Abstract
Fibrotic diseases occur in virtually every tissue of the body and are a major cause of mortality, yet they remain largely untreatable and poorly understood on a mechanistic level. The development of anti-fibrotic agents has been hampered, in part, by the insufficient fibrosis biomimicry provided by traditional in vitro platforms. This review focuses on recent advancements toward creating 3-D platforms that mimic key features of fibrosis, as well as the application of novel imaging and sensor techniques to analyze dynamic extracellular matrix remodeling. Several opportunities are highlighted to apply new tools from the fields of biomaterials, imaging, and systems biology to yield pathophysiologically-relevant in vitro platforms that improve our understanding of fibrosis and may enable identification of potential treatment targets.
Graphical abstract
Fibrosis: When a scar goes too far
Fibrotic disease can occur in virtually any tissue in the body, spanning both systemic diseases (e.g., systemic sclerosis, multifocal fibrosclerosis) and organ-specific disorders (e.g., pulmonary fibrosis, liver cirrhosis, cardiac fibrosis) [1]. Combined, fibrotic diseases contribute to an estimated 45% of deaths in the industrialized world [2]. The common feature that ties together these diseases is the deposition of disordered and excessive extracellular matrix (ECM), particularly collagen types I and III [3]. The primary cellular mediators of this process are myofibroblasts, also known as ‘activated fibroblasts’, which are commonly identified by their expression of alpha-smooth muscle actin (αSMA) and contractile behavior [4]. The ECM accumulation and crosslinking promoted by myofibroblasts leads to tissue stiffening, which then acts in a positive feedback loop to sustain pathological levels of myofibroblast activity [5]. Although ECM upregulation and scar formation can be normal components of tissue repair, these events are dysregulated in fibrosis, where the heavily crosslinked, collagen-rich matrix progressively supplants the original functional tissue, eventually leading to organ dysfunction and failure.
Despite the prevalence of fibrotic diseases and the wide spectrum of tissues that they affect, there are few, if any, treatment options available [6••]. The poor biomimcry of traditional cell culture platforms is one reason for the slow progress toward identifying anti-fibrotic treatments [7]. However, recent years have seen the development of numerous approaches to create 3-D environments that possess improved pathophysiological relevance and enable precise and dynamic control over microenvironmental variables. The importance of specifically using 3-D conditions was illustrated by recent work demonstrating large differences in myofibroblast gene expression across 2-D vs. 3-D platforms possessing identical stiffness and composition [8•]. Of particular interest in designing these scaffold systems are three features that comprise the etiological backbone of fibrosis: 1) ECM composition, 2) TGF-β1 presence, and 3) mechanical stiffness (Figure 1). The use of biomaterials to study fibrosis-related behaviors has been broadly reviewed elsewhere [9•,10], while herein we focus on recent biological discoveries related to the three aforementioned features in fibrosis, advancements in the design of engineered 3-D platforms for studying these phenomena, and how this work may be combined with other emerging engineering-based approaches to further our understanding and treatment of fibrosis.
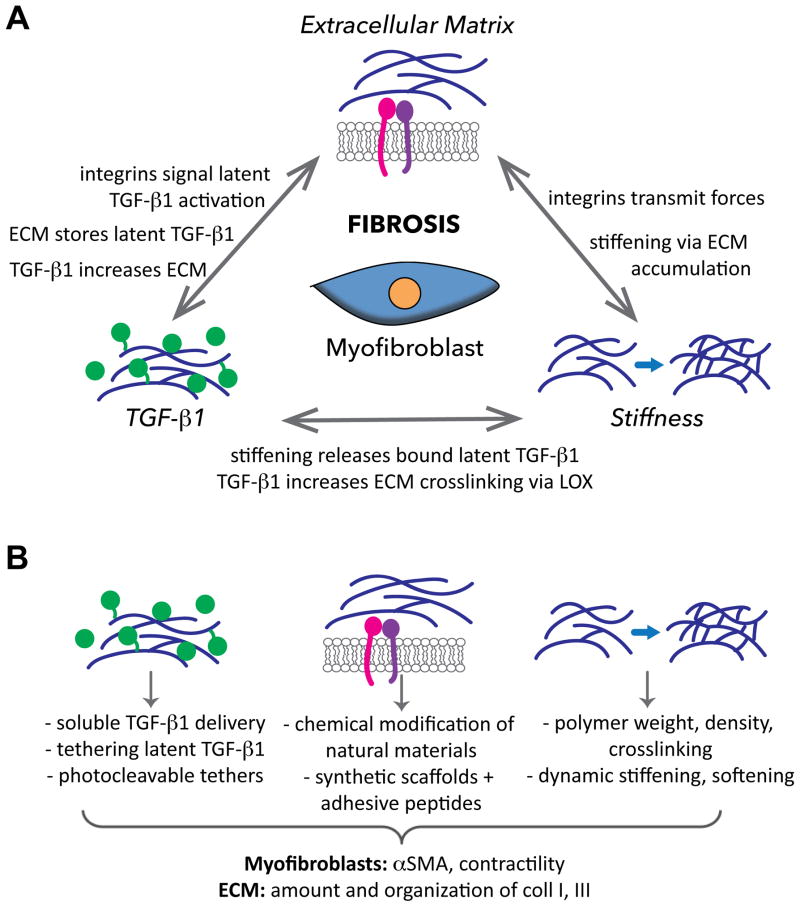
Figure 1.
A) Fibrosis is strongly influenced by extracellular matrix cues, presence of soluble and ECM-bound TGF-β1, and matrix stiffness. All three of these features are regulated by myofibroblastic activity, in addition to being able to modulate one another. B) Examples of 3-D in vitro approaches used to mimic the three aforementioned features, and outcomes that signify fibrosis.
Engineering the Microenvironment
ECM Cues
Myofibroblasts interact with the ECM via integrins and other non-integrin receptors, and these specific receptor-ligand interactions are responsible for transmitting information to the cell about the composition, structure, and mechanics of their extracellular environment, all of which are altered in fibrosis. Specific integrin-ligand binding events have been found to govern fibrogenic behaviors; for instance, binding via the α2β1 integrin exerts a protective effect in cardiac and valve fibrosis, while other β1-containing integrins (e.g., α1β1, α5β1) increase fibrotic events in these same tissues [5]. Thus, the type of ECM ligand presented is a critically important consideration in the construction of engineered constructs, as it may influence the disease state of the system.
As reviewed elsewhere [9•–12], various native materials have been employed as 3-D constructs in the study of fibrosis, with the use of collagen-based hydrogels being the most common [7]. Other innovations in naturally-derived 3-D fibrosis culture platforms have included the development of scaffold-free 3-D strategies, such as layer-by-layer seeding [13] or the stacking of intact cell sheets [14]. However, while these systems offer environments rich in complex biological cues, they are severely limited in their ability to be mechanically tailored or to withstand extended culture times, which is particularly problematic in the context of fibrosis, a gradual disease that is highly dependent upon matrix mechanics. These systems also do not permit controlled variation of ECM identity.
One approach to merge the need for ECM ligands, physical tailorability, and long-term culture has been the chemical modification of natural materials, such as methacrylation of gelatin (Gel-MA) or hyaluronic acid (HA-MA) [15,16]. Culture of encapsulated valvular interstitial cells (VICs, a myofibroblast precursor) was supported for several weeks in Gel-MA and HA-MA scaffolds. Moreover, the differentiation of VICs into myofibroblasts was dependent upon ECM identity, with combinations of HA-MA and Gel-MA yielding a more quiescent VIC phenotype than either component individually [15,16]. More controlled investigations of the role of ECM ligand identity in fibrosis can be achieved by modification of “blank slate” materials, such as polyethylene glycol (PEG), with adhesive peptides. While such hydrogels have been used extensively in the fabrication of engineered tissues [17], they have only recently been applied to explicitly examine fibrosis. The strong ligand-dependence of myofibroblast differentiation was illustrated by a recent study of VICs cultured within PEG-peptide hydrogels [18]; intriguingly, the α2β1-binding peptide used in this work was associated with low αSMA expression, which is consistent with in vivo observations, but contrary to previous 2-D work using similar materials [19]. Together, these studies highlight not only the powerful influence that ECM identity exerts on fibrotic disease phenomena, but also the importance of controlling these cues in the in vitro context.
Delivery of TGF-β1
Transforming growth factor-beta1 (TGF-β1) is a molecular cornerstone in the pathogenesis of fibrosis. In simplified terms, TGF-β1 promotes fibrogenesis by increasing ECM production and decreasing its degradation [20]. Thus, delivery of this molecule to in vitro fibrosis platforms is a critical element in studying fibrotic mechanisms.
The delivery of TGF-β1 in vitro is generally performed by simple addition of soluble TGF-β1 to the culture platform. However, in vivo, TGF-β1 is secreted as part of a latent complex that binds to the ECM, making its presentation in a tethered form highly relevant to fibrosis. Several approaches have been described to covalently tether growth factors to biomaterial scaffolds [21], and significant increases in the production of collagen and overall ECM have been reported when smooth muscle cells or chondrocytes were cultured in scaffolds containing tethered TGF-β1 [22,23]. Recent advancements have yielded additional ways to better mimic the in vivo sequestration and release of TGF-β1 from the ECM. For example, latent TGF-β1 complexes were tethered to polymer scaffolds for release by encapsulated cells, resulting in increased protein and ECM synthesis [24]. Photodegradable sequences have also been used to attach TGF-β1 to polymer scaffolds, allowing temporally controlled release of tethered TGF-β1 [25,26]. TGF-β1-binding peptide sequences may also be used to achieve reversible, non-covalent sequestration [27]. To date, these systems have not been used in the study of fibrosis. However, given the importance of TGF-β1 sequestration in modulating interactions with both integrins and mechanosensing mechanisms, systems that better mimic its in vivo bioavailability may greatly advance the physiological relevance of in vitro culture platforms.
Tailoring Matrix Mechanics
The poor success rate of anti-fibrotic drugs has also been attributed to their failure to address the mechanical events that drive fibrosis progression [5]. Fibroblasts are highly sensitive to tissue stiffness [28•–30], and there is a close connection between matrix mechanics and TGF-β1 activation, where matrix stiffening increases the release of TGF-β1 from the ECM [20], a finding with profound implications for fibrosis.
Following Engler’s seminal publication describing control of cell fate by substrate stiffness [31], there has been a rapid acceleration in the use of mechanically tunable substrates to study fibrotic behaviors. A recent development in this area is the synthesis of biomaterial systems that enable dynamic, in situ scaffold stiffening and/or softening [28•,29,32–34]. Importantly, these systems allow cell morphology, viability, and phenotype to be consistent across all conditions at the experiment start, yielding the ability to more accurately correlate changes in cell behavior with induced changes in matrix mechanics. However, most investigations of matrix mechanics in fibrosis have been limited to 2-D, presenting a significant limitation for the study of this disease, where 3-D ECM structure and cell contractility play critical roles. A particularly interesting development is that recent 3-D studies have shown increased αSMA expression within softer materials [16,35•], a finding which is contrary to numerous 2-D studies [28–30].
Merging Cues from the ECM, TGF-β1, and Matrix Mechanics
The cues provided by ECM identity, TGF-β1, and matrix mechanics are closely intertwined (Figure 1); the latent TGF-β1 complex can directly bind to certain integrins, while integrins can sense matrix stiffness, and matrix stiffening can induce the release of TGF-β1 from the ECM [36]. Additionally, all three of these variables can directly influence myofibroblast differentiation. Combined variation of adhesive peptide sequence and stiffness has suggested that ligand identity is more influential than stiffness in regulating valvular myofibroblast differentiation [19], while another study has reported that sensitivity of pulmonary fibroblasts to exogenous TGF-β1 is increased with increasing stiffness [37]; these combinatorial approaches are needed to decipher the ‘decision-making’ process of cells during fibrosis. Although no system has yet combined the delivery of specific ECM cues with tethered TGF-β1 and modulation of scaffold stiffness, this is theoretically feasible with the merger of existing technologies. This approach could yield valuable information on the hierarchy and relationship of fibrotic behaviors, thus informing the development of anti-fibrotic treatments.
New Directions for the Evaluation of in vitro Fibrosis Platforms
In vitro fibrogenesis is typically characterized via evaluation of cell phenotype and ECM synthesis. Myofibroblasts may be identified by their expression of αSMA, as well as their contractility and activation of related signaling pathways [4] via standard biological techniques. Meanwhile, ECM synthesis is primarily monitored via the quantification of collagen I, and, less frequently, collagen III. However, these analysis approaches do not yield information about ECM organization, a critical feature in fibrosis. The recent development of molecular imaging tools also offers an opportunity to gain insight into fibrogenic processes through the non-invasive visualization of ECM dynamics in living cultures.
Second Harmonic Generation Microscopy
In second harmonic generation (SHG) microscopy, the supramolecular assembly of collagen fibers in 3-D is imaged in the absence of exogenous molecular markers or dyes [38]. SHG yields a rich data set that quantitatively describes collagen organization; a greatly truncated set of such measurements can be seen in Figure 2. The acquisition of these data may also be automated to diagnose fibrotic conditions. For instance, multiple studies have combined SHG imaging of collagen alterations with machine learning algorithms to not only characterize the extent of fibrosis in clinical samples, but also provide automated, high-throughput diagnoses of fibrotic disease [38–40].
Figure 2.
A) Histological sections of a healthy (left) and diseased/fibrotic (right) human aortic heart valve stained with Movat’s pentachrome. Healthy leaflets possess a trilayered ECM structure, consisting of the fibrosa (F), spongiosa (S), and ventricularis (V). Yellow staining indicates collagen. B) Second harmonic generation (SHG) images of healthy (left) and fibrotic (right) leaflets and brief example of fiber characteristics that can be quantified by automated analysis of SHG images, illustrating the advanced ECM characterization that can be performed with alternative imaging methods. *p<0.0001; Scale bar = 50 μm.
Other imaging advancements that may benefit the analysis of in vitro fibrosis platforms include a novel pairing of SHG with fluorescence lifetime imaging microscopy (FLIM), which was able to distinguish between collagen type I and type III, the two main ECM markers of fibrosis [41••]. Meanwhile, the combination of SHG with spectral lifetime imaging microscopy (SLIM) provides the ability to image real-time changes in cellular metabolites in the context of ECM alterations [42,43]. The application of these techniques to study fibrotic microenvironments has the potential to yield similar insight into the interactions between the ECM, inflammatory signals, and cellular behavior.
Molecular Probes
Multiple types of molecular probes have been developed to perform non-invasive, real-time imaging of both ECM dynamics and mechanotransduction events. For instance, collagen-binding fluorescent probes [44,45•] can enable high-resolution imaging of collagen assembly and remodeling in living tissues, producing similar quality of information as SHG [46], but without the need for specialized imaging instrumentation. Molecular sensors have also been designed to non-invasively image the real-time activity of enzymes involved in fibrotic ECM remodeling, such as matrix metalloproteinases (MMPs) [47], as well as lysyl oxidase (LOX) [48], which crosslinks fibers of collagen type I and type III, as well as elastin [49]. Also relevant to fibrosis is the recent development of molecular tension sensors [50•] which enable dynamic visualization of cell-generated mechanical forces. In combination with other imaging modalities, probes that provide such real-time measurement of ECM dynamics in living tissues may be used to track the same specimen over different stages of fibrosis and ultimately enable a more holistic characterization and understanding of fibrogenesis. Molecules that participate in ECM crosslinking and organization may also serve as targets for anti-fibrotic treatments [51].
Conclusions and Future Directions
Fibrotic diseases affect a wide range of tissues and are a significant cause of mortality, but they remain poorly understood on a mechanistic level, with few treatment options available. The development of in vitro platforms to recapitulate fibrotic events opens the door to not only improving our understanding of these diseases but also identifying and testing novel therapeutic approaches. Recent years have seen the generation of complex 3-D culture systems that may be used to independently vary microenvironmental properties and elucidate disease etiologies, thereby advancing our understanding of how myofibroblasts interact with their microenvironment. However, there remain numerous existing scaffold design and evaluation strategies that have yet to be applied to the study of fibrosis. As noted above, reversible tethering of latent TGF-β1 to scaffolds may permit a physiologically relevant presentation of this important pro-fibrotic molecule. Additionally, with few exceptions [52], current 3-D in vitro fibrosis platforms have not incorporated co-culture, despite numerous 2-D studies and advancements toward ‘organ-on-a-chip’ systems emphasizing the importance of other cell types in the progression of this disease [53,54].
With respect to fibrosis evaluation, new imaging techniques have been developed to improve the clinical diagnosis of fibrosis [38–40,55], but most of these efforts have yet to crossover to the execution of in vitro fibrosis studies. The application of techniques that enable non-invasive, real-time imaging of collagen assembly and remodeling in engineered fibrosis platforms has the potential to greatly advance the quality of information gained from these studies and their relevance to in vivo conditions. Furthermore, there are continued efforts to identify improved cellular markers for fibrosis beyond the traditional, yet non-specific, αSMA. Following the hypothesis that αSMA-positive myofibroblasts arise from the differentiation of local epithelial progenitors via epithelial-to-mesenchymal transition (EMT), transcription factors related to EMT have been quantified to monitor fibrosis [56]; however, some studies have cast doubt on this EMT-myofibroblast relationship [57]. Specific microRNAs (e.g., miR-29) are also emerging as key biological regulators in fibrosis [58••], with strong potential as therapeutic targets.
Finally, despite the interconnected nature of fibrotic events and need to improve the identification and efficacy of potential treatments, computational modeling tools have rarely been applied to study fibrosis. An agent-based model was recently generated to describe cell-cell interactions in liver fibrosis and predict response to treatment [59], but most other applications of computational methods to study fibrosis have involved analysis of large datasets for gene expression or signaling, rather than prediction of cellular- and tissue-level behaviors. In addition to helping decipher the cellular decision-making processes that regulate fibrogenesis, systems biology-based approaches have the capacity to generate models that identify potential targets [60] or assess and predict drug sensitivity and resistance [61], which could yield significant advancements in the treatment of this complex disease.
Highlights.
Development of anti-fibrotic treatments is hampered by culture system limitations
Fibrosis involves ECM disarray, TGF-β1 signaling, and matrix stiffening
Tunable 3-D engineered structures can recapitulate the aforementioned features
Novel imaging and sensor tools can allow visualization of matrix dynamics in vitro
Acknowledgments
This work was supported by the National Heart, Lung, and Blood Institute and the National Institute of Biomedical Imaging and Bioengineering at the National Institutes of Health (R01-HL093281 and R21-EB019508 to K.S.M.), as well as a Predoctoral Fellowship from the American Heart Association (15PRE 22170006 to A.M.P.), the Translational Cardiovascular Sciences Training Program (T32 HL07936 to H.N.H.), and a National Science Foundation Graduate Research Fellowship (to A.J.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Cited References
- 1.Rosenbloom J, Castro SV, Jimenez SA. Narrative review: fibrotic diseases: cellular and molecular mechanisms and novel therapies. Ann Intern Med. 2010;152:159–166. doi: 10.7326/0003-4819-152-3-201002020-00007. [DOI] [PubMed] [Google Scholar]
- 2.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klingberg F, Hinz B, White ES. The myofibroblast matrix: implications for tissue repair and fibrosis. J Pathol. 2013;229:298–309. doi: 10.1002/path.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroer AK, Merryman WD. Mechanobiology of myofibroblast adhesion in fibrotic cardiac disease. J Cell Sci. 2015;128:1865–1875. doi: 10.1242/jcs.162891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••6.Friedman SL, Sheppard D, Duffield JS, Violette S. Therapy for fibrotic diseases: nearing the starting line. Sci Transl Med. 2013;5:167sr161. doi: 10.1126/scitranslmed.3004700. This paper presents a wide-ranging overview of fibrotic diseases, their shared features, and a critical analysis of opportunities for therapeutic targeting. [DOI] [PubMed] [Google Scholar]
- 7.Chen CZ, Raghunath M. Focus on collagen: in vitro systems to study fibrogenesis and antifibrosis state of the art. Fibrogenesis Tissue Repair. 2009;2:7. doi: 10.1186/1755-1536-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •8.Mabry KM, Payne SZ, Anseth KS. Microarray analyses to quantify advantages of 2D and 3D hydrogel culture systems in maintaining the native valvular interstitial cell phenotype. Biomaterials. 2015;74:31–41. doi: 10.1016/j.biomaterials.2015.09.035. Although researchers often assert that 3-D culture is superior to, and differs greatly from, 2-D culture, it is quite rare to actually perform such a quantitative comparison, especially while controlling for stiffness and composition. With over 1,000 genes differentially regulated across 2-D vs. 3-D cultures and 3-D better mimicking in vivo conditions, this study provides compelling reason to focus research efforts on 3-D platforms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •9.Smithmyer ME, Sawicki LA, Kloxin AM. Hydrogel scaffolds as in vitro models to study fibroblast activation in wound healing and disease. Biomater Sci. 2014;2:634–650. doi: 10.1039/c3bm60319a. This paper provides an excellent overview of various hydrogel environments, both 2-D and 3-D, that have been used to study fibroblast activation and/or wound healing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desrochers TM, Palma E, Kaplan DL. Tissue-engineered kidney disease models. Adv Drug Deliv Rev. 2014;69–70:67–80. doi: 10.1016/j.addr.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanta J. Collagen matrix as a tool in studying fibroblastic cell behavior. Cell Adh Migr. 2015;9:308–316. doi: 10.1080/19336918.2015.1005469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Broek LJ, Limandjaja GC, Niessen FB, Gibbs S. Human hypertrophic and keloid scar models: principles, limitations and future challenges from a tissue engineering perspective. Exp Dermatol. 2014;23:382–386. doi: 10.1111/exd.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosoya H, Kadowaki K, Matsusaki M, Cabral H, Nishihara H, Ijichi H, Koike K, Kataoka K, Miyazono K, Akashi M, et al. Engineering fibrotic tissue in pancreatic cancer: a novel three-dimensional model to investigate nanoparticle delivery. Biochem Biophys Res Commun. 2012;419:32–37. doi: 10.1016/j.bbrc.2012.01.117. [DOI] [PubMed] [Google Scholar]
- 14.Corriveau MP, Boufaied I, Lessard J, Chabaud S, Senecal JL, Grodzicky T, Chartier S, Raymond Y, Moulin VJ. The fibrotic phenotype of systemic sclerosis fibroblasts varies with disease duration and severity of skin involvement: reconstitution of skin fibrosis development using a tissue engineering approach. J Pathol. 2009;217:534–542. doi: 10.1002/path.2482. [DOI] [PubMed] [Google Scholar]
- 15.Hjortnaes J, Camci-Unal G, Hutcheson JD, Jung SM, Schoen FJ, Kluin J, Aikawa E, Khademhosseini A. Directing valvular interstitial cell myofibroblast-like differentiation in a hybrid hydrogel platform. Adv Healthc Mater. 2015;4:121–130. doi: 10.1002/adhm.201400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan B, Hockaday LA, Kapetanovic E, Kang KH, Butcher JT. Stiffness and adhesivity control aortic valve interstitial cell behavior within hyaluronic acid based hydrogels. Acta Biomater. 2013;9:7640–7650. doi: 10.1016/j.actbio.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J. Bioactive modification of poly(ethylene glycol) hydrogels for tissue engineering. Biomaterials. 2010;31:4639–4656. doi: 10.1016/j.biomaterials.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould ST, Anseth KS. Role of cell-matrix interactions on VIC phenotype and tissue deposition in 3D PEG hydrogels. J Tissue Eng Regen Med. 2013 doi: 10.1002/term.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gould ST, Darling NJ, Anseth KS. Small peptide functionalized thiol-ene hydrogels as culture substrates for understanding valvular interstitial cell activation and de novo tissue deposition. Acta Biomater. 2012;8:3201–3209. doi: 10.1016/j.actbio.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinz B. The extracellular matrix and transforming growth factor-beta1: Tale of a strained relationship. Matrix Biol. 2015;47:54–65. doi: 10.1016/j.matbio.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Masters KS. Covalent growth factor immobilization strategies for tissue repair and regeneration. Macromol Biosci. 2011;11:1149–1163. doi: 10.1002/mabi.201000505. [DOI] [PubMed] [Google Scholar]
- 22.Sridhar BV, Doyle NR, Randolph MA, Anseth KS. Covalently tethered TGF-beta1 with encapsulated chondrocytes in a PEG hydrogel system enhances extracellular matrix production. J Biomed Mater Res A. 2014;102:4464–4472. doi: 10.1002/jbm.a.35115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann BK, Schmedlen RH, West JL. Tethered-TGF-beta increases extracellular matrix production of vascular smooth muscle cells. Biomaterials. 2001;22:439–444. doi: 10.1016/s0142-9612(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 24.Place ES, Nair R, Chia HN, Szulgit G, Lim EH, Stevens MM. Latent TGF-beta hydrogels for cartilage tissue engineering. Adv Healthc Mater. 2012;1:480–484. doi: 10.1002/adhm.201200038. [DOI] [PubMed] [Google Scholar]
- 25.Griffin DR, Schlosser JL, Lam SF, Nguyen TH, Maynard HD, Kasko AM. Synthesis of photodegradable macromers for conjugation and release of bioactive molecules. Biomacromolecules. 2013;14:1199–1207. doi: 10.1021/bm400169d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grim JC, Marozas IA, Anseth KS. Thiol-ene and photo-cleavage chemistry for controlled presentation of biomolecules in hydrogels. J Control Release. 2015 doi: 10.1016/j.jconrel.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belair DG, Le NN, Murphy WL. Design of growth factor sequestering biomaterials. Chem Commun (Camb) 2014;50:15651–15668. doi: 10.1039/c4cc04317k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •28.Kirschner CM, Alge DL, Gould ST, Anseth KS. Clickable, photodegradable hydrogels to dynamically modulate valvular interstitial cell phenotype. Adv Healthc Mater. 2014;3:649–657. doi: 10.1002/adhm.201300288. In this study, the authors describe a novel hydrogel platform that allows in situ modulation of both stiffness and topographical features. This approach is significant because it enables sequential, dynamic changes to the physical scaffold environment during cell culture, which may be used to mimic stages of disease processes. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Tibbitt MW, Langer SJ, Leinwand LA, Anseth KS. Hydrogels preserve native phenotypes of valvular fibroblasts through an elasticity-regulated PI3K/AKT pathway. Proc Natl Acad Sci U S A. 2013;110:19336–19341. doi: 10.1073/pnas.1306369110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yip CY, Chen JH, Zhao R, Simmons CA. Calcification by valve interstitial cells is regulated by the stiffness of the extracellular matrix. Arterioscler Thromb Vasc Biol. 2009;29:936–942. doi: 10.1161/ATVBAHA.108.182394. [DOI] [PubMed] [Google Scholar]
- 31.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 32.Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- ••33.Stowers RS, Allen SC, Suggs LJ. Dynamic phototuning of 3D hydrogel stiffness. Proc Natl Acad Sci U S A. 2015;112:1953–1958. doi: 10.1073/pnas.1421897112. Unlike most other dynamically-tunable biomaterials, the material described in this paper can undergo both dynamic stiffening and softening. Temporal modulation of material stiffness was also achieved following in vivo implantation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosales AM, Mabry KM, Nehls EM, Anseth KS. Photoresponsive elastic properties of azobenzene-containing poly(ethylene-glycol)-based hydrogels. Biomacromolecules. 2015;16:798–806. doi: 10.1021/bm501710e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •35.Mabry KM, Lawrence RL, Anseth KS. Dynamic stiffening of poly(ethylene glycol)-based hydrogels to direct valvular interstitial cell phenotype in a three-dimensional environment. Biomaterials. 2015;49:47–56. doi: 10.1016/j.biomaterials.2015.01.047. This work provides a clear illustration of the benefits of using dynamically-stiffened materials, rather than separate scaffolds with static stiffness; use of static stiffness gels results in differences in cell morphology at the time of seeding, while dynamic stiffening allows all cells to reach the same starting point prior to experiencing a change in matrix mechanics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duscher D, Maan ZN, Wong VW, Rennert RC, Januszyk M, Rodrigues M, Hu M, Whitmore AJ, Whittam AJ, Longaker MT, et al. Mechanotransduction and fibrosis. J Biomech. 2014;47:1997–2005. doi: 10.1016/j.jbiomech.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chia HN, Vigen M, Kasko AM. Effect of substrate stiffness on pulmonary fibroblast activation by TGF-beta. Acta Biomater. 2012;8:2602–2611. doi: 10.1016/j.actbio.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 38.Pena A, Fabre A, Debarre D, Marchal-Somme J, Crestani B, Martin J, Beaurepaire E, Schanne-Klein M-C. Three-Dimensional INvestigation and Scoring of Extracellular Matrix Remodeling During Lung Fibrosis Using Multiphoton Microscopy. Microscopy research and technique. 2007;70:162–170. doi: 10.1002/jemt.20400. [DOI] [PubMed] [Google Scholar]
- 39.Tilbury K, Hocker J, Wen BL, Sandbo N, Singh V, Campagnola PJ. Second harmonic generation microscopy analysis of extracellular matrix changes in human idiopathic pulmonary fibrosis. Journal of biomedical optics. 2014;19:086014. doi: 10.1117/1.JBO.19.8.086014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strupler M, Pena a-M, Hernest M, Tharaux P-L, Martin J-L, Beaurepaire E, Schanne-Klein M-C. Second harmonic imaging and scoring of collagen in fibrotic tissues. Optics express. 2007;15:4054–4065. doi: 10.1364/oe.15.004054. [DOI] [PubMed] [Google Scholar]
- ••41.Ranjit S, Dvornikov A, Stakic M, Hong S-H, Levi M, Evans RM, Gratton E. Imaging Fibrosis and Separating Collagens using Second Harmonic Generation and Phasor Approach to Fluorescence Lifetime Imaging. Scientific Reports. 2015;5:13378. doi: 10.1038/srep13378. A combination of two label-free imaging methods (SHG and FLIM) is applied to obtain fingerprints of different collagens. This approach allowed automated quantification of collagen types I and III, which could greatly advance the characterization of fibrosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Provenzano PP, Eliceiri KW, Keely PJ. Multiphoton microscopy and fluorescence lifetime imaging microscopy (FLIM) to monitor metastasis and the tumor microenvironment. Clinical & Experimental Metastasis. 2009;26:357–370. doi: 10.1007/s10585-008-9204-0. [DOI] [PubMed] [Google Scholar]
- 43.Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, White JG, Keely PJ. Collagen density promotes mammary tumor initiation and progression. BMC medicine. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biela E, Galas J, Lee B, Johnson GL, Darzynkiewicz Z, Dobrucki JW. Col-F, a fluorescent probe for ex vivo confocal imaging of collagen and elastin in animal tissues. Cytometry A. 2013;83:533–539. doi: 10.1002/cyto.a.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •45.Aper SJ, van Spreeuwel AC, van Turnhout MC, van der Linden AJ, Pieters PA, van der Zon NL, de la Rambelje SL, Bouten CV, Merkx M. Colorful protein-based fluorescent probes for collagen imaging. PLoS One. 2014;9:e114983. doi: 10.1371/journal.pone.0114983. A set of molecular probes that enable real-time, multicolor visualization of living tissue in situ was developed. Tracking of collagen dynamics in engineered tissues could be used to better understand both developmental and disease processes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boerboom RA, Krahn KN, Megens RT, van Zandvoort MA, Merkx M, Bouten CV. High resolution imaging of collagen organisation and synthesis using a versatile collagen specific probe. J Struct Biol. 2007;159:392–399. doi: 10.1016/j.jsb.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Cai Y, Zhu L, Zhang F, Niu G, Lee S, Kimura S, Chen X. Noninvasive monitoring of pulmonary fibrosis by targeting matrix metalloproteinases (MMPs) Mol Pharm. 2013;10:2237–2247. doi: 10.1021/mp300613x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aslam T, Miele A, Chankeshwara SV, Megia-Fernandez A, Michels C, Akram AR, McDonald N, Hirani N, Haslett C, Bradley M, et al. Optical molecular imaging of lysyl oxidase activity – detection of active fibrogenesis in human lung tissue. Chem Sci. 2015;6:4946–4953. doi: 10.1039/c5sc01258a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez C, Rodriguez-Sinovas A, Martinez-Gonzalez J. Lysyl oxidase as a potential therapeutic target. Drug News Perspect. 2008;21:218–224. doi: 10.1358/dnp.2008.21.4.1213351. [DOI] [PubMed] [Google Scholar]
- •50.LaCroix AS, Rothenberg KE, Berginski ME, Urs AN, Hoffman BD. Construction, imaging, and analysis of FRET-based tension sensors in living cells. Methods Cell Biol. 2015;125:161–186. doi: 10.1016/bs.mcb.2014.10.033. This review provides an overview of FRET-based tension sensors, which have recently been developed to study mechanotransduction events in living cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ratner M. Landmark approvals in idiopathic pulmonary fibrosis. Nat Biotechnol. 2014;32:1069–1070. doi: 10.1038/nbt1114-1069. [DOI] [PubMed] [Google Scholar]
- 52.Tseng H, Balaoing LR, Grigoryan B, Raphael RM, Killian TC, Souza GR, Grande-Allen KJ. A three-dimensional co-culture model of the aortic valve using magnetic levitation. Acta Biomater. 2014;10:173–182. doi: 10.1016/j.actbio.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Q, Patel D, Kwa T, Haque A, Matharu Z, Stybayeva G, Gao Y, Diehl AM, Revzin A. Liver injury-on-a-chip: microfluidic co-cultures with integrated biosensors for monitoring liver cell signaling during injury. Lab Chip. 2015 doi: 10.1039/c5lc00874c. [DOI] [PubMed] [Google Scholar]
- 54.Gould ST, Matherly EE, Smith JN, Heistad DD, Anseth KS. The role of valvular endothelial cell paracrine signaling and matrix elasticity on valvular interstitial cell activation. Biomaterials. 2014;35:3596–3606. doi: 10.1016/j.biomaterials.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Won S, Davies-Venn C, Liu S, Bluemke DA. Noninvasive imaging of myocardial extracellular matrix for assessment of fibrosis. Curr Opin Cardiol. 2013;28:282–289. doi: 10.1097/HCO.0b013e32835f5a2b. [DOI] [PubMed] [Google Scholar]
- 56.Lovisa S, LeBleu VS, Tampe B, Sugimoto H, Vadnagara K, Carstens JL, Wu CC, Hagos Y, Burckhardt BC, Pentcheva-Hoang T, et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med. 2015;21:998–1009. doi: 10.1038/nm.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kriz W, Kaissling B, Le Hir M. Epithelial-mesenchymal transition (EMT) in kidney fibrosis: fact or fantasy? J Clin Invest. 2011;121:468–474. doi: 10.1172/JCI44595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••58.Montgomery RL, Yu G, Latimer PA, Stack C, Robinson K, Dalby CM, Kaminski N, van Rooij E. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med. 2014;6:1347–1356. doi: 10.15252/emmm.201303604. MicroRNAs are highly influential in multiple diseases and have great potential to be used as therapeutics, but strategies to restore or increase miRNA function are limited. This work describes the development of a synthetic mimic of miR-29 and its injection into an animal model of pulmonary fibrosis, where it restored endogenous miR-29 function and reversed pulmonary fibrosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dutta-Moscato J, Solovyev A, Mi Q, Nishikawa T, Soto-Gutierrez A, Fox IJ, Vodovotz Y. A Multiscale Agent-Based in silico Model of Liver Fibrosis Progression. Front Bioeng Biotechnol. 2014;2:18. doi: 10.3389/fbioe.2014.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schoeberl B, Pace EA, Fitzgerald JB, Harms BD, Xu L, Nie L, Linggi B, Kalra A, Paragas V, Bukhalid R, et al. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci Signal. 2009;2:ra31. doi: 10.1126/scisignal.2000352. [DOI] [PubMed] [Google Scholar]
- 61.Faratian D, Goltsov A, Lebedeva G, Sorokin A, Moodie S, Mullen P, Kay C, Um IH, Langdon S, Goryanin I, et al. Systems biology reveals new strategies for personalizing cancer medicine and confirms the role of PTEN in resistance to trastuzumab. Cancer Res. 2009;69:6713–6720. doi: 10.1158/0008-5472.CAN-09-0777. [DOI] [PubMed] [Google Scholar]