer activity through steric hindrance or by blocking passage through the cell membrane[32–35]. Within a few minutes of illumination, the inactivating conjugate is released, allowing the inducers to enter the cell or enabling the dimerizing agent to catalyze dimerization. This strategy has been used to control the location of membrane ruffling[32,33], extension of cell processes[34] and protein localization[35]. An attractive feature of this approach is its potential for multiplexing, in that photocleavage occurs over a fairly narrow window of wavelengths (which can itself be tuned to different wavelengths), thus allowing for orthogonal activation of dimerization of more than one set of proteins[34]. Microfluidic devices present an alternative method for spatial control over inducer delivery. Lin et al. cultured cells containing rapamycin-activatable Rac constructs in fluidically engineered cell-length rapamycin gradients and found that asymmetry in Rac activation could direct cell polarity and migration[36].
An alternative approach to chemical induction of protein activity is to design proteins that recognize exogenous ligands. Park et al. expressed in motile cells an engineered G protein receptor that responds solely to clozapine-N-oxide. This molecule acted as a chemotactic cue, directing cell migration towards areas of higher concentration. T-lymphocytes expressing this construct honed to CNO-releasing beads implanted in mice[37].
Optically Induced Protein Activation
The integration of light-sensitive domains in protein engineering has given rise to the field of optogenetics. These protein domains undergo conformational changes in response to absorption of light of a particular wavelength. These photoreceptors vary in their wavelength sensitivity, photochemical kinetics and activation mechanism. Some photoreceptors heterodimerize in response to the light-induced conformational change, allowing similar engineering approaches seen with CID. For example, RhoGTPase clustering through optically stimulated Cry2 produces spatially controlled changes in cytoskeletal organization[38]. Light oxygen voltage (LOV) photoreceptors operate through steric hindrance. For instance, a photoactivatable Rac1 construct operates by fusing CA Rac1 to the LOV C-terminal alpha helix. A light-induced conformational change permits Rac1 to activate its effectors and induce cell protrusions and motility.[39] Light-gated ion channels, notably channelrhodopsins, open to allow the flow of ions in response to illumination. Non-channel classes of light actuators and their application to cell motility have been recently reviewed[40].
As with chemical induction of protein activity, light induction is rapid; depending on the photoreceptor system used, activation occurs on the order of one second. Importantly, many light-inducible systems are reversible, with light-induced states having a half-life of 10 seconds to 10 minutes. Optical induction also allows for precise spatial control over protein activation, and can target single cells or a specific region within a cell. Optical signals are minimally invasive and can reach cells embedded in three-dimensional materials and living tissue[41–43].
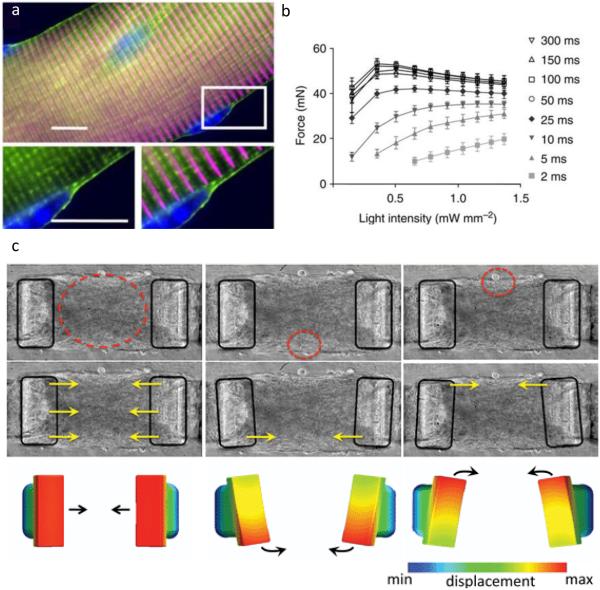
Light-gated ion channels offer a particularly elegant system for studying calcium-induced mechanobiological phenotypes such as muscle cell contraction. Bruegmann and coworkers expressed EYFP-tagged channelrhodopsin-2 (ChR2) in mice[44,45]. Illumination with blue light induced membrane depolarization and calcium influx in isolated cardiomyocytes and depolarized cardiomyocytes in live mouse hearts[44]. Skeletal muscle explants contracted, and the magnitude of force generated could be modulated based on the intensity and duration of the light pulses (Figure 2) [45]. Sakar et al. differentiated skeletal muscle myoblasts expressing ChR2 in matrigel-collagen gels formed around Polydimethylsiloxane (PDMS) cantilever pairs[46]. Optically stimulated myotubes generated contractile forces, quantified through cantilever deflection. This activation could be spatially restricted to single myotubes or expanded to target multiple myotubes, allowing for control over axial and rotational force generation in these devices (Figure 2).
Figure 2. Control of cell contractile forces using optogenetics.
(a) Skeletal muscle fiber isolated from mice expressing EYFP-tagged channelrhodopsin-2 (green), which localizes to the cell membrane, including the T-tubule membrane invaginations that surround sarcomeric alpha-actinin (magenta). Scale bar shows 10 µm. (b) Explanted soleus muscles contracted in response to blue light with dependence on light pulse duration and intensity. “Functional expression of ChR2 in skeletal muscle.” by Bruegmann et al. [45], licensed under CC BY 4.0 (c) Devices comprised of skeletal myotubules expressing channelrhodopsin-2 in matrigel-collagen gels surrounding PDMS cantilevers can be induced to contract axially or rotationally by controlling the region of illumination (circled in red). Reproduced with permission from Sakar et al.[46]
An offshoot of this approach is to use protein engineering to create novel calcium-dependent signaling. Mills et al. fused calmodulin, which is activated when bound to calcium ions, to the GTPases Cdc42, Rac1 and RhoA[47,48]. By controlling calcium influx with an engineered light-sensitive calcium channel[49], the authors could direct filopodial extension and cell migration.
Magnetically Induced Protein Activity
In general, the above strategies all require some degree of gene/protein engineering, which may not be appropriate or even feasible in all systems. An appealing alternative is the use of functionalized magnetic nanoparticles (MNP) to activate signaling cascades by physically clustering signaling molecules. Originally restricted to cell-surface receptors[50,51], recent efforts have used microinjection and other methods to place the MNP inside the cell[52,53]. These particles can then be localized to an area of interest by application of a field with a magnetic tip[50,54], or can been arranged into a concentration gradient[55,56]. Importantly, this process is rapid, occurring over seconds to minutes, and reversible, with particles dispersing after removal of the magnetic field.
In vitro functionalization of MNPs is challenging for large protein complexes that cannot be easily expressed and isolated from bacterial cultures. Etoc and coworkers addressed this issue by functionalizing MNPs in situ by injecting MNPs coated with a HaloTag ligand into cells expressing a protein of interest fused with HaloTag[53]. HaloTag binds to its ligand irreversibly, recruiting the target protein to the MNP. Cdc42-functionalized MNPs concentrated with a magnetic field localized actin polymerization while membrane-bound Rac1 signaling was dynamically modulated by bringing MNPs functionalised with the Rac1 activator TIAM1 into and out of membrane proximity.
Conclusion
Mechanobiological behaviors are regulated by temporally and spatially precise cues. Depending on the phenotype of interest, a variety of strategies are available for controlling cell force generation and signaling. Small-molecule induction of gene expression represents one approach but only produces behavioural changes on the order of hours to days and does not offer subcellular resolution. On the other hand, direct control of protein activity allows for much more rapid actuation of signaling events, and incorporation of exogenous cues such as magnetic probes or light allows for subcellular control over protein activity. MNPs may also present translational advantages, since clinical applications of certain magnetic nanoparticles are already approved[57].
Rewiring or tuning mechanobiological signaling circuits could have useful medical applications. For example, this approach could allow correction or control of mechanically-driven cell behavior in instances where inserting a scaffold or otherwise modifying the extracellular environment is impractical or impossible. Proof-of-principle experiments have shown that genetic[18], chemical[37] and optical[44,45] actuation of signals can control cell mechanobiology in animal models to achieve clinically relevant goals, such as targeted immune cell recruitment or reduced cancer cell invasion. A challenging but critical next step will be to explore the use of these approaches in living organisms, with an eye towards eventual clinical use. Important considerations include safe and targeted delivery of any necessary genetic constructs.
HIGHLIGHTS.
Mechanobiological phenotype is regulated by spatially and temporally precise cues.
Mechanobiological signaling can be “rewired” to accept non-mechanical triggers.
Conditional promoters enable reversible, graded control over cell mechanobiology.
Proteins can be activated directly with small molecules, light, and magnetic fields.
Acknowledgements
J.H.H. thanks the Natural Science and Engineering Research Council (NSERC) of Canada for their support. S.K. gratefully acknowledges the support of the NSF (CMMI 1055965, CBET PESO 1105539), the NIH (1R21EB016359, 1R01NS074831, IMAT 1R21CA174573), and the W. M. Keck Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors do not disclose any conflicts of interest.
References
- 1.Na S, Collin O, Chowdhury F, Tay B, Ouyang M, Wang Y, Wang N. Rapid signal transduction in living cells is a unique feature of mechanotransduction [Internet] Proc. Natl. Acad. Sci. 2008;105:6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vartak N, Bastiaens P. Spatial cycles in G-protein crowd control [Internet] EMBO J. 2010;29:2689–2699. doi: 10.1038/emboj.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinkhabwala A, Bastiaens PI. Spatial aspects of intracellular information processing [Internet] Curr. Opin. Genet. Dev. 2010;20:31–40. doi: 10.1016/j.gde.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Inder K, Harding A, Plowman SJ, Philips MR, Parton RG, Hancock JF. Activation of the MAPK module from different spatial locations generates distinct system outputs. [Internet] Mol. Biol. Cell. 2008;19:4776–84. doi: 10.1091/mbc.E08-04-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. [Internet] Cell. 2013;155:1422–34. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isenberg BC, DiMilla PA, Walker M, Kim S, Wong JY. Vascular Smooth Muscle Cell Durotaxis Depends on Substrate Stiffness Gradient Strength [Internet] Biophys. J. 2009;97:1313–1322. doi: 10.1016/j.bpj.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D-H, Han K, Gupta K, Kwon KW, Suh K-Y, Levchenko A. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients [Internet] Biomaterials. 2009;30:5433–5444. doi: 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. [Internet] Nat. Methods. 2012;9:266–9. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 9.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation [Internet] Nat. Chem. Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin K-J, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang J-I, Rebres R, Roach T, Seaman W, Simon MI, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression [Internet] Proc. Natl. Acad. Sci. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fussenegger M, Morris RP, Fux C, Rimann M, von Stockar B, Thompson CJ, Bailey JE. Streptogramin-based gene regulation systems for mammalian cells. [Internet] Nat. Biotechnol. 2000;18:1203–8. doi: 10.1038/81208. [DOI] [PubMed] [Google Scholar]
- 12.Braselmann S, Graninger P, Busslinger M. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins. [Internet] Proc. Natl. Acad. Sci. U. S. A. 1993;90:1657–61. doi: 10.1073/pnas.90.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.No D, Yao TP, Evans RM. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. [Internet] Proc. Natl. Acad. Sci. U. S. A. 1996;93:3346–51. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitzinger M, Kemmer C, Fluri DA, Daoud El-Baba M, Weber W, Fussenegger M. The food additive vanillic acid controls transgene expression in mammalian cells and mice [Internet] Nucleic Acids Res. 2011;40:e37–e37. doi: 10.1093/nar/gkr1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu HS, Lee CH, Lee CF, Su IJ, Chang TY. Lac/Tet dual-inducible system functions in mammalian cell lines. [Internet] Biotechniques. 1998;24:624–8. doi: 10.2144/98244st03. 630–2. [DOI] [PubMed] [Google Scholar]
- 16.MacKay JL, Keung AJ, Kumar S. A genetic strategy for the dynamic and graded control of cell mechanics, motility, and matrix remodeling. [Internet] Biophys. J. 2012;102:434–42. doi: 10.1016/j.bpj.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacKay JL, Kumar S. Simultaneous and independent tuning of RhoA and Rac1 activity with orthogonally inducible promoters. [Internet] Integr. Biol. (Camb) 2014;6:885–94. doi: 10.1039/c4ib00099d. ** Orthogonal conditional promoters allowed for simultaneous and independent control of expression levels of two key mechanotransductive proteins that exhibit antagonistic crosstalk. By varying the concentration of two small molecule inducers over time, the authors were able to switch between mechanobiological phenotypes, including cell spreading and migration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong SY, Ulrich TA, Deleyrolle LP, MacKay JL, Lin J-MG, Martuscello RT, Jundi MA, Reynolds BA, Kumar S. Constitutive activation of myosin-dependent contractility sensitizes glioma tumor-initiating cells to mechanical inputs and reduces tissue invasion. [Internet] Cancer Res. 2015;75:1113–22. doi: 10.1158/0008-5472.CAN-13-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Javaherian S, Anesiadis N, Mahadevan R, McGuigan AP. Design principles for generating robust gene expression patterns in dynamic engineered tissues [Internet] Integr. Biol. 2013;5:578. doi: 10.1039/c3ib20274g. ** The authors provided key insights into the impacts of cell migration and promoter temporal dynamics on cell population pattern resolution. They modelled pattern blurriness as a function of cell speed and persistance as well as pattern feature size. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Gazit Z, Pelled G, Gazit D, Vunjak-Novakovic G. Patterning osteogenesis by inducible gene expression in microfluidic culture systems. [Internet] Integr. Biol. (Camb) 2011;3:39–47. doi: 10.1039/c0ib00053a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deans TL, Singh A, Gibson M, Elisseeff JH. Regulating synthetic gene networks in 3D materials. [Internet] Proc. Natl. Acad. Sci. U. S. A. 2012;109:15217–22. doi: 10.1073/pnas.1204705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baraniak PR, Nelson DM, Leeson CE, Katakam AK, Friz JL, Cress DE, Hong Y, Guan J, Wagner WR. Spatial control of gene expression within a scaffold by localized inducer release [Internet] Biomaterials. 2011;32:3062–3071. doi: 10.1016/j.biomaterials.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue T, Do Heo W, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. [Internet] Nat. Methods. 2005;2:415–8. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Engineered allosteric activation of kinases in living cells. [Internet] Nat. Biotechnol. 2010;28:743–7. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karginov AV, Tsygankov D, Berginski M, Chu P-H, Trudeau ED, Yi JJ, Gomez S, Elston TC, Hahn KM. Dissecting motility signaling through activation of specific Src-effector complexes [Internet] Nat. Chem. Biol. 2014;10:286–290. doi: 10.1038/nchembio.1477. * This paper presents a powerful tool for dissecting and controlling signaling pathways. Insertion of FKBP into a highly conserved kinase domain allowed rapamycin-induced allosteric activation of Src, and this approach may be useful for other kinases. The binding partner of this activated Src was specifically controlled by fusing the target effector with FBP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapp GT, Liu S, Stein A, Wong DT, Reményi A, Yeh BJ, Fraser JS, Taunton J, Lim WA, Kortemme T. Control of protein signaling using a computationally designed GTPase/GEF orthogonal pair. [Internet] Proc. Natl. Acad. Sci. U. S. A. 2012;109:5277–82. doi: 10.1073/pnas.1114487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP·Rapamycin·FRB Ternary Complex [Internet] J. Am. Chem. Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 28.Feng S, Laketa V, Stein F, Rutkowska A, MacNamara A, Depner S, Klingmüller U, Saez-Rodriguez J, Schultz C. A rapidly reversible chemical dimerizer system to study lipid signaling in living cells. [Internet] Angew. Chem. Int. Ed. Engl. 2014;53:6720–3. doi: 10.1002/anie.201402294. [DOI] [PubMed] [Google Scholar]
- 29.Liu P, Calderon A, Konstantinidis G, Hou J, Voss S, Chen X, Li F, Banerjee S, Hoffmann J-E, Theiss C, et al. A bioorthogonal small-molecule-switch system for controlling protein function in live cells. [Internet] Angew. Chem. Int. Ed. Engl. 2014;53:10049–55. doi: 10.1002/anie.201403463. [DOI] [PubMed] [Google Scholar]
- 30.Miyamoto T, DeRose R, Suarez A, Ueno T, Chen M, Sun T, Wolfgang MJ, Mukherjee C, Meyers DJ, Inoue T. Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. [Internet] Nat. Chem. Biol. 2012;8:465–70. doi: 10.1038/nchembio.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y-C, Nihongaki Y, Liu T-Y, Razavi S, Sato M, Inoue T. Rapidly reversible manipulation of molecular activity with dual chemical dimerizers. [Internet] Angew. Chem. Int. Ed. Engl. 2013;52:6450–4. doi: 10.1002/anie.201301219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karginov AV, Zou Y, Shirvanyants D, Kota P, Dokholyan NV, Young DD, Hahn KM, Deiters A. Light regulation of protein dimerization and kinase activity in living cells using photocaged rapamycin and engineered FKBP. [Internet] J. Am. Chem. Soc. 2011;133:420–3. doi: 10.1021/ja109630v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Umeda N, Ueno T, Pohlmeyer C, Nagano T, Inoue T. A photocleavable rapamycin conjugate for spatiotemporal control of small GTPase activity. [Internet] J. Am. Chem. Soc. 2011;133:12–4. doi: 10.1021/ja108258d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright CW, Guo Z-F, Liang F-S. Light control of cellular processes by using photocaged abscisic acid. [Internet] Chembiochem. 2015;16:254–61. doi: 10.1002/cbic.201402576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballister ER, Aonbangkhen C, Mayo AM, Lampson MA, Chenoweth DM. Localized light-induced protein dimerization in living cells using a photocaged dimerizer. [Internet] Nat. Commun. 2014;5:5475. doi: 10.1038/ncomms6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin B, Holmes WR, Wang CJ, Ueno T, Harwell A, Edelstein-Keshet L, Inoue T, Levchenko A. Synthetic spatially graded Rac activation drives cell polarization and movement. [Internet] Proc. Natl. Acad. Sci. U. S. A. 2012;109:E3668–77. doi: 10.1073/pnas.1210295109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park JS, Rhau B, Hermann A, McNally KA, Zhou C, Gong D, Weiner OD, Conklin BR, Onuffer J, Lim WA. Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. [Internet] Proc. Natl. Acad. Sci. U. S. A. 2014;111:5896–901. doi: 10.1073/pnas.1402087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells [Internet] Nat. Methods. 2013;10:249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 39.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. [Internet] Nature. 2009;461:104–8. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weitzman M, Hahn KM. Optogenetic approaches to cell migration and beyond. [Internet] Curr. Opin. Cell Biol. 2014;30:112–20. doi: 10.1016/j.ceb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Q, Wang X, Tibbitt MW, Anseth KS, Montell DJ, Elisseeff JH. Light activated cell migration in synthetic extracellular matrices. [Internet] Biomaterials. 2012;33:8040–6. doi: 10.1016/j.biomaterials.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryson JB, Machado CB, Crossley M, Stevenson D, Bros-Facer V, Burrone J, Greensmith L, Lieberam I. Optical control of muscle function by transplantation of stem cell-derived motor neurons in mice. [Internet] Science. 2014;344:94–7. doi: 10.1126/science.1248523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Hyun Y-M, Lim K, Lee H, Cummings RJ, Gerber SA, Bae S, Cho TY, Lord EM, Kim M. Optogenetic control of chemokine receptor signal and T-cell migration. [Internet] Proc. Natl. Acad. Sci. U. S. A. 2014;111:6371–6. doi: 10.1073/pnas.1319296111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruegmann T, Malan D, Hesse M, Beiert T, Fuegemann CJ, Fleischmann BK, Sasse P. Optogenetic control of heart muscle in vitro and in vivo. [Internet] Nat. Methods. 2010;7:897–900. doi: 10.1038/nmeth.1512. [DOI] [PubMed] [Google Scholar]
- 45.Bruegmann T, van Bremen T, Vogt CC, Send T, Fleischmann BK, Sasse P. Optogenetic control of contractile function in skeletal muscle. [Internet] Nat. Commun. 2015;6:7153. doi: 10.1038/ncomms8153. * This paper demonstrated that a dynamic range of force generation is achievable in skeletal muscle explants expressing optically-gated cation channels. The authors also performed a proof-of-principle experiment for the clinically-relevant adeno-associated viral (AAV) delivery of optogenetic ion channels that would allow for light-stimulated control of skeletal muscle force generation in mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakar MS, Neal D, Boudou T, Borochin MA, Li Y, Weiss R, Kamm RD, Chen CS, Asada HH. Formation and optogenetic control of engineered 3D skeletal muscle bioactuators. [Internet] Lab Chip. 2012;12:4976–85. doi: 10.1039/c2lc40338b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mills E, Pham E, Truong K. Structure based design of a Ca2+-sensitive RhoA protein that controls cell morphology [Internet] Cell Calcium. 2010;48:195–201. doi: 10.1016/j.ceca.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 48.Mills E, Pham E, Nagaraj S, Truong K. Engineered Networks of Synthetic and Natural Proteins To Control Cell Migration [Internet] ACS Synth. Biol. 2012;1:211–220. doi: 10.1021/sb3000172. [DOI] [PubMed] [Google Scholar]
- 49.Pham E, Mills E, Truong K. A synthetic photoactivated protein to generate local or global Ca(2+) signals. [Internet] Chem. Biol. 2011;18:880–90. doi: 10.1016/j.chembiol.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Mannix RJ, Kumar S, Cassiola F, Montoya-Zavala M, Feinstein E, Prentiss M, Ingber DE. Nanomagnetic actuation of receptor-mediated signal transduction. [Internet] Nat. Nanotechnol. 2008;3:36–40. doi: 10.1038/nnano.2007.418. [DOI] [PubMed] [Google Scholar]
- 51.Bharde AA, Palankar R, Fritsch C, Klaver A, Kanger JS, Jovin TM, Arndt-Jovin DJ. Magnetic nanoparticles as mediators of ligand-free activation of EGFR signaling. [Internet] PLoS One. 2013;8:e68879. doi: 10.1371/journal.pone.0068879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen O, Riedemann L, Etoc F, Herrmann H, Coppey M, Barch M, Farrar CT, Zhao J, Bruns OT, Wei H, et al. Magneto-fluorescent core-shell supernanoparticles [Internet] Nat. Commun. 2014;5:5093. doi: 10.1038/ncomms6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Etoc F, Lisse D, Bellaiche Y, Piehler J, Coppey M, Dahan M. Subcellular control of Rac-GTPase signalling by magnetogenetic manipulation inside living cells. [Internet] Nat. Nanotechnol. 2013;8:193–8. doi: 10.1038/nnano.2013.23. ** Magnetic nanoparticles (MNPs) functionalized with Cdc42 or with a Rac1-activating guanine nucleotide exchange factor were used to concentrate protein signaling to target subcellular locations and control mechanobiological processes such as actin polymerization and process extention. Importantly, the authors showed that this signaling process was context-dependent; actin polymerization mediated by Rac1 activation was limited when the MNPs were concentrated within the cell body compared to the lamellipodia. [DOI] [PubMed] [Google Scholar]
- 54.Hoffmann C, Mazari E, Lallet S, Le Borgne R, Marchi V, Gosse C, Gueroui Z. Spatiotemporal control of microtubule nucleation and assembly using magnetic nanoparticles. [Internet] Nat. Nanotechnol. 2013;8:199–205. doi: 10.1038/nnano.2012.246. [DOI] [PubMed] [Google Scholar]
- 55.Etoc F, Vicario C, Lisse D, Siaugue J-M, Piehler J, Coppey M, Dahan M. Magnetogenetic Control of Protein Gradients Inside Living Cells with High Spatial and Temporal Resolution [Internet] Nano Lett. 2015;15:3487–3494. doi: 10.1021/acs.nanolett.5b00851. [DOI] [PubMed] [Google Scholar]
- 56.Bonnemay L, Hostachy S, Hoffmann C, Gautier J, Gueroui Z. Engineering spatial gradients of signaling proteins using magnetic nanoparticles. [Internet] Nano Lett. 2013;13:5147–52. doi: 10.1021/nl402356b. * The authors established magnetic nanoparticle (MNP) gradients in Xenopus egg extracts. MNPs functionalized with the GTPase Ran nucleated microtubule aster formation, however in the presence of the gradient the asters formed off-center. This strategy could be applied to further tease apart how protein signaling distributions can impact cytoskeletal asymmetries. [DOI] [PubMed] [Google Scholar]
- 57.Goodwill PW, Saritas EU, Croft LR, Kim TN, Krishnan KM, Schaffer DV, Conolly SM. X-Space MPI: Magnetic Nanoparticles for Safe Medical Imaging [Internet] Adv. Mater. 2012;24:3870–3877. doi: 10.1002/adma.201200221. [DOI] [PubMed] [Google Scholar]