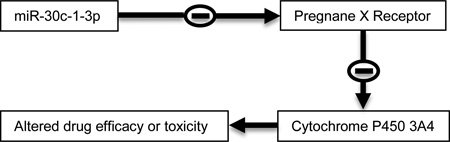
Abstract
The pregnane X receptor (PXR) is a master regulator of genes involved in drug elimination. Recently, activation of PXR has also been linked to the development of many disease conditions such as metabolic disorders and malignancies. MicroRNAs (miRs) emerge as important molecular species involved in these conditions. This study was undertaken to test a large number of miRs for their ability to regulate PXR expression. As many as 58 miRs were tested and miR-30c-1-3p was identified to suppress PXR expression. The suppression was achieved by targeting the 3’-untranslated region, 438 nucleotides from the stop codon. The suppression was detected in multiple cell lines from different organ origins. In addition, miR-30c-1-3p altered basal and induced expression of cytochrome P450 3A4 (CYP3A4), a prototypical target gene of PXR. The alteration varied depending on the time and amounts of miR-30c-1-3p. CYP3A4 is responsible for the metabolism of more than 50% medicines. The interconnection between miR-30c-1-3p and PXR signifies a role of miRs in drug-drug interactions and chemosensitivity.
Graphical abstract
1. Introduction
All organisms are exposed constantly to toxic chemicals from both foreign and endogenous sources. Organisms such as humans have evolved several defensive systems against chemical insults [1]. In mammals, these systems are generally referred to as phase I [2, 3], phase II [4] and phase III [5, 6]. Phase I and II consist of drug-metabolizing enzymes, whereas phase III of drug transporters. The expression of these genes undergoes constant changes in response to chemical stimuli. The pregnane X receptor (PXR, NR1I2) is established as a master transcription factor intimately involved in the regulated expression of these genes [7–9]. Structurally, PXR belongs to the nuclear hormone receptor superfamily [10, 11]. Like other nuclear receptors, PXR consists of a variable N-terminal domain, a highly conserved DNA-binding domain, a hinge region and a multifunctional C-terminal ligand-binding domain [10]. The DNA-binding domain recognizes conical sequence AGG/TTCA [12, 13]. The major portion of the ligand-binding domain is helical in structure, and the C-terminal helix (helix 12) is directly involved in switching from repressing to activating status of a target gene [14]. Binding to an agonist induces conformational changes of this helix, leading to a platform favoring association with coactivators, namely transactivation.
The expression of PXR itself, like its target genes, is regulated by certain xenobiotics and disease conditions [8, 15–18]. For example, the hypolipidemic agent clofibrate and synthetic glucocorticoid dexamethasone have been shown to induce PXR [17, 18]. The induction synergistically increased the expression of cytochrome P450 3A genes (CYP3A) [17, 18], the prototypical targets of PXR [10]. Dexamethasone induced PXR in both rodents and humans [16, 18, 19]. Proinflammatory stimuli, on the other hand, have been shown to suppress the expression of PXR [20, 21]. The level of PXR mRNA was rapidly decreased in rodents treated with lipopolysaccharide, a potent immune stimulant [15]. In human hepatocytes, proinflammatory cytokine interleukin-6 markedly reduced the levels of PXR mRNA [20]. The suppression was accompanied by significant reduction of the induction of PXR-regulated genes such as CYP3A23 [15, 20, 21].
While transactivation and repression are recognized as major mechanisms for the regulated expression of PXR [17, 18, 20], post-transcriptional mechanisms have been increasingly implicated. MicroRNA (miR)-148a reportedly down-regulated PXR post-transcriptionally [22]. On the other hand, miRs are important regulators in a wide spectrum of diseases including malignances and metabolic disorders [23–25]. Interestingly, PXR has been linked to the development of these very conditions [26, 27]. Expression of constitutively activated PXR markedly increased hepatic lipids in mice and the steatotic phenotype was recapitulated in human primary hepatocytes by rifampicin, a prototypical activator of human PXR [26]. The expression of PXR is dysregulated in a wide range of cancerous tissues such as breast cancer, endometrial cancer and colon cancer [27].
In this study, we took a comprehensive approach and tested a large number of miRs for their ability to regulate PXR expression. As many as 58 miRs were tested and miR-30c-1-3p was identified to suppress PXR. The suppression was achieved by targeting the 3’-untranslated region (UTR) and detected in multiple cell lines from different organ origin. Importantly, miR-30c-1-3p was shown to alter the expression of CYP3A4, a prototypical target gene of PXR. CYP3A4 is responsible for the metabolism of more than 50% medicines. Therefore, the PXR and miR-30c-1-3p connection likely constitutes a major determinant in drug-drug interactions and chemosensitivity.
2. Materials and methods
2.1. Plasmid constructs
All miR precursor clones were purchased from System Biosciences Inc (Mountain View, CA). The CYP3A4-DP-Luc reporter was described in our previous publication [28]. The PXR cDNA reporters harboring a 3’-UTR segment were prepared with the pGL3 promoter vector (Promega, Madison, WI) through Xba I and Fse I restriction endonuclease sites. The 3’-UTR segments were amplified by PCR with high fidelity Platinum Taq DNA polymerase (Life Technology Co., Carlsbad, CA). A cDNA clone encoding human PXR, used as the PCR template, was described elsewhere [16]. The PXR 3142/3691Luc and 3690/4408Luc reporters were prepared initially and the PXR 3142/3691Luc reporter was used as the template for preparing deletion mutants at 5’ end. The primers for PCR amplification are listed in Table I. All reporter constructs were subjected to sequence analysis.
Table I.
Sequences of Oligonucleotides
| Oligonucleotide | Sequence |
|---|---|
| PXR3142/3691Luc (XbaI) | 5’-tgagcggctgcccttggg-3’ |
| PXR3142/3691Luc (FseI) | 5’-agaggactcccacagata-3’ |
| PXR3690/4408Luc (XbaI) | 5’-ggagtcctctagagagatgagaagccagga-3’ |
| PXR3690/4408Luc (FseI) | 5’-gtacattatttaattcct-3’ |
| PXR3192/3691Luc (XbaI) | 5’-gccctctgagccgccact-3’ |
| PXR3242/3691Luc (XbaI) | 5’-gacaatgccctgctggcc-3’ |
| PXR3292/3691Luc (XbaI) | 5’-ggctagcattcctcagga-3’ |
| PXR3342/3691Luc (XbaI) | 5’-ctgtagggagtgaagcca-3’ |
| PXR3392/3691Luc (XbaI) | 5’-aggtcaggaccatcagag-3’ |
| PXR3442/3691Luc (XbaI) | 5’-tgtggtctggggagaaat-3’ |
| PXR3492/3691Luc (XbaI) | 5’-aagggaccaagcgaccaa-3’ |
| PXR3542/3691Luc (XbaI) | 5’-ccacgtttgttcgcttcc-3’ |
| PXR3592/3691Luc (XbaI) | 5’-gtctcccacttcccactc-3’ |
| PXR3642/3691Luc (XbaI) | 5’-tccaggcctgtactcatc-3’ |
Numbered according to NM_003889.
2.2. Cell transfection and luciferase assay
Three cell lines were used in this study including 293T (human embryonic kidney), HepG2 (human hepatocellular carcinoma) and LS180 (human colon adenocarcinoma). HepG2 and LS180 lines were purchased from American Type Culture Collection (Manassas, VA), but the 293T line was from GenHunter Corporation (Nashville, TN). All cell lines were maintained in Dulbecco’s modified eagle medium (DMEM) containing 10% fetal bovine serum, penicillin and streptomycin, 1× non-essential amino acids. Unless otherwise indicated, cells were plated in 48 well-plates and transiently transfected by GenJet version II from SignaGen Laboratories (Rockville, MD). For reporter assays, the transfection mixture typically contained 50 ng of a reporter, 50 ng of a miR construct and 0.2 ng of the CMV-Renilla luciferase plasmid. After incubation at 37°C for 24 h, cells were extensively washed, collected and assayed for luciferase activities with the Dual-Luciferase Reporter Assay System as described previously [6, 16]. The reporter luciferase activity was normalized with Renilla luciferase activity, and the vector-transfected cells served as the basal reporter activity for miRs-transfected cells. It should be noted that the same amount of total plasmids were used in all reporter assays.
2.3. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The LS180 cell line was primarily used for the RT-qPCR because this cell line has been shown to robustly support PXR-directed transactivation [29]. Cells were typically plated in 24-well plates for overnight and then transfected with the miR-30c-1-3p construct or the corresponding vector. Cells were harvested 72 or 96 h after the transfection. In some cases, cells were treated with DMSO or rifampicin (10 µM) after transfection. Harvested cells were used for the preparation of total RNA. For the determination of PXR or CYP3A4 mRNA, total RNA (1 µg) was subjected to the synthesis of the first strand cDNA as described previously [30]. cDNAs were then diluted 8 times and RT-qPCR was conducted with TaqMan Gene Expression Assay (Applied Biosystems, Foster City, CA). The TaqMan probes were: PXR, Hs00243666_m1; CYP3A4, Hs00604506_m1; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 4352934E; and RNA polymerase II, Hs00172187_m1. The PCR amplification was conducted in a total volume of 20 µl containing universal PCR master mixture (10 µl), gene-specific TaqMan assay mixture (1 µl), and cDNA template (6 µl). The mRNA levels were normalized according to the level of GAPDH and the normalization of selected samples was confirmed based on the signal of RNA polymerase II. Amplification and quantification were done with the Applied Biosystems 7500 Real-Time PCR System.
3. Results
3.1. Identification of miR-30c-1-3p as a suppressive miR of PXR
PXR is a master regulator of the expression of genes involved in drug metabolism, metabolic disorders and tumor growth behaviors [7–9, 26, 27]. Like its target genes, the expression of PXR is regulated by factors such as age, disease mediators and therapeutic agents [17, 18, 31, 32]. While transactivation and repression are common mechanisms in regulated expression of PXR, emerging evidence suggests an involvement of post-transcriptional mechanisms in PXR expression, particularly through miR species [22]. miRs constitute a superfamily of small RNA species, and many of them are implicated in the development of a wide range of diseases [23–27]. To shed light on the missing link between PXR expression and miRs, we tested a large number of miRs for their ability to regulate the expression of PXR. We took the advantage of miRs as predominantly post-transcriptional regulators [24, 25], mRNA-based PXR reporters (cDNA reporters) were used for the initial study.
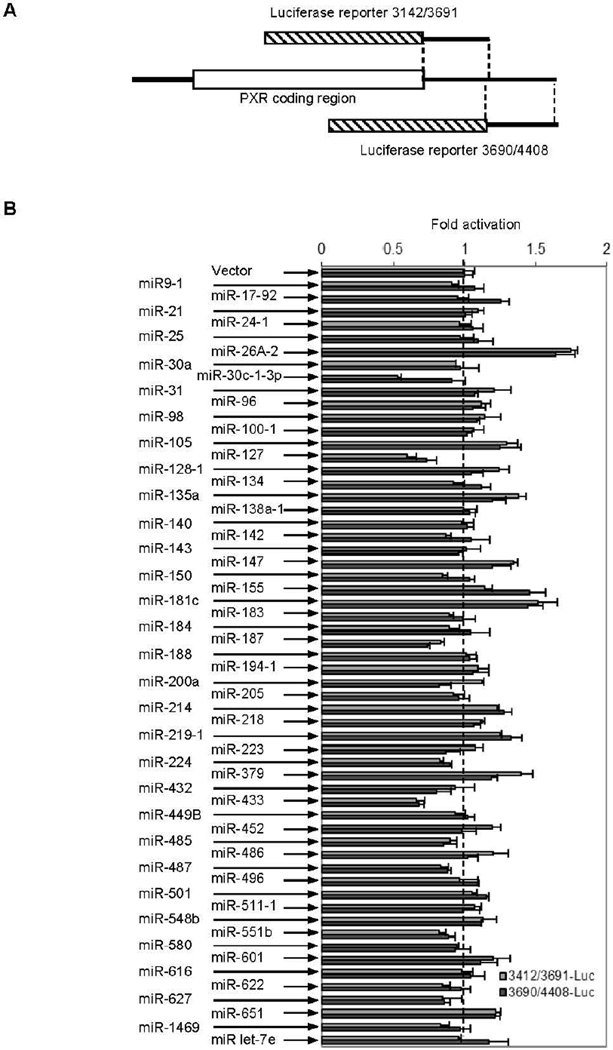
While there are exceptions, miRs usually target the 3’-UTR sequences. Therefore, we constructed two luciferase reporters that together harbor the entire 3’-UTR sequence of the dominant human PXR transcripts (Fig. 1A) (NM_003889). These two reporters were designated PXR3142/3691Luc and PXR3690/4408Luc, respectively. These two reporters were prepared by ligating the respective 3’-UTR fragments into the XbaI and FseI sites. It should be noted that the full-length 3’ UTR region of PXR has an XbaI site, and two reporters were prepared to avoid internal XbaI site. Cotransfection was performed in 293T cells to determine the effect of a miR on the reporter activity. In addition, a renilla luciferase construct was included in the transfection. As shown in Fig. 1B, all miRs, with an exception of miR-30c-1-3p, affected the activity of both reporters to a similar extent. Transfection of miR-30c-1-3p, compared with the vector, resulted in decreased activity of PXR3142/3691Luc by 50%, but only 10% decrease on the activity of PXR3690/4408Luc. Some miRs such as miR-26A-2, compared with the vector, caused significantly increases in the activity of both reporters. Others such as miR-433 and miR-127 significantly decreased the activity of both reporters (Fig. 1B). These miRs were not investigated further because of the following reasons: (1) they showed no selectivity toward two reporters, (2) bioinformatics analysis predicted no binding sites for these miRs, and/or (3) they affected the renilla luciferase activity, which was used for normalizing transfection efficiency.
Fig. 1. Suppression of PXR reporters derived from the 3’-untranslated region (UTR).
(A) Diagrammatic presentation of PXR reporters The PXR3142/3691Luc reporter contains the cDNA segment from nucleotide 3142 to 3691, whereas the PXR3690/4408Luc reporter contains from nucleotide 3690 to 4408. The hatched box represents the luciferase coding sequence. (B) Identification of PXR suppressive miR(s) Cells (293T) were transiently transfected by GenJet version II with a mixture containing 50 ng of a miR precursor construct, 50 ng of a reporter, or the vector along with 5 ng of the Renilla luciferase plasmid. The transfected cells were cultured for 24 h, harvested and analyzed for luciferase activities with a Dual-Luciferase Reporter Assay System. The results were from one of two experiments in triplicate.
3.2. Sequence-specific targeting by miR-30c-1-3p in both hepatic and intestinal cell lines
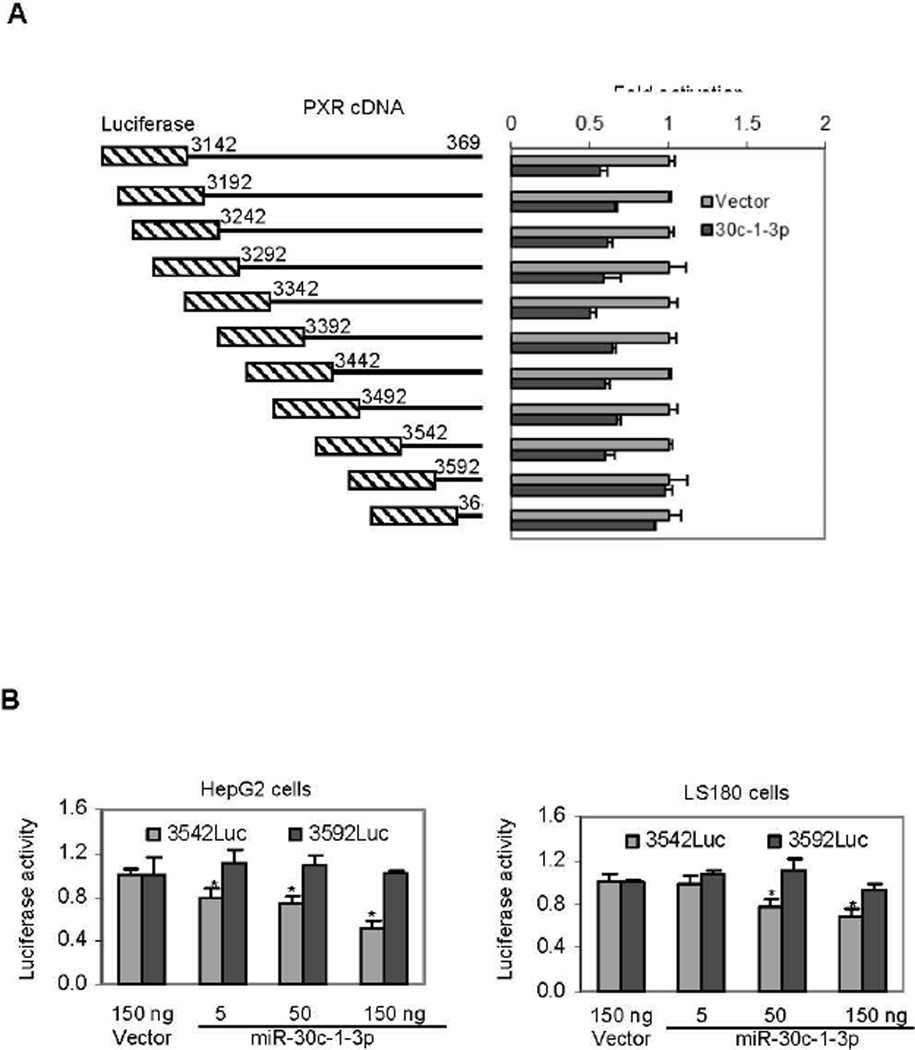
The screening study clearly demonstrated that human PXR mRNA is a sequence-specific target of miR-30c-1-3p. To locate the sequence that supports the action of this miR, a serial of deletions from the 5’ end were made on the PXR3142/3691Luc reporter and the resultant reporters were tested for the lost activity toward miR-30c-1-3p. As shown in Fig. 2A, all deletion mutants, except PXR3592Luc and PXR3642Luc, were repressed by miR-30c-1-3p. These results established that this 50 base sequence, namely from 3542 to 3592 support the repression of miR-30c-1-3p on PXR expression. We are in the process of specifying the precise sequence and nucleotides that support the action of miR-30c-1-3p by site-directed mutagenesis.
Fig. 2. Characterization of miR-30c-1-3p on the suppression of PXR.
(A) Dissection of the PXR3142/2691Luc reporter for the identification of miR-30c-1-3p response Cells (293T) were transiently transfected by GenJet version II with a mixture containing 50 ng of the miR-30c-1-3p construct, 50 ng of a reporter, or the vector along with 5 ng of the Renilla luciferase plasmid. The transfected cells were cultured for 24 h, harvested and analyzed for luciferase activities with a Dual-Luciferase Reporter Assay System. (B) Suppression of PXR3542/3691Luc by miR-30c-1-3p in HepG2 and LS180 cell lines Cells were transfected by GenJet version II with a mixture containing 5-150 ng of the miR-30c-1-3p construct, 50 ng of a reporter (PXR3542/3691 or PXR3592/3691Luc, or the vector along with 5 ng of the Renilla luciferase plasmid. The vector plasmid was used to equalize the total amount of constructs. The transfected cells were cultured for 24 h, harvested and analyzed for luciferase activities as described above. Asterisk signs indicate statistical significance from vector-transfected cells (P < 0.05). Significant differences were made according to One-way ANOVA followed by a DUNCAN’s multiple comparison test.
It is well known that some miRs are processed in a cell-specific manner [33]. We next tested whether the repression by miR-30c-1-3p occurs in HepG2 and LS180 cell line. HepG2 was derived from hepatocellular carcinoma whereas LS180 from colon adenocarcinoma. Importantly, both organs abundantly express PXR [34, 35]. In addition, the cotransfection was performed with various amounts of miR-30c-1-3p to establish the concentration-response relationship. As expected, the PXR3542Luc but not PXR3592Luc reporter was repressed in both cell lines (Fig. 2B). HepG2 cells supported greater repression than LS180 cells. Overall, the magnitude of the repression occurred in a concentration-dependent manner (Fig. 2B).
3.3. Effect of miR-30c-1-3p on the expression of CYP3A4
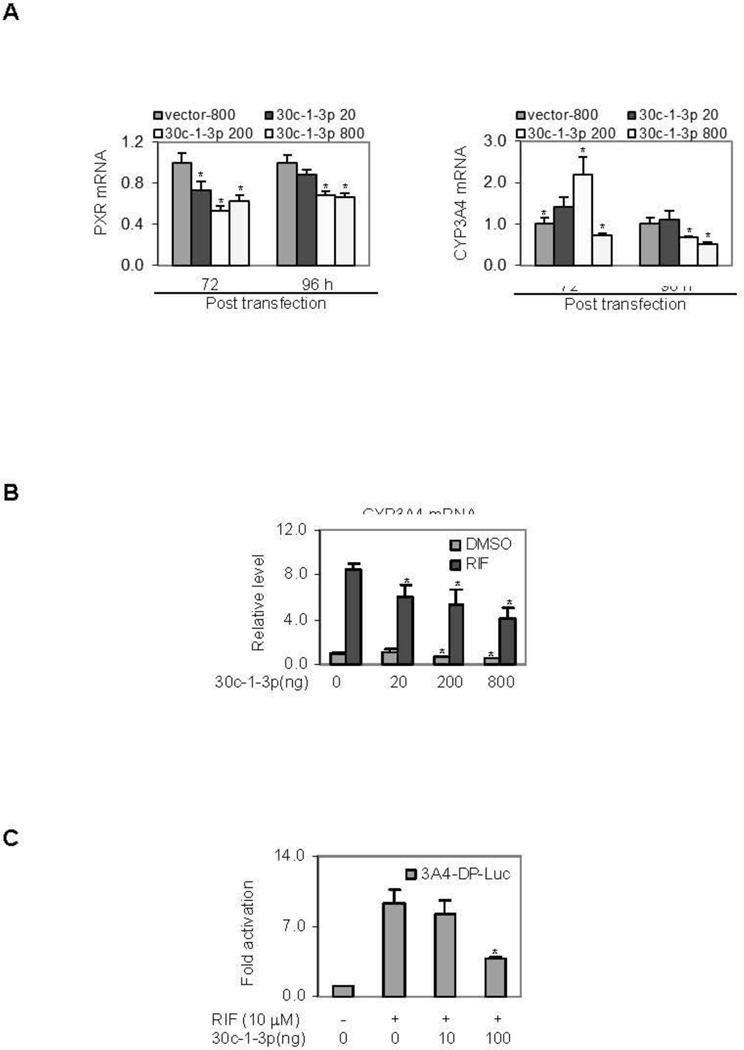
The study with the reporters established that the action of miR-30c-1-3p on PXR suppression is sequence-specific [24, 25]. Next we tested whether the repressive activity of the reporter can be recapitulated on the expression of the endogenous gene of PXR. LS180 but not HepG2 cells were used for the induction study because LS180 cells supported higher induction of CYP3A4 than HepG2 cells. Initially, cells were transfected with miR-30c-1-3p or the corresponding vector, cultured for various lengths of time, and the levels of PXR mRNA were determined. Fig. 3A shows representative results of this study. The level of PXR mRNA was decreased in miR-30c-1-3p transfected cells, and the 96 h time-point showed a relatively less decrease than the 72 h time-point (Left of Fig. 3A). The decrease in cells transfected with 20 ng plasmid of miR-30c-1-3p was less profound than that in cells transfected with 200 and 800 ng. Nonetheless, 200 and 800 ng caused a comparable decrease in both time-points. Next we tested whether miR-30c-1-3p alters the mRNA level of CYP3A4, a prototypical target of PXR [36]. While miR-30c-1-3p caused changes in the level of CYP3A4 mRNA, the changes varied depending on the amount of miR-30c-1-3p plasmid used for the transfection as well as the time after the transfection (Fig. 3A, Right). At the 72 h time-point, cells transfected with the miR-30c-1-3p plasmid at 20 and 200 ng exhibited increases in CYP3A4 mRNA, whereas a slight decrease was detected in cells transfected with 800 ng. At the 96 h time-points, significant decreases were detected in cells transfected with 200 and 800 ng plasmid (Fig. 3A, Right).
Fig. 3. Effect of miR-30c-1-3p on the expression of CYP3A4.
(A) Effect of miR-30c-1-3p on the expression of PXR and CYP3A4 Cells (LS180) were transfected by GenJet version II with the miR-30c-1-3p construct at 20, 200 and 800 ng. The vector plasmid was used to equalize the total amount of constructs. Cells were harvested 72 or 96 h post-transfection. Total RNA was isolated and the mRNA levels of PXR (Left) and CYP3A4 (Right) were determined by RT-qPCR. All experiments were performed three times in triplicate. Asterisk signs in the data indicate statistical significance from vector-transfected cells (P < 0.05). (B) Effect of miR-30c-1-3p on the induction of CYP3A4 Cells (LS180) were transfected as described above and treated with DMSO or 10 µM rifampicin (RIF) 72 h post-transfection. The treated cells were collected 24 h thereafter and the level of CYP3A4 mRNA was determined by RT-qPCR. All experiments were performed three times in triplicate. Asterisk signs in the data indicate statistical significance from vector-transfected cells or cells treated with DMSO (P < 0.05). (C) Effect of miR-30c-1-3p on the activation of CYP3A4-DP-Luc reporter Cells (LS180) were plated in 48 well-plates and transfected with a mixture containing 50 ng of the CYP3A4-DP-Luc reporter, 50 ng of miR-30c-1-3p or the vector along with 5 ng of the Renilla luciferase plasmid. The vector was used to equalize the total amount of constructs. After incubation at 37°C for 24 h, the transfected cells were treated with 10 µM rifampicin (RIF) or the same volume of DMSO for 48 h. Luciferase activities were determined with a Dual-Luciferase Reporter Assay System and the reporter activity was normalized based on the Renilla luminescence signal. All experiments were performed three times in triplicate. Asterisk signs in the data indicate statistical significance from vector-transfected and RIF-treated cells (P < 0.05).
We next tested whether miR-30c-1-3p also alters the induction of CYP3A4. Cells were transfected with miR-30c-1-3p or the vector, and then treated with DMSO or rifampicin, a prototypical activator of human PXR. As shown in Fig. 3B (Left), the overall expression of CYP3A4 mRNA was decreased at both basal and induced level with an exception of the basal level in cells transfected with 20 ng plasmid of miR-30c-1-3p. As described above, marked decreases in basal expression were detected in cells transfected with miR-30c-1-3p at 200 and 800 ng. As a result, these cells showed no changes in terms of fold of induction. To further establish the role in reduced induction of CYP3A4, a CYP3A4 reporter was tested for the reduced activation in response to miR-30c-1-3p [28]. Consistent with the result on the level of CYP3A4 mRNA, cotransfection of miR-30c-1-3p decreased the activation of this reporter by as much as 60% (Fig. 3C).
4. Discussion
PXR has been established to play a major role in the expression of genes involved in drug elimination [7, 9, 11]. However, emerging evident has suggested that this nuclear receptor, in addition to drug elimination, is integrally connected with endobiotic signaling and homeostasis of energy balance [11]. While PXR is recognized as a master regulator of gene expression, we and other investigators have reported that the expression of PXR is regulated by many factors such as age, disease mediators and therapeutic agents [17, 18, 31, 32]. Multiple mechanisms are reportedly involved in the regulated expression of PXR including transactivation, repression and miR-148a silencing [22]. This study identified and characterized miR-30c-1-3p as a silencer of PXR. A set of experiments have shown that miR-30c-1-3p targeted the PXR 3’-UTR and decreased the level of PXR mRNA, suggesting that miR-30c-1-3p downregulates PXR expression, at least partially by reducing the mRNA stability of this nuclear receptor. Importantly, this miR altered the expression of CYP3A4, a prototypical target of PXR [36], pointing to a functional role of miR-30c-1-3p in drug elimination.
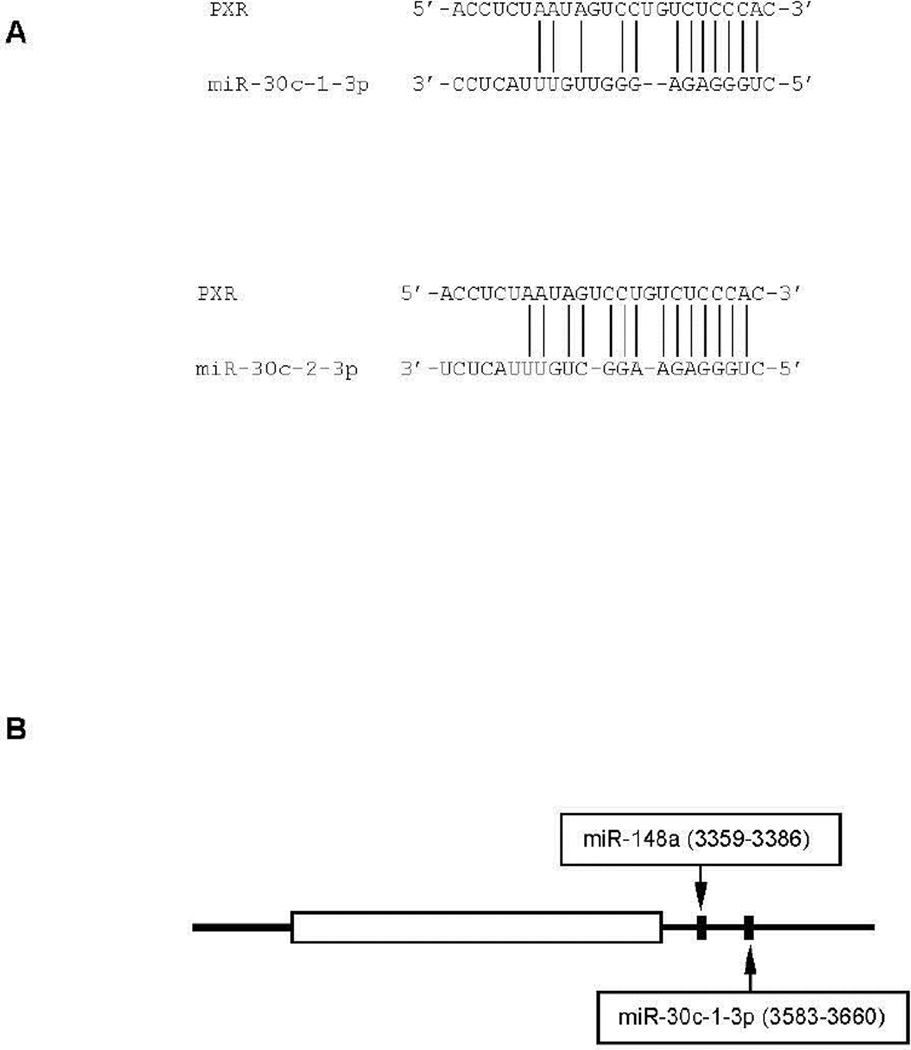
It has been reported that miR-30c-1-3p belongs to the miR-30 family, and the human genome has six miR-30 genes [37, 38]. However, these genes produce only five distinct mature guide strands. Importantly, these guide strands have the identical seed sequence, allowing them to regulate the expression of the same target genes, at least through the guide strand. This study, however, has demonstrated that miR-30a, a member of this family, failed to suppress the PXR reporters (Fig. 1B), suggesting that PXR is a target of the passenger strand (i.e., miR-30c-1-3p). In addition to miR-30c-13p, the miR-30c class has another member, namely miR-30c-2-3p [39]. Both miRs are derived from intronic sequences of other genes. The miR-30c-1-3p gene is located at chromosome 1 and the miR-30c-2-3p gene is located at chromosome 6. Importantly, miR-30c-13p and miR-30c-2-3p differ in the sequence (Fig. 4A). Interestingly, miR-30c-2-3p matches better than miR-30c-1-3p with the PXR 3-UTR region (Fig. 4A). Therefore, it is likely that miR-30c-2-3p is more potent than miR-30c-1-3p in silencing PXR, although the relative expression of these two miRs likely determines their contribution to PXR silencing. In addition, miR-148a, another miR, reportedly silenced PXR [22]. Based on the recognition sequences (Fig. 4B), both miR-30c and miR-148a target the same RNA species of PXR, although their recognition elements are 218 nucleotides apart (Fig. 4B). Nevertheless, it remains to be determined whether and how miR-148a networks with miR-30c in terms of regulating PXR expression.
Fig. 4. Sequence targeted by miR-30c-1-3p and its location in the 3’-UTR of PXR transcript.
(A) Sequence targeted by mir-30c The sequence targeted by miR-30c is shown as pair-matching for both miR-30c-1-3p and miR-30c-2-3p. (B) Location of the sequence targeted by miR-30c and miR-148a In addition to the location of the sequence targeted by miR-30c, the location of the sequence targeted by miR-148a is also shown. Specifically, miR-30c (both 30c-1-3p and -2-3p) likely targets the sequence from nucleotide 3583 to nucleotide 3660, whereas miR148a from 3359 to 3386, respectively. The sequence is numbered according to NM_003889.
The silencing of PXR by miR-30c-1-3p may have important clinical consequences. In this study, we have shown that miR-30c-1-3p decreased the level of PXR mRNA accompanied by altered expression of CYP3A4, a prototypical gene of PXR [36]. The altered expression of CYP3A4, however, varied depending on time and the amounts of miR-30c-1-3p. The overall expression of CYP3A4 mRNA was decreased in both basal and induction conditions (Fig. 3B, Left), but the decrease was not evident until later time-point (Fig. 3A, Right). It has been reported that miR-mediated suppression was delayed [40]. The altered expression of CYP3A4 by miR-30c-1-3p was in particular as it represented a mechanism secondarily to the suppression of PXR. Such an indirect mechanism required longer time to achieve the anticipated effect. On the other hand, the basal level of CYP3A4 mRNA at the early time-point was actually increased when small amounts of miR-30c-1-3p were used (Fig. 3A, Right). The precise mechanism on the increase remains to be determined. Nuclear receptors including PXR are known to interact with co-repressors and coactivators [14]. It is the presence of a ligand that induces conformational changes from repressing to activating status of a target gene. It should be not emphasized that our observation, increased expression of CYP3A4 by PXR knockdown, was consistent with previous report that knockout of pxr (in mice) increased the expression of cyp3a11 (the counterpart of human CYP3A4) [41].
The functionality of PXR has been linked to a wide range of behavior changes of malignancies and some of them are opposing to each other [27]. In some cases, activation of PXR up-regulates the expression of proapoptotic genes thus shows anti-tumor activity. In other cases, PXR is linked to upregulation of antiapoptotic genes and favors tumor progression. Nevertheless, up-regulated expression of genes involved in drug eliminations is generally considered to be a major contributing factor to the development of chemoresistance. Interestingly, miR-30c has been shown to promote cell apoptosis, inhibit cell proliferation, reduce tumor clonogenicity and suppress metastatic potentials [42, 43]. In addition, the expression of miR-30c was significantly decreased in many chemoresistant cell lines [44]. It remains to be determined whether miR-30c members can overcome PXR-directed chemoresistance. Interestingly, miR-148a, another silencer of PXR, has also been downregulated in advanced cancer [45].
In summary, our study presents several important conclusions. Firstly, identification of miR-30c-1-3p as a negative regulator of PXR, along with the previous reporter on miR-148a, points the existence of miR-networks in regulating the functionality of PXR. Secondly, miR-30c-1-3p but not miR-30a suppressed PXR, underscoring the importance of the passenger strand in gene silencing. Thirdly, activation of PXR has been closely linked to the development of a spectrum of tumor behaviors, particularly in chemoresistance. miR-30c and miR-148a, on the other hand, have been associated with less aggressive behaviors of malignancy. These findings suggest that miR-30c/miR-148a-PXR represents favorable outcomes of chemotherapy, particularly chemotherapeutic agents with a potent activating activity of PXR.
Highlights.
The pregnane X receptor (PXR) is a master regulator of genes for drug metabolism.
miR-30c-1-3p is a silencer of PXR by targeting the 3’ untranslated region.
The silencing led to decreased induction of cytochrome P450 3A4 (CYP3A4).
CYP3A4 is involved in the metabolism of more than half of therapeutic agents.
Acknowledgments
This work was supported by NIH grants R01GM61988, R01ES07965 and R15AT007705 to BY.
Abbreviation
- CYP3A4
cytochrome P450 3A4
- DMEM
Dulbecco’s modified eagle medium
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- miR
microRNA
- PXR
pregnane X receptor
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors indicate no potential conflict of interest.
REFERENCE
- 1.Parkinson A, Ogilvie BW, Buckley DB, Kazmi F, Czerwinski M, Parkinson O O. In: Biotransformation of xenobiotics. Klaassen CD, editor. New York: the Casarett & Doull’s Toxicology, the Basic Science of Poisons McGraw-Hill; 2013. pp. 185–366. [Google Scholar]
- 2.Lewis DF. Human cytochromes P450 associated with the phase 1 metabolism of drugs and other xenobiotics: a compilation of substrates and inhibitors of the CYP1, CYP2 and CYP3 families. Curr Med Chem. 2003;10:1955–1972. doi: 10.2174/0929867033456855. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Yan B. Photochemotherapeutic agent 8-methoxypsoralen induces the expression of cytochrome P450 3A4 and carboxylesterase HCE2: evidence on a differential involvement of the pregnane X receptor. Toxicol Sci. 2007;95:13–22. doi: 10.1093/toxsci/kfl120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deenen MJ, Cats A, Beijnen JH, Schellens JH. Part 3: Pharmacogenetic variability in phase II anticancer drug metabolism. Oncologist. 2011;16:992–1005. doi: 10.1634/theoncologist.2010-0260. (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Waterschoot RA, Schinkel AH. A critical analysis of the interplay between cytochrome P450 3A and P-glycoprotein: recent insights from knockout and transgenic mice. Pharmacol Rev. 2011;63:390–410. doi: 10.1124/pr.110.002584. [DOI] [PubMed] [Google Scholar]
- 6.Yang D, Yang J, Shi JD, Deng R, Yan B. Scoparone potentiates transactivation of the bile salt export pump gene and this effect is enhanced by cytochrome P450 metabolism but abolished by a PKC inhibitor. Brit J Pharmacol. 2011;164:1547–1557. doi: 10.1111/j.1476-5381.2011.01522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolution of the pregnane x receptor: adaptation to cross-species differences in biliary bile salts. Mol Endocrinol. 2005;19:1720–1739. doi: 10.1210/me.2004-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vachirayonsti T, Ho KW, Yang D, Yan B. Suppression of the pregnane X receptor during endoplasmic reticulum stress is achieved by down-regulating hepatocyte nuclear factor-4a and up-regulating liver-enriched inhibitory protein. Toxicol Sci. 2015;144:382–392. doi: 10.1093/toxsci/kfv008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omiecinski CJ, Vanden Heuvel JP, Perdew GH, Peters JM. Xenobiotic metabolism, disposition, and regulation by receptors: from biochemical phenomenon to predictors of major toxicities. Toxicol Sci. 2011;120:S49–S75. doi: 10.1093/toxsci/kfq338. (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timsit YE, Negishi M. CAR and PXR: the xenobiotic-sensing receptors. Steroids. 2007;72:231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ihunnah CA, Jiang M, Xie W. Nuclear receptor PXR, transcriptional circuits and metabolic relevance. Biochim Biophys Acta. 2011;1812:956–963. doi: 10.1016/j.bbadis.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modica S, Bellafante E, Moschetta A. Master regulation of bile acid and xenobiotic metabolism via the FXR, PXR and CAR trio. Front Biosci. 2009;14:4719–4745. doi: 10.2741/3563. [DOI] [PubMed] [Google Scholar]
- 13.Liu F, Song X, Yang D, Deng R, Yan B. The far and distal enhancers in the CYP3A4 gene coordinates the proximal promoter in responding similarly to the pregnane X receptor similarly but differentially to hepatocyte nuclear factor-4a. Biochem J. 2008;409:243–250. doi: 10.1042/BJ20070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carnahan VE, Redinbo MR. Structure and function of the human nuclear xenobiotic receptor PXR. Curr Drug Metab. 2005;6:357–367. doi: 10.2174/1389200054633844. [DOI] [PubMed] [Google Scholar]
- 15.Sachdeva K, Yan B, Chichester CO. Lipopolysaccharide and cecal ligation/puncture differentially affect the subcellular distribution of the pregnane X receptor but consistently cause suppression of its target gene CYP3A. Shock. 2003;19:469–474. doi: 10.1097/01.shk.0000048903.46342.ec. [DOI] [PubMed] [Google Scholar]
- 16.Song X, Xie M, Zhang H, Li Y, Sachdeva K, Yan B. The pregnane X receptor binds to response elements in a genomic context-dependent manner, and PXR activator rifampicin selectively alters the bindings among target genes. Drug Metab Dispos. 2004;32:35–42. doi: 10.1124/dmd.32.1.35. [DOI] [PubMed] [Google Scholar]
- 17.Ma Y, Song X, Sachdeva K, Liu J, Li Y, Yang D, Deng R, Chichester CD, Yan B. Clofibrate and perfluorodecanoate both up-regulate the expression of the pregnane X receptor but only clofibrate enhances its ligand-dependent induction of cytochrome P4503A23. Biochem Pharmacol. 2005;69:1363–1371. doi: 10.1016/j.bcp.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Shi D, Yang D, Yan B. Dexamethasone transcriptionally increases the expression of the pregnane X receptor and synergistically enhances pyrethroid deltamethrin in the induction of cytochrome P450 3A23. Biochem Pharmacol. 2010;80:1274–1283. doi: 10.1016/j.bcp.2010.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper BW, Cho TM, Thompson PM, Wallace AD. Phthalate induction of CYP3A4 is dependent on glucocorticoid regulation of PXR expression. Toxicol Sci. 2008;103:268–277. doi: 10.1093/toxsci/kfn047. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Hao C, Yang D, Shi D, Song X, Luan X, Hu G, Yan B. Pregnane X receptor is required for interleukin-6-mediated down-regulation of cytochrome P450 3A4 in human hepatocytes. Toxicol Lett. 2010;197:219–226. doi: 10.1016/j.toxlet.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beigneux AP, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. Reduction in cytochrome P-450 enzyme expression is associated with repression of CAR (constitutive androstane receptor) and PXR (pregnane X receptor) in mouse liver during the acute phase response. Biochem Biophys Res Commun. 2002;293:145–149. doi: 10.1016/S0006-291X(02)00196-1. [DOI] [PubMed] [Google Scholar]
- 22.Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem. 2008;283:9674–9980. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- 23.Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim T, Reitmair A. Non-Coding RNAs: Functional Aspects and Diagnostic Utility in Oncology. Int J Mol Sci. 2013;14:4934–4968. doi: 10.3390/ijms14034934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu HW, Sze DM, Cho WC. MicroRNAs Involved in Anti-Tumour Immunity. Int J Mol Sci. 2013;14:5587–5607. doi: 10.3390/ijms14035587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, Ren S, Evans RM, Xie W. A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem. 2006;281:15013–15020. doi: 10.1074/jbc.M511116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pondugula SR, Mani S. Pregnane xenobiotic receptor in cancer pathogenesis and therapeutic response. Cancer Lett. 2013;328:1–9. doi: 10.1016/j.canlet.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song X, Li Y, Liu J, Mukundan M, Yan B. Simultaneous substitution of phenylalaine-305 and aspartate-318 of rat PXR by the corresponding human residues abolishes the ability to transactivate the cytochrome P450 3A23 promoter. J Pharmacol Exp Ther. 2005;312:571–582. doi: 10.1124/jpet.104.074971. [DOI] [PubMed] [Google Scholar]
- 29.Zheng XE, Wang Z, Liao MZ, Lin YS, Shuhart MC, Schuetz EG, Thummel KE. Human PXR-mediated induction of intestinal CYP3A4 attenuates 1α,25-dihydroxyvitamin D3 function in human colon adenocarcinoma LS180 cells. Biochem Pharmacol. 2011;84:391–401. doi: 10.1016/j.bcp.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao D, Chen YZ, Yang D, Yan B. Age-related inducibility of carboxylesterases by the antiepileptic agent phenobarbital and implications in drug metabolism and lipid accumulation. Biochem Pharmacol. 2012;84:232–239. doi: 10.1016/j.bcp.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pascussi JM, Drocourt L, Fabre JM, Maurel P, Vilarem MJ. Dexamethasone induces pregnane X receptor and retinoid X receptor-alpha expression in human hepatocytes: synergistic increase of CYP3A4 induction by pregnane X receptor activators. Mol Pharmacol. 2000;58:361–372. doi: 10.1124/mol.58.2.361. [DOI] [PubMed] [Google Scholar]
- 32.Vyhlidal CA, Gaedigk R, Leeder JS. Nuclear receptor expression in fetal and pediatric liver: correlation with CYP3A expression. Drug Metab Dispos. 2006;34:131–137. doi: 10.1124/dmd.105.005967. [DOI] [PubMed] [Google Scholar]
- 33.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1988;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, LeCluyse E, Liu L, Hu M, Matoney L, Zhu W, Yan B. Rat pregnane X receptor: molecular cloning, tissue distribution and xenobiotic regulation. Arch Biochem Biophys. 1999;368:14–22. doi: 10.1006/abbi.1999.1307. [DOI] [PubMed] [Google Scholar]
- 36.Klein K, Zanger UM. Pharmacogenomics of Cytochrome P450 3A4: Recent Progress Toward the "Missing Heritability" Problem. Front Genet. 2013;4:12. doi: 10.3389/fgene.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 38.Bridge G, Monteiro R, Henderson S, Emuss V, Lagos D, Georgopoulou D, Patient R, Boshoff C. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood. 2012;120:5063–5072. doi: 10.1182/blood-2012-04-423004. [DOI] [PubMed] [Google Scholar]
- 39.Karbiener M, Neuhold C, Opriessnig P, Prokesch A, Bogner-Strauss JG, Scheideler M. MicroRNA-30c promotes human adipocyte differentiation and co-represses PAI-1 and ALK2. RNA Biol. 2011;8:850–860. doi: 10.4161/rna.8.5.16153. [DOI] [PubMed] [Google Scholar]
- 40.Haenisch S, Laechelt S, Bruckmueller H, Werk A, Noack A, Bruhn O, Remmler C, Cascorbi I. Down-regulation of ATP-binding cassette C2 protein expression in HepG2 cells after rifampicin treatment is mediated by microRNA-379. Mol Pharmacol. 2011;80:314–320. doi: 10.1124/mol.110.070714. [DOI] [PubMed] [Google Scholar]
- 41.Staudinger JL JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li XH, Ha CT, Fu D, Xiao M. Micro-RNA30c negatively regulates REDD1 expression in human hematopoietic and osteoblast cells after gamma-irradiation. PLoS One. 2012;7:e48700. doi: 10.1371/journal.pone.0048700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bockgorn J, Yee K, Chang YF, Prat A, Huo D, Nwachukwu C, Dalton R, Huang S, Swanson KE, Perou CM, Olopade OI, Clarke MF, Greene GL, Liu H, Greene GL, Liu H. MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Res Treat. 2013;137:373–382. doi: 10.1007/s10549-012-2346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;111:478–486. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi M, Cuatrecasas M, Balaguer F, Hur K, Toiyama Y, Castells A, Boland CR, Goel A. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PLoS One. 2012;7:e46684. doi: 10.1371/journal.pone.0046684. [DOI] [PMC free article] [PubMed] [Google Scholar]