Abstract
Post-translational modification (PTM) of nuclear receptor superfamily members regulates various aspects of their biology to include sub-cellular localization, the repertoire of protein-binding partners, as well as their stability and mode of degradation. The nuclear receptor pregnane X receptor (PXR, NR1I2) is a master-regulator of the drug-inducible gene expression in liver and intestine. The PXR-mediated gene activation program is primarily recognized to increase drug metabolism, drug transport, and drug efflux pathways in these tissues. The activation of PXR also has important implications in significant human diseases including inflammatory bowel disease and cancer. Our recent investigations reveal that PXR is modified by multiple PTMs to include phosphorylation, SUMOylation, and ubiquitination. Using both primary cultures of hepatocytes and cell-based assays, we show here that PXR is modified through acetylation on lysine residues. Further, we show that increased acetylation of PXR stimulates its increased SUMO-modification to support active transcriptional suppression. Pharmacologic inhibition of lysine de-acetylation using trichostatin A (TSA) alters the sub-cellular localization of PXR in cultured hepatocytes, and also has a profound impact upon PXR transactivation capacity. Both the acetylation and SUMOylation status of PXR is affected by its ability to associate with the lysine de-acetylating enzyme histone de-acetylase (HDAC)3 in a complex with silencing mediator of retinoic acid and thyroid hormone receptor (SMRT). Taken together, our data support a model in which a SUMO-acetyl ‘switch’ occurs such that acetylation of PXR likely stimulates SUMO-modification of PXR to promote the active repression of PXR-target gene expression.
Keywords: Pregnane X receptor, acetylation, SUMOylation, ubiquitination, post-translational modification, nuclear receptor, histone deacetylase 3, silencing mediator of retinoic acid and thyroid hormone receptor
Graphical Abstract
1. Introduction
Pregnane x Receptor (PXR, NR1I2) was initially described as a master-regulator of drug-inducible xenobiotic detoxification pathways in the enterohepatic system [1, 2]. However, there is increasing recognition that PXR activation has a multiplicity of ‘non-canonical’ roles. For example, recent evidence indicates that PXR activation in liver impacts regulation of glucose and lipid metabolism [3, 4], and may affect the development of multi-drug resistance in certain solid tumor types [5]. Moreover, a fundamental role for PXR activation in ameliorating pro-inflammatory signals and loss of intestinal barrier permeability in the inflamed condition has been identified [6, 7].
Numerous studies reveal that acetylation of transcriptional regulatory proteins blends together with phosphorylation, ubiquitination, SUMOylation to form complex programs of gene activation [8]. More recently, attention has been given to the notion that a complex interplay between post-translational modifications (PTMs) occurs to allow alterations of PXR biology depending upon the physiological context [9, 10]. It is abundantly clear that different PTMs form a complex regulatory network with interactions and integrated features that resemble a refined language. We feel that such a complex and interwoven regulatory program of gene expression is likely to play a pivotal role in disease pathogenesis and progression. The discovery of the repertoire of PTMs that target the PXR protein and how they interact with each other is thus an important and newly emerging field of research.
Our recent efforts combined with that of others indicate that phosphorylation, ubiquitylation, and SUMOylation of PXR play a pivotal and likely interactive role in regulating the biological function of this nuclear receptor protein [11-18]. However, where the PTMs occur on the protein and how they might interact with each other in live cells to regulate its complex biology in liver and intestine is only beginning to be understood. There is a clear recognition that both SUMOylation and acetylation of numerous transcriptional regulatory proteins and histone proteins occurs in a coordinated and unified manner [8, 19, 20]. We therefore sought to investigate whether SUMOylation and acetylation have the ability to determine aspects of PXR biology, and whether they might interact with each other to affect PXR activity in hepatocytes.
Using an immunoprecipitation approach, we detect acetylation of PXR in primary hepatocytes and this is reduced by treatment with the PXR ligand rifampicin (Rif). The acetylation of PXR is increased in hepatocytes following treatment with the class I and class II lysine de-acetylation inhibitor TSA. Further, we identify the lysine/histone deacetylase HDAC3-SMRT co-repressor multi-protein complex as a likely regulator of ligand-dependent PXR acetylation. The co-expression of fluorescently tagged HDAC3 and PXR proteins in hepatocytes indicate that PXR and HDAC3 co-localize in mouse hepatocytes, and that the increased acetylation of PXR produced by treatment with TSA alters their sub-cellular localization. Treatment of transfected cells with TSA produces synergistic transactivation of a PXR-dependent reporter gene when combined with Rif. Enzymatic de-acetylation of PXR with the HDAC3-SMRT co-repressor complex inhibits SUMO-modification of PXR, while pharmacological promotion of acetylation with TSA promotes high levels of SUMO-modification of PXR. The acetylated and SUMOylated forms of PXR differentially associate with the HDAC3-SMRT co-repressor complex. The covalent attachment of SUMO proteins to PXR produces a strong repressive function that is functionally separate from its interaction with the HDAC3-SMRT co-repressor complex. Taken together, our data presented here support a model in which a SUMO-acetyl ‘switch’ occurs at the level of PXR, such that acetylation is prerequisite to promote SUMO-modification of PXR to support active repression of PXR-target gene expression.
2. Materials and Methods
2.1 Chemicals and Plasmids
Rifampicin (Rif), Trichostatin A (TSA), and Pregnenolone-16α-carbonitrile (PCN) were purchased from Sigma-Aldrich. All other reagents including culture medium for primary hepatocytes and mammalian cell lines were purchased from standard sources. The expression vectors encoding FLAG-tagged full length human PXR, (His)6-tagged SUMO3, and protein inhibitor of activated STAT-1 (PIAS1) were as previously described (Cui et al., 2015). The expression vector encoding FLAG-tagged HDAC3 is a kind gift from Dr. Eric Verdin and purchased from Addgene (plasmid #13819) [21]. The expression vector encoding HDAC3-GFP was a kind gift of Dr. Eric Olson [22]. The expression vector encoding full length SMRT was a kind gift of Dr. J.D. Chen [23]. The RFP-PXR expression vector was constructed by using the following primers to amplify human PXR to add a HindIII site: Left Primer- 5’ GACGGCCAAGCTTCGATGGAGGTGAGACCCAAAG 3’; Right Primer: 5’ GACGGCAAGCTTTCAGCTACCTGTGATGCCG 3’. The resulting amplimer was inserted into the pM-Cherry-C1 expression vector using the HindIII site (ClonTech). To generate (His)6-PXR-SUMO1 and (His)6-PXR-SUMO3 linear fusion construct, we used the following PCR primers that introduce XhoI restriction sites and both adds a STOP codon and removes one C-terminal glycine from the SUMO1 and SUMO3: SUMO1- left primer- 5’ GACGGCCTCGAGGCCATGTCTGACCAGGAGGCAAAA 3’; SUMO1- right primer- 5’ GACGGCCTCGAGCTACCCCGTTTGTTCCTGATAAAC 3’; SUMO3- left primer- 5’ GACGGCCTCGAGCCATGTCCGAGGAGAAGCCCAAG 3’; SUMO3- right primer 5’ GACGGCCTCGAGCTATCCCGTCTGCTgCTGGAACAC 3’. The modified SUMO1 and SUMO3 sequences were amplified and then sub-cloned into pShuttle-(His)6-PXR-(FLAG)3 expression vector using the XhoI restriction site that exists in between the last amino acid of PXR and the triple FLAG tag in this expression vector [11]. The FLAG-SUMO3-PXR construct was generated using the following PCR primers to introduce EcoRI restriction sites and removes the STOP codon and one glycine residue from SUMO3: SUMO3- left primer- 5’ GACGGCGAATTCATGTCCGAGGAGAAGCCCAAG 3’; SUMO3- right primer- 5’ GACGGCGAATTCTCCCGTCTGCTGCTGGAACAC 3’. The resulting amplimer was digested with EcoRI and inserted into the EcoRI site that exists between the FLAG epitope and PXR in the previously described pCMV-Tag human PXR expression construct [11]. All expression vectors were sequenced on both strands to ensure the integrity of the resulting open-reading frames.
2.2 Isolation and Culturing of Primary Mouse Hepatocytes
Primary hepatocytes were isolated from C57BL6 mice at age 6-10 weeks with a classic collagenase perfusion procedure as previously described (Staudinger, 2003). Potential sex differences were determined throughout the study, and identical results were acquired from both male and female mice. The representative results were obtained from male mice.
2.3 Total RNA Isolation, Reverse Transcription, and Real-Time Quantitative-Polymerase Chain Reaction Analysis
Real-time (RT) quantitative polymerase chain reaction (qPXR) was executed as previously described (Ding, 2005).
2.4 Immunoprecipitation Assay
Primary hepatocytes or Hepa1-6 cells cultured in 10 cM dishes and were harvested in 1 mL of a Lysis Buffer containing 150 mM NaCl, 50 mM Tris-Cl (pH 8.0), 1% Triton X-100, 20 mM N-Ethylmaleimide (NEM), and 1% Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). Samples were disrupted through sonication and subsequently centrifuged at 18,000 × g for 10 minutes to remove the insoluble substances. A fraction of the supernatant (5%) was saved as a loading control for western blot analysis, while the rest of sample was subjected to pre-clearing with 5% Protein A/G Sepharose beads at 4 °C. The pre-cleared cell lysates were separated from beads by centrifugation and then incubated with the appropriate antibody. The anti-acetylated lysine antibody cocktail was previously described and consisted of equal masses of four separate monoclonal antibodies from Novus Biologicals-15G10, Santa Cruz-AKL5C1, Cell Signaling-Ac-K-103, and Thermo Scientific-1C6 [24]. An equal amount of each antibody was added to the mixture, and the final antibody concentration was 4 μg per 1 mL of total cell lysate. The antibody cocktail was mixed together with a 5% volume of Protein A/G Sepharose beads and was applied to cell lysates for immunoprecipitation of acetylated proteins overnight at 4 °C with shaking. FLAG-tagged-HDAC3 and associated proteins were immunoprecipitated using antibody-agarose conjugated beads (Anti-FLAG M2 Affinity Gel, Sigma Aldrich, A2220). Subsequently, the beads were pelleted gently and were washed 3 times with Lysis Buffer containing 20 mM NEM, and 1% Halt™ Protease and Phosphatase Inhibitor Cocktail. Immunoprecipitated proteins were eluted in 30 μL 2X-Laemmli buffer and heated at 95 °C for 10 minutes for western blot analysis.
2.5 Cell-Based Immobilized Metal Affinity Pull-Down Assay
SUMO-modified proteins were enriched with cobalt beads using a modification of a previously described approach [25]. Briefly, 10 cM dishes containing primary cultures of mouse hepatocytes or Hepa1-6 cells were harvested using 1 mL of a strong denaturing lysis buffer 6M Guanidine-Cl (pH 8) according to specified experimental treatment. The whole cell lysates were then applied to 30 μL of cobalt beads and incubated on a rotor at room temperature for two hours with shaking. The gathered proteins were collected via centrifugation and washed twice with lysis buffer, three times with 8M Urea buffer (pH 6.5), and once with 1 × PBS. Proteins were removed from the beads using 30 μL of 2 × Laemmli buffer and heated at 95 °C for 10 minutes for subsequent western blot analysis.
2.6 Fluorescence Microscopy
Primary cultured mouse hepatocytes were transfected using Lipofecamine 2000 (Invitrogen) and maintained in William's E Media prior to image analysis. Twenty-four hours post-transfection, hepatocytes were washed once with 1x PBS and subsequently stained with Hoechst 33432 for an additional 30 minutes. To visualize, mouse hepatocytes were washed three times with 1x PBS and then maintained in Opti-MEM® I Reduced Serum Media during fluorescence protein image analysis. Fluorescent proteins were imaged with a 30x air objective, and excited at either 400 nm (GFP) or 561 nm (RFP). The nuclei were visualized using Hoechst 33432 staining under ultraviolet light.
2.7 Luciferase Reporter Gene Assay
CV-cells were seeded at 1 × 104 cells per well (n=8) in 96-well plate and transfected using lipofectamine 2000. Twenty-four hours post-transfection, cells were treated with either 10 μM rifampicin (Rif), 0.5 μM trichostatin A (TSA), or both for an additional 24 hours. Cells were lysed using standard conditions at 32 μL per well of lysis buffer (100 mM KPO4, pH 7.8; 0.2% Triton X-100; 1 mM DTT) and subjected to luciferase activity analysis (12 μL) using a standard manufacturer's protocol (Promega). The transfection efficiency was quantified according to the activity of β-galactosidase (20 μL). Fold change among experimental groups were normalized to control group with relative luciferase units/ β-galactosidase readouts.
2.8 Western Blot Analysis
Western blot analysis was conducted as described previously (Xu et al., 2009). Purchased antibodies include the mouse monoclonal anti-PXR antibody (H-11, Santa Cruz), rabbit monoclonal anti-SUMO1 (C9H1, Cell Signaling) and anti-SUMO2/3 antibodies (18H8, Cell Signaling). The mouse monoclonal anti-acetylated lysine antibody cocktail consisted of cat# 15G10 from Novus Biologicals, Cat# AKL5C1 from Santa Cruz, Cat # Ac-K-103 from Cell Signaling, and Cat # 1C6 from Thermo Scientific.
2.9 Liquid Chromatography-Tandem Mass Spectrometry Analysis
The LC-MS/MS analysis for identification of PXR post-translational modification was performed as described previously (Cui et al., 2015).
2.10 Statistical Analysis
The statistical analysis was executed wherever required. Statistical differences among one experimental group were determined using a one-way ANOVA followed by the Duncan's multiple range post hoc test. Moreover, statistical differences between experimental groups were determined using the Student's t test.
3. Results
3.1 The Interface between PXR Acetylation and SUMOylation
A study involving the farnesoid X receptor, a close relative of PXR, revealed the existence of a SUMO-acetyl ‘switch’ [20]. PXR has previously been identified as a likely substrate for the acetylation and SUMOylation signal transduction pathways [11, 25-27]. Furthermore, a recent study indicates that PXR physically associates with the lysine/histone de-acetylating enzyme HDAC3 in cell-based assays, and it has been suggested that this enzyme can de-acetylate PXR [17]. In vivo, HDAC3 forms an obligate and stable complex with the well-known nuclear receptor co-repressor protein SMRT, and the SMRT-HDAC3 co-repressor complex exhibits strong lysine de-acetylase activity [28, 29]. An additional study indicates that the SMRT protein is the preferred co-repressor protein-partner of PXR [23]. Therefore, we sought to determine the extent to which potential de-acetylation of PXR by HDAC3-SMRT co-repressor complex affects the SUMOylation level of PXR in cells. The murine hepatoma cell line, Hepa1-6, can support high levels of PXR SUMO-modification [11]. Cultures were therefore transfected with expression plasmids encoding PXR, (His)6-SUMO3, the SUMO-E3 ligase enzyme- PIAS1, HDAC3, and SMRT as indicated (Figure 1A). Forty-eight hours post-transfection, total SUMOylated proteins were gathered using strong denaturing conditions in an immobilized metal affinity pull-down assay [25]. The level of SUMOylated PXR was determined using western blotting analysis. Forced over-expression of the HDAC3-SMRT lysine de-acetylase enzyme complex strongly reduced the overall SUMOylation level of PXR.
Figure 1. The Interface between Acetylation and SUMOylation of PXR.
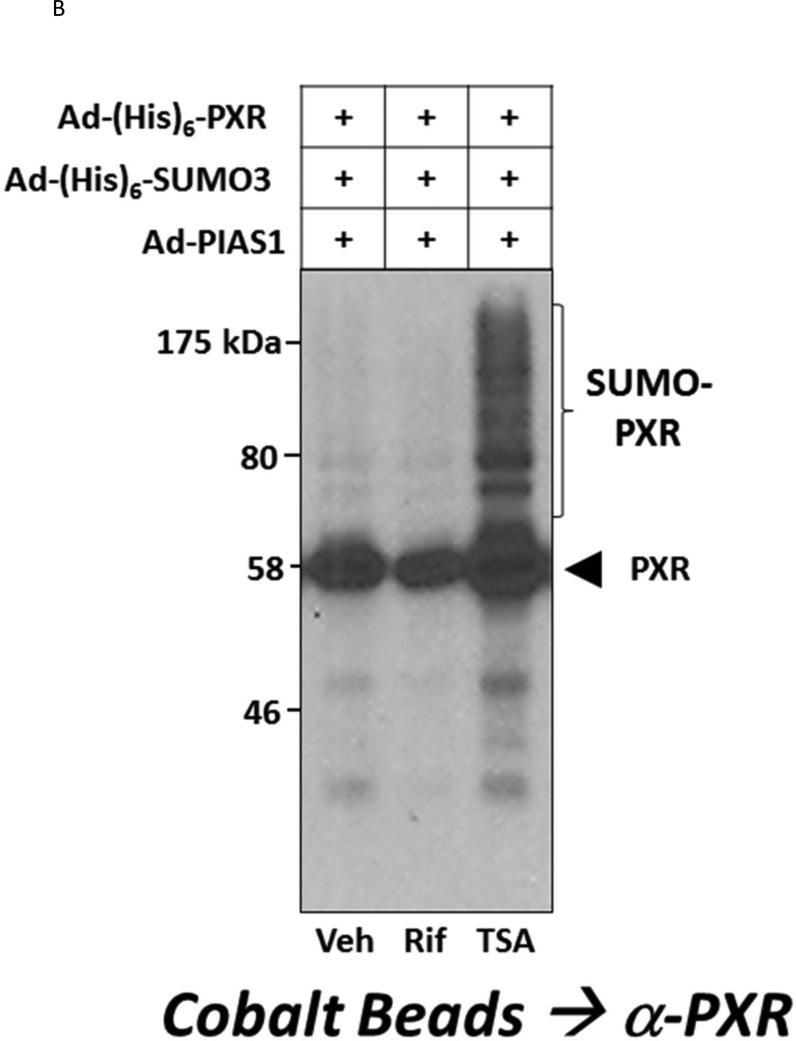
(A) Hepa1-6 cells were transfected with the indicated plasmid-based expression vectors. Cell lysates were produced using strong denaturing conditions to inhibit de-SUMOylation enzymes. SUMOylated proteins were enriched using cobalt beads and were washed sequentially using both guanidine-HCl and urea-based wash buffers. Proteins were eluted using 2X-Laemmli buffer and were then resolved using 10% SDS-PAGE. Western blot analysis was performed with an anti-PXR antibody that detects all modified forms of the protein (Santa Cruz, H-11 monoclonal Ab). (B) Hepa1-6 cells were transduced with the indicated adenoviral expression vectors. Twenty-four hr post-transduction, cells were treated with vehicle (0.1% DMSO) rifampicin (Rif, 10 μM) or Trichostatin A (TSA, 0.5 μM) for an additional twenty-four hr. SUMOylated proteins were gathered as in (A) and western blot with the anti-PXR antibody was used to analyze the extent of SUMO-modification.
To investigate whether increased acetylation of PXR alters its SUMOylation status we used TSA, a pharmacological inhibitor of the class I and II mammalian HDAC enzymes. Cultures of Hepa1-6 cells were co-transduced with adenoviral vectors encoding Ad-(His)6-PXR, Ad-(His)6-SUMO3, and Ad-PIAS1 (Figure 1B). Twenty-four hours post-transduction, cells were treated with vehicle (0.1% DMSO), 10 μM Rif, or 0.5 μM TSA for an additional 24 hours. SUMO-modified PXR proteins were enriched and the level of SUMO-modified PXR protein was determined using western blotting. Inhibition of lysine de-acetylase activity with TSA promoted the formation of high levels of SUMO-modified PXR, while both vehicle- and Rif-treated cells exhibited comparatively low levels of SUMO-modified PXR. Taken together, the data in Figure 1 suggest that acetylation and SUMOylation interface with each other at the level of the PXR protein and that acetylation of PXR likely promotes its subsequent SUMOylation in a cooperative and coincident manner.
3.2 Acetylation of PXR is altered during the Transactivation Process in Hepatocytes
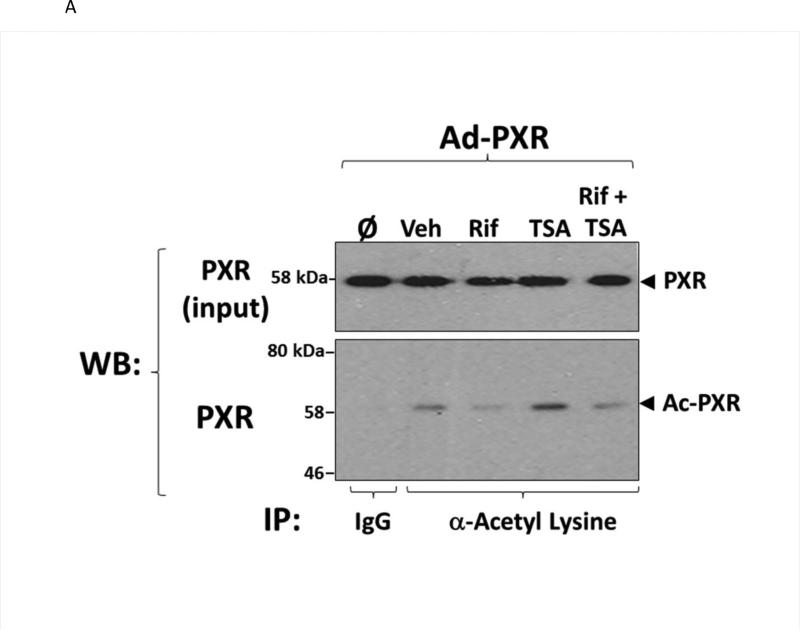
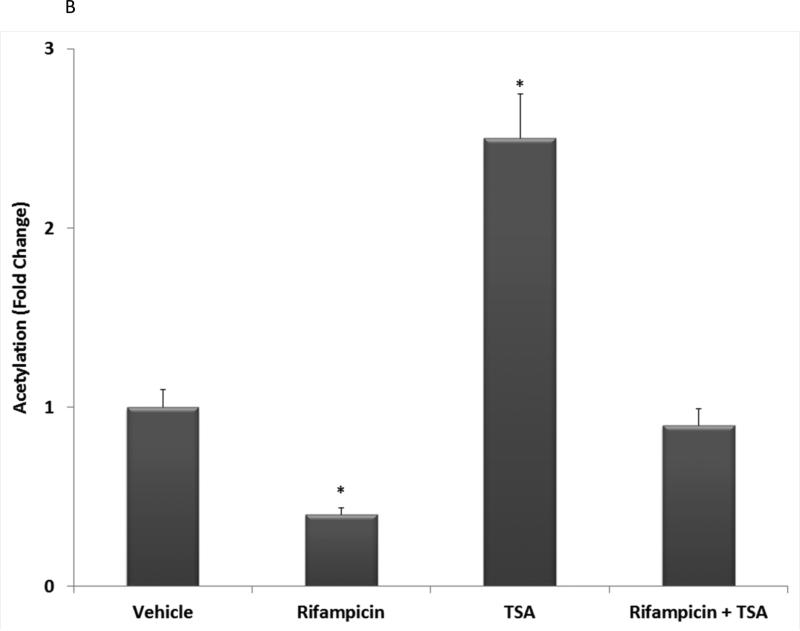
To examine the extent to which PXR acetylation status is altered in liver in response to ligand activation, we transduced hepatocytes isolated from wild type C57BL/6 mice with an adenoviral expression vector encoding a (His)6-tagged form of human PXR [Ad-(His)6-PXR]. Twenty-four hours post-transduction, hepatocytes were treated with vehicle (0.1% DMSO), 10 μM Rif, 0.5 μM TSA, or were co-treated with Rif and TSA together for an additional 24 hours. Acetylated proteins were immuno-purified from the respective cell extracts using a cocktail consisting of four commercially available anti-acetylated lysine monoclonal antibodies as described in Materials and Methods. A non-immune mouse IgG antibody was used as a negative control. Acetylated human PXR protein was detected in the acetylated lysine-enriched protein extracts by western blot analysis (Figure 2A). Note the slight decrease in electrophoretic mobility that is typical of acetylated nuclear receptor proteins. Treatment with TSA produced a significant increase in the level of PXR acetylation. In contrast, ligand activation of PXR by Rif tempered the TSA-induced acetylation of PXR when compared with the vehicle treated cells (Figure 2B). These results reveal that PXR is the molecular target of the acetylation signaling pathway in hepatocytes, and suggest that PXR transactivation capacity is inversely correlated with this PTM.
Figure 2. Acetylation of PXR is Altered during Transactivation by Ligand.
(A) Primary hepatocytes isolated from wild type C57BL/6 mice were isolated and transduced with an adenoviral expression vector encoding a FLAG-tagged form of human PXR (Left Panel). Total acetylated proteins were immunoprecipitated from cell extracts using a cocktail of four anti-acetylated lysine monoclonal antibodies as described in Materials and Methods. A non-immune antibody was also used as a negative control (IgG). Acetylated PXR was identified by western blotting with anti-PXR polyclonal antibody. (B) Western Blot images were quantitated by densitometric scanning of the X-ray films with the UVP Biodoc-It 220 image analysis system and 1D Gel Analysis Software. The numbers represent the relative densitometric image intensity of acetylated PXR divided by the image intensity of input levels of PXR, where vehicle treated control group was set to equal 1. Asterisks indicate a statistical difference from vehicle-treated samples (n = 3, and P < 0.05).
3.3 The Transactivation Capacity of PXR is modified by the Acetylation Signaling Pathway in Hepatocytes
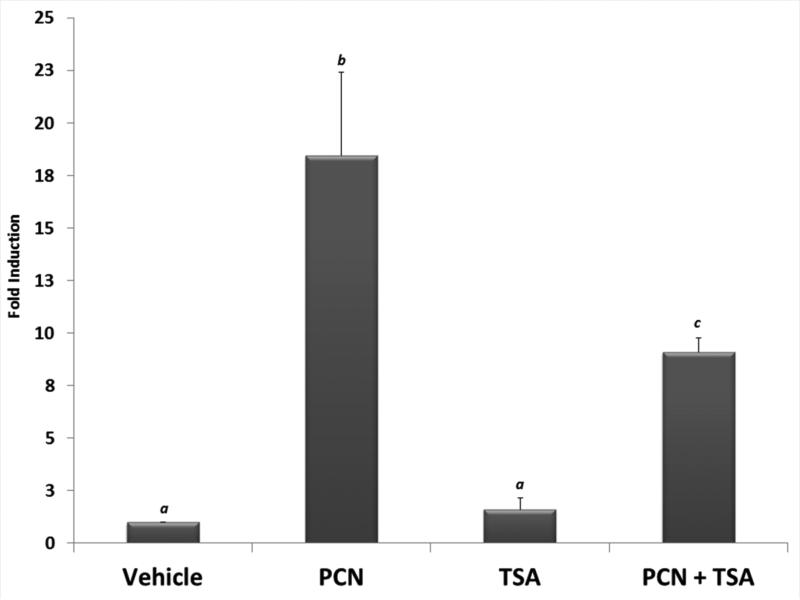
We next sought to examine the functional consequence of increased acetylation upon drug-inducible PXR activity. Primary hepatocytes isolated from wild type C57BL/6 mice were cultured overnight. The cultures were then treated for 24 hours with either vehicle (0.1% DMSO), pregnenolone 16α-carbonitrile (10 μM, PCN), 0.5 μM TSA, or were co-treated with PCN and TSA. Total RNA was isolated and the expression of the prototypical PXR-target gene cytochrome P450 family 3 subfamily a polypeptide 11 (Cyp3a11) was measured using real-time quantitative PCR analysis (Figure 3). As expected, significant induction of Cyp3a11 was observed following treatment with PCN. Treatment with TSA by itself did not produce significant alterations in Cyp3a11 gene expression. In contrast, the co-treatment of PCN together with TSA significantly diminished PCN-inducible Cyp3a11 gene expression levels when compared with the PCN alone treatment group. These data indicate that the canonical PXR transactivation capacity is suppressed by acetylation in cultures of primary hepatocytes.
Figure 3. PXR Transactivation Capacity is the Molecular Target of Acetylation in Hepatocytes.
Primary hepatocytes isolated from C57BL/6 mice were cultured overnight. Following twenty-four hr treatment with pregnenolone 16α-carbonitrile (PCN, 10 μM), trichostatin A (TSA, 0.5 μM), or both together, total RNA was isolated and the relative expression level of the Cyp3a11 gene was determined. Data are normalized to β-actin levels and are presented as fold induction. Letters different from each other indicate a statistical difference between treatment groups (n=3, and p<0.05).
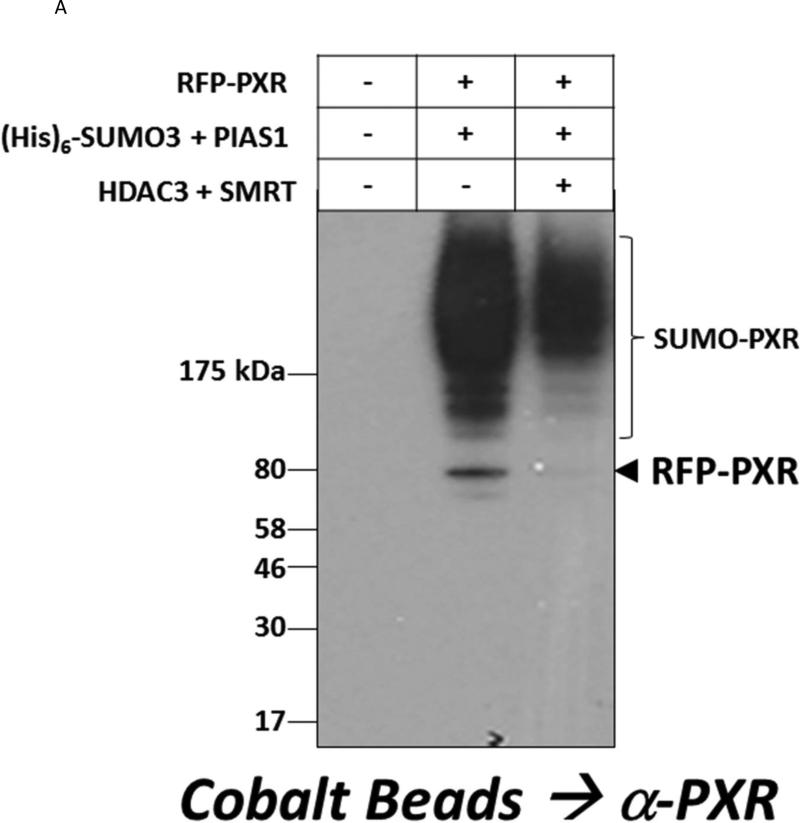
3.4 Acetylation Affects the Sub-cellular Localization of PXR in Hepatocytes
Like other nuclear receptors, PXR can shuttle between the cytoplasmic and nuclear compartment to modulate its transcriptional activity in response to different cellular signaling pathways [30]. The translocation of PXR to the nucleus is thought to be tightly regulated by various PTMs [31]. We first determined whether the engineered fusion between the red fluorescent protein and PXR (RFP-PXR) is able to undergo SUMO-modification, and whether RFP-PXR SUMOylation is subject to regulation by the HDAC3-SMRT lysine de-acetylase corepressor complex. Cultured hepatoma cells were transfected with expression vectors encoding RFP-PXR, (His)6-SUMO3, PIAS1, HDAC3, and SMRT as indicated (Figure 4A). Forty-eight hours post-transfection, SUMOylated proteins were gathered and SUMOylated RFP-PXR proteins were detected by western blotting. The RFP-PXR fusion protein indeed retained its ability to be targeted by SUMO, and like wild type PXR, the HDAC3-SMRT co-repressor deacetylation complex significantly inhibited SUMO-modification of RFP-PXR.
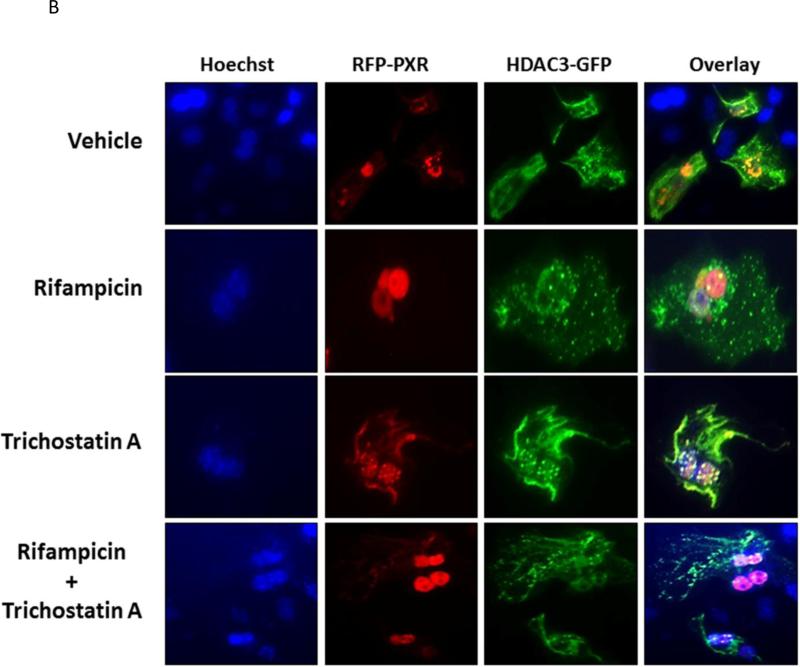
Figure 4. The HDAC3 Deacetylase Enzyme Affects PXR-SUMOylation and Co-localizes with PXR in Hepatocytes.
(A) Hepa1-6 cells were transfected with the indicated plasmid expression vectors. SUMOylated proteins were captured using cobalt beads and the extent of RFP-PXR modification was analyzed using western blotting with an anti-PXR antibody. (B) Primary cultures of mouse hepatocytes were transfected with RFP-PXR and HDAC3-GFP. Twenty-four hr post-transfection, hepatocytes were treated with rifampicin (10 μM), trichostatin A (0.5 μM), or both for an additional 24 hr. Fluorescent cells were imaged as described under Materials and Methods. To facilitate the visualization of the nucleus Hoechst 33342 (3 μM) was added to the live cells thirty min prior to imaging.
We next sought to investigate the extent to which acetylation might alter the sub-cellular localization of PXR. Primary hepatocytes were transfected with expression vectors that encode RFP-PXR and green fluorescent-tagged HDAC3 (HDAC3-GFP) proteins, respectively (Figure 4B). Twenty-four hours post-transfection, cultures were treated with vehicle (0.1% DMSO), 10 μM Rif, 0.5 μM TSA, or both together for an additional 24 hours. Fluorescence image analysis was performed as described in Materials and Methods. Hoechst 33342 (3μM) was used to visualize the nucleus by staining for 30 minutes immediately prior to imaging. Under vehicle-treated conditions, PXR was distributed roughly equally between the nuclear periphery and cytoplasmic compartments, and also exhibited strong co-localization with HDAC3-GFP. Treatment with Rif produced nearly complete translocation of RFP-PXR to the nucleus and also produced much less co-localization with HDAC3-GFP, save for some low levels of punctate sub-nuclear localization. Treatment with produced a high level of RFP-PXR localization to the aforementioned punctate sub-nuclear architecture. Interestingly, additional co-localization of RFP-PXR and HDAC3-GFP was observed at the cell periphery following TSA treatment. Co-treatment with Rif and TSA together produced broad and diffuse RFP-PXR fluorescence in the nucleus with a somewhat diminished presence of the punctate nuclear pattern. In contrast to the Rif-treated hepatocytes, the co-treated cells exhibited a significant amount of PXR cytoplasmic co-localization with HDAC3-GFP. These data suggest that acetylation of PXR can likely affect its transactivation capacity, in part, through altering its sub-nuclear and sub-cellular localization.
3.5 Acetylated PXR interacts with HDAC3-SMRT Co-repressor Complex
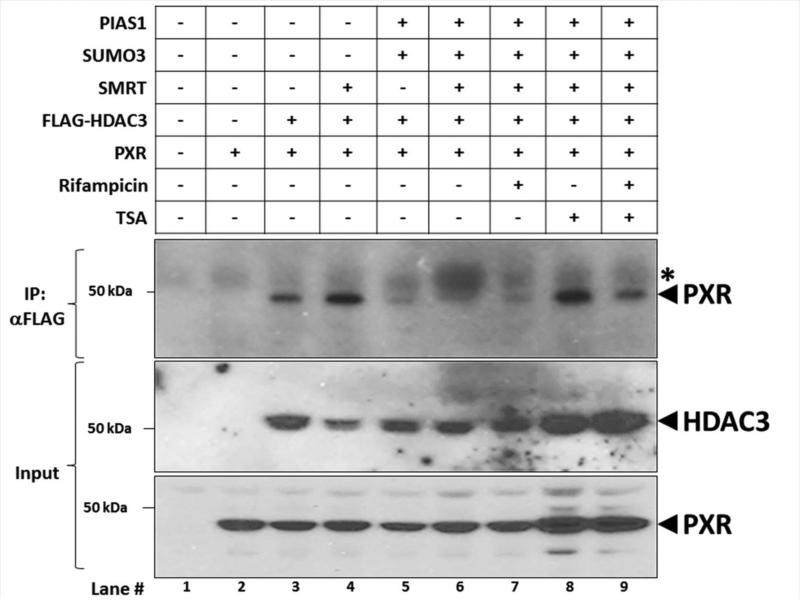
We next investigated the extent of PXR-HDAC3-SMRT protein-protein interactions using an immunoprecipitation approach. Cultures of Hepa1-6 cells were transfected with expression vectors encoding PXR, FLAG-HDAC3, SMRT, SUMO3, and PIAS1 as indicated (Figure 5). Twenty-four hours post-transfection, cells were treated with vehicle (0.1% DMSO), 10 μM Rif, 0.5 μM TSA, or both for an additional 24 hours. Immunoprecipitation was performed using a monoclonal anti-FLAG antibody that recognizes FLAG-tagged HDAC3. A non-immune mouse IgG antibody was used as negative control. The ability of PXR to associate with HDAC3 was determined using western blot analysis. The PXR protein interacts with HDAC3 in Hepa1-6 cell extracts (Lane 3), and over-expression of SMRT further enhanced this interaction (Lane 4). Interestingly, forced over-expression of the SUMO signaling machinery abolished interaction between PXR and the HDAC3-SMRT de-acetylation co-repressor complex (Lanes 5-7). In contrast, treatment of cells with TSA restored the interaction of PXR with the HDAC3-SMRT corepressor complex (Lane 8). As expected, the interaction between PXR and the HDAC3-SMRT was reduced by addition of Rif (Lane 9). These data support the concept that acetylated PXR interacts with HDAC3-SMRT while SUMOylated PXR has a greatly diminished capacity to bind to this co-repressor complex.
Figure 5. Acetylated PXR Interacts with HDAC3-SMRT Corepressor Complex.
Hepa1-6 cells were transfected with the indicated expression vectors. Twenty-four hr post-transfection, cells were treated with rifampicin (10 μM), trichostatin A, or both for an additional 24 hr. Cell extracts were subjected to immunoprecipitation with an antibody that recognizes FLAG-HDAC3 (α-FLAG). A non-immune antibody was also used as a negative control (IgG). Western blot analysis was conducted using an anti-PXR polyclonal antibody to detect interaction between HDAC3-SMRT corepressor protein complex and PXR. The asterisk (*) indicates a non-specific background band.
3.6 Covalent Attachment of SUMO Represses Ligand-dependent Transactivation Capacity
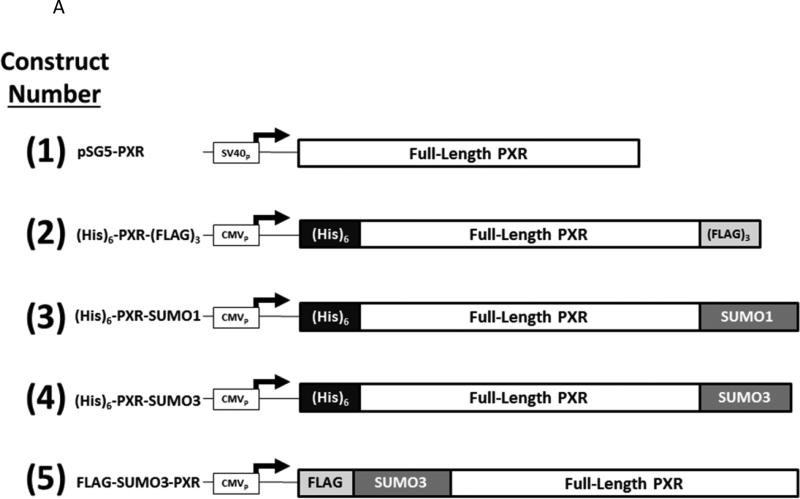
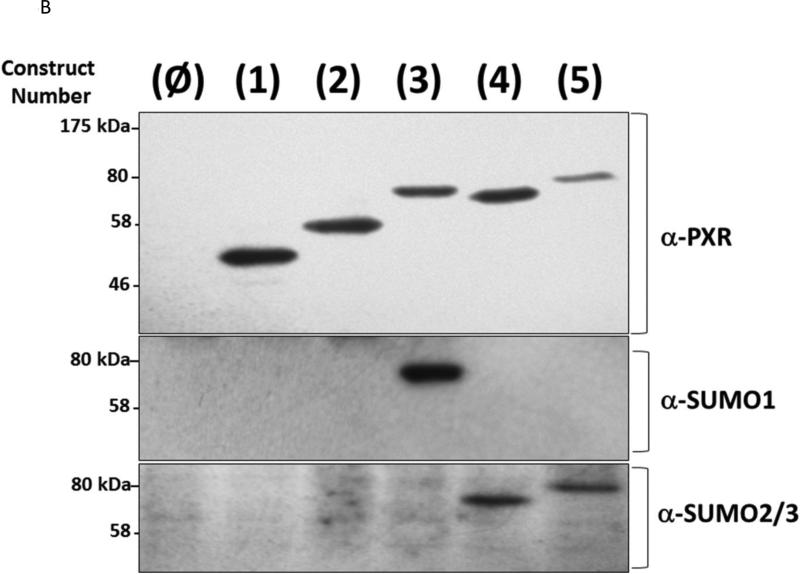
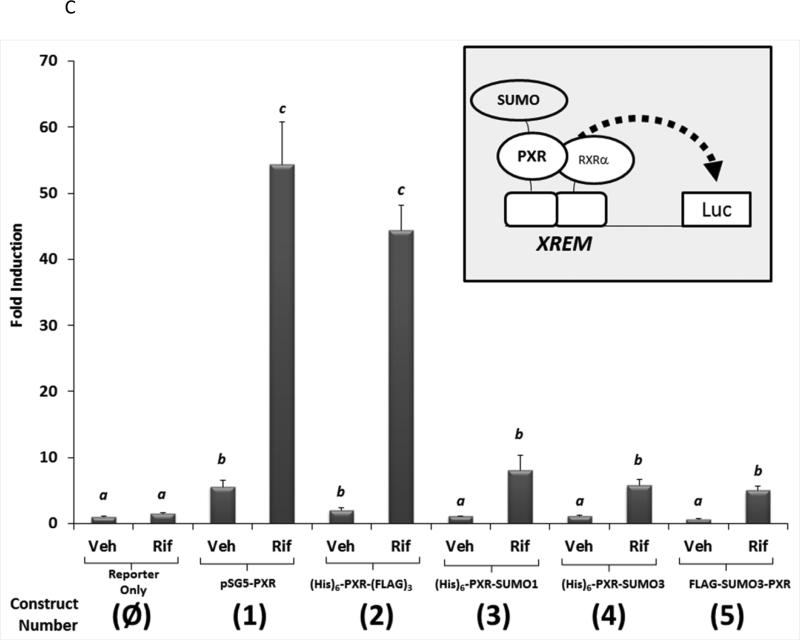
It is widely held that SUMOylation of transcriptional factors leads to transcriptional repression via alteration of sub-nuclear localization, or perhaps by increasing the interactions with transcription co-repressor proteins [32]. However, a recent publication indicates that PXR activity is increased following SUMO-modification [15]. We have previously found that co-expression of the SUMO E3-ligase enzyme PIAS1 modestly increases drug-inducible Cyp3a11 gene expression in hepatocytes [11]. However, in the same study we also found that PXR is required for PIAS1-dependent rifampicin-mediated suppression of pro-inflammatory cytokine gene expression. One strategy utilized to determine whether SUMOylation suppresses transactivation capacity has been to utilize a gene fusion approach [33]. Thus, to more precisely determine how SUMO-modification of PXR affects its ability to activate gene expression we generated a series of expression vectors that encode a variety of linear PXRSUMO fusion proteins as depicted in Figure 6A. The relative protein expression level of the five forms of PXR was roughly equal as verified using antibodies that recognize PXR, SUMO1, and SUMO3 in western blot analysis (Figure 6B). We next examined the effect of covalent attachment of SUMO on regulating drug-inducible PXR-dependent transactivation capacity using a reporter gene derived from the CYP3A4 promoter to drive luciferase activity (XREM-Luc) in cell based assays [34]. Twenty-four hours post-transfection, CV-1 cells were treated with Rif for an additional 24 hours. Treatment of cells expressing wild-type PXR (construct number 1) with Rif increased reporter gene activity by approximately 50-fold. To determine if covalently attaching additional protein sequences to the N-terminus or C-terminus of PXR had non-specific inhibitory effects upon its transactivation capacity, we fused the six-histidine tag [(His6)] to the N-terminus and the triple FLAG-tag [(FLAG3)] to the C-terminus of PXR, respectively, as shown in construct number 2. Cells expressing this form of PXR did not significantly alter its Rifinducible transactivation capacity. In contrast, cells expressing either the PXR-SUMO1 (construct number 3), the PXR-SUMO3 (construct number 4), or the SUMO3-PXR (construct number 5) exhibited significantly diminished Rif-inducible reporter gene activity (Figure 6C). These data indicate that covalent attachment of SUMO to PXR, even in the absence of isopeptide linkage and irrespective of the location of SUMO at the N- or C-terminus, produces a strong repression of its drug-inducible transactivation capacity.
Figure 6. Linear SUMO-fusion Proteins are Deficient in Transactivation Capacity.
(A) A series of expression vectors were constructed (Left Panel) encoding various forms of PXR, with some fused to SUMO proteins, as described in Materials and Methods. (B) Western blot analysis was performed using the indicated antibodies that recognize PXR, SUMO1, and SUMO3 using cell extracts from CV-1 cells that were transfected with the plasmid-based expression vectors as indicated. (C) CV-1 cells were transfected with the XREM-Luc reporter gene together with the indicated construct. The reporter gene alone was used as a negative control (Reporter Only). Twenty-four hr post-transfection, cells were treated with either vehicle (0.1% DMSO) or rifampicin (Rif, 10 μM) for an additional 24 hours. Luciferase activity was normalized to β-galactosidase controls and data are presented as fold induction + SEM. Letters different from each other indicate statistically significant differences between relevant treatment groups (p<0.05).
3.7 SUMOylation of PXR is Dominant over HDAC3-mediated Inhibition of PXR Activity
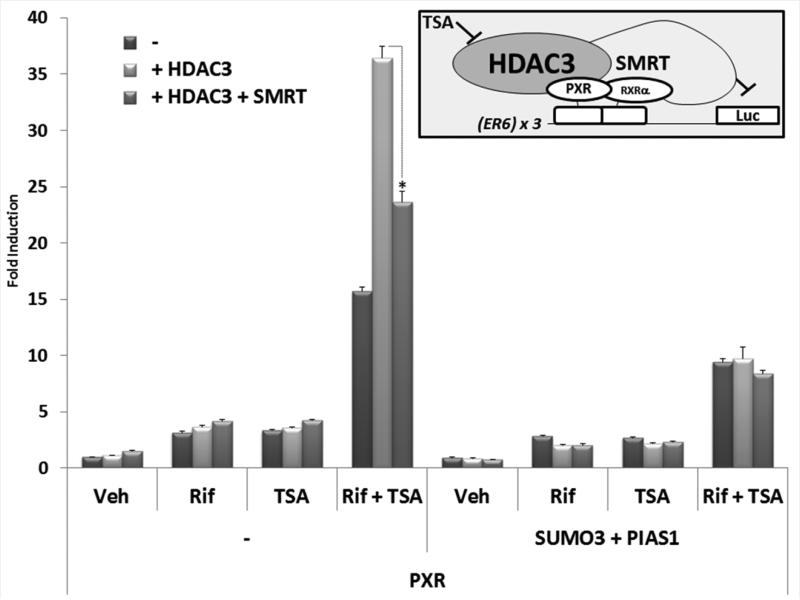
Under normal conditions, the co-repressor protein SMRT is required for the lysine-deacetylase activity of HDAC3 in vivo [28, 29]. The SMRT co-repressor also serves as a molecular scaffold for additional regulatory proteins to modulate metabolic and inflammatory processes in liver [35]. Using a multimerized PXR-response element driving the luciferase reporter gene [(ER6) × 3-Luc] we sought to determine the extent to which HDAC3-SMRT co-repressor complex contributes to the repression of PXR transactivation capacity following SUMO-modification. We chose the multimerized PXR-response element reporter gene because it lacks the additional complex and over-lapping enhancer elements associated with the CYP3A4-derived XREM-Luc reporter gene [34], and it likely represents a more direct readout of PXR transactivation capacity in cell-based assays versus the XREM-Luc reporter gene. Treatment of cells with Rif that were transfected with either PXR alone, PXR together with HDAC3, or PXR together with HDAC3 and SMRT produced approximately 3-4-fold increase in reporter gene activity (Figure 7). Treatment with TSA also produced very modest levels of reporter gene activity in these three experimental groups (3-4-fold induction). In contrast, co-treatment of PXR-transfected cells with Rif and TSA together produced an approximately 16-fold increase in reporter gene activity. The addition of HDAC3 dramatically increased reporter gene activity to approximately 36-fold, while the addition of SMRT significantly reduced the reporter gene activity to approximately 24-fold. Similar experiments performed in the absence of PXR did not produce synergistic reporter gene activity in the absence of the receptor (data not shown). These data confirm that HDAC3 and SMRT have a profound impact upon ligand-dependent PXR transactivation capacity such that pharmacological inhibition of HDAC3-SMRT co-repressor complex activity with TSA produces a synergistic activation of this PXR-dependent reporter gene. When the same experimental groups were used in the presence of expression vectors encoding SUMO3 and PIAS1, the overall ligand-dependent synergistic transactivation capacity produced by co-treatment with Rif and TSA was strongly diminished, independent of the presence of HDAC3 and SMRT. These data indicate that SUMOylation likely represses PXR transactivation capacity in a manner that is distinct from that mediated by the HDAC3-SMRT co-repressor complex.
Figure 7. SUMOylation of PXR Represses PXR Transactivation.
CV-1 cells were transfected with the (ER6)×3-Luc reporter gene together with the indicated expression vectors. Twenty-four hr post-transfection, cells were treated with either vehicle (Veh, 0.1% DMSO), rifampicin (Rif, 10 μM), trichostatin A (TSA, 0.5 μM), or both together for an additional twenty-four hours. Luciferase activity was normalized to β-galactosidase controls and data are presented as fold induction above vehicle control + SEM. Asterisks indicate statistically significant differences between relevant treatment groups (p<0.05).
3.8 Ubiquitination of PXR on Lysine 109 in Hepatocytes is TSA-Dependent
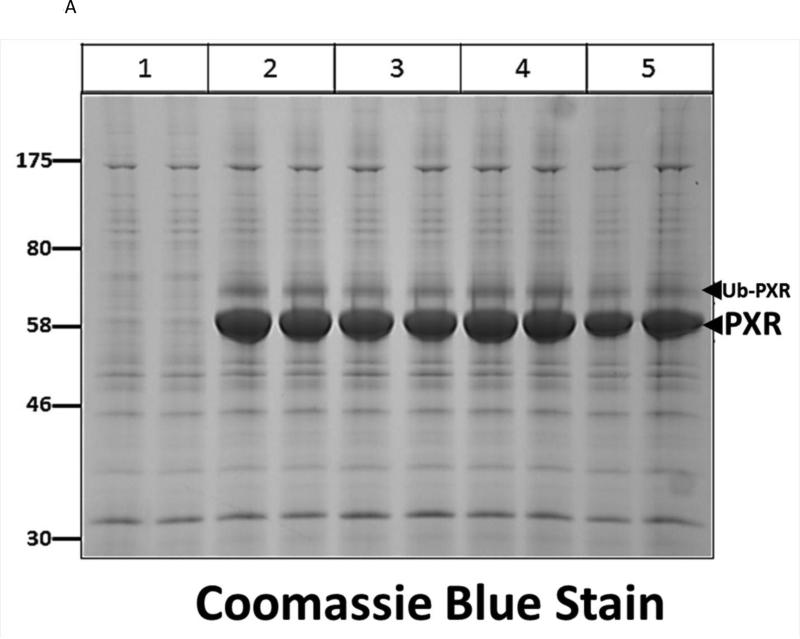
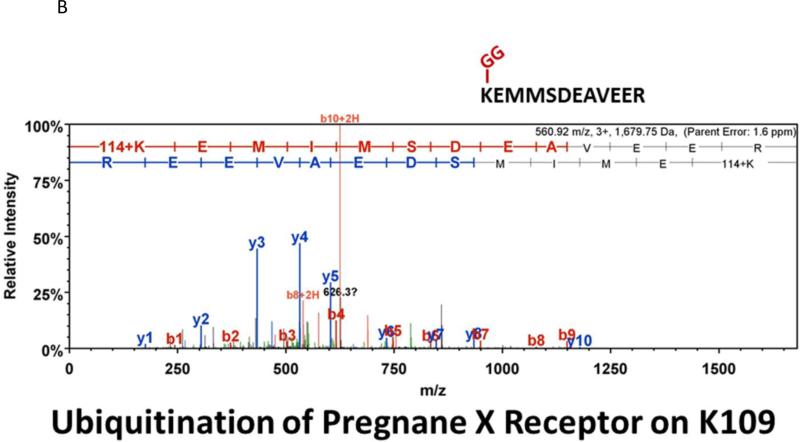
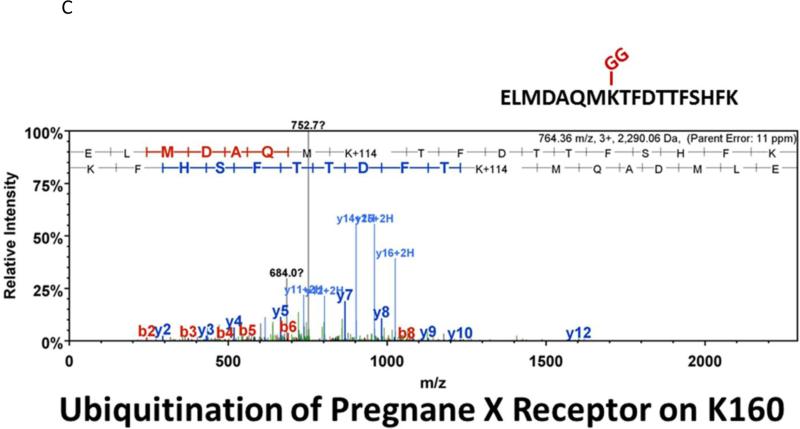
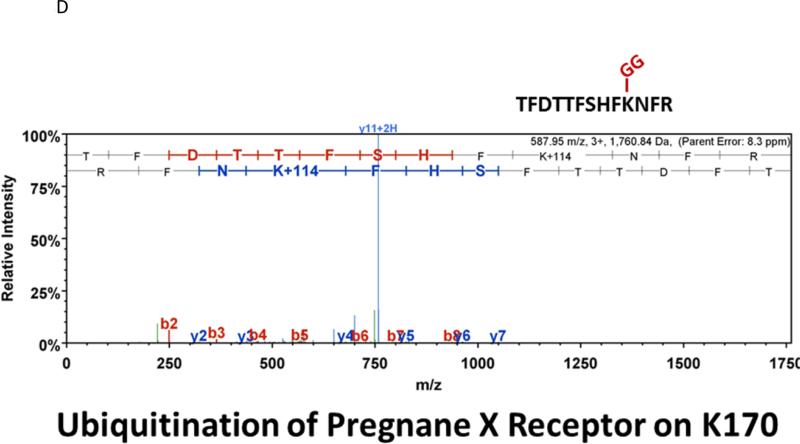
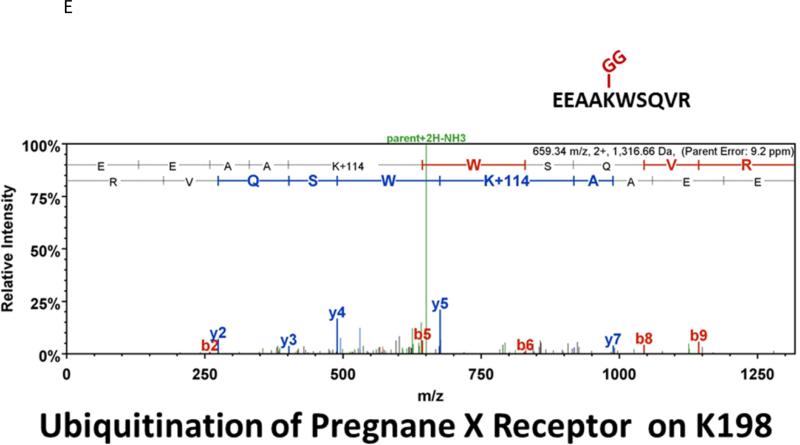
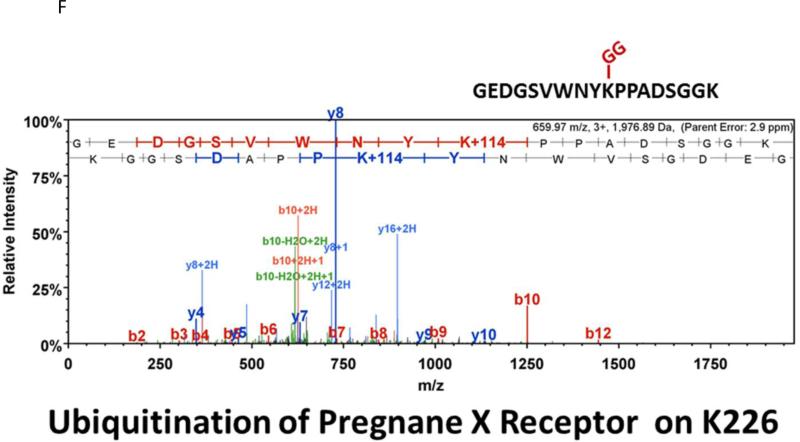
Many laboratories have identified potential PTMs in PXR through an in vitro approach using either LC-MS/MS, or by using a biochemical approach with purified components [12, 13, 17]. These experimental approaches are encumbered by the relative absence of meaningful biology when performed purely in vitro. In an effort to identify a specific site of acetylation in vivo, we took advantage of our adenoviral system to express and purify the (His)6-tagged form of PXR from primary cultures of rat hepatocytes following treatment with either vehicle (0.1% DMSO) or TSA (0.5 μM). Using this experimental approach we routinely achieve high expression and robust purification of the Ad-PXR protein from cultured hepatocytes as demonstrated using coomassie blue staining (Figure 8A). Our overall coverage of the Ad-PXR protein following digestion with trypsin using LC-MS/MS methods was approximately 60%, to include fifteen out of twenty-eight total lysine residues contained within the human PXR protein. We failed to detect acetylation on observable lysine residues following trypsin digestion. However, we found that vehicle treated PXR is heavily multi-mono ubiquitinated on K109, K160, K170, K198, and K226 (Figure 8B-F). Following treatment with TSA, both K160 and K170 were still heavily ubiquitinated. In contrast, ubiquitin modification at K198 and K226 was absent. Moreover, the PTM-status at the K109 position was ambiguous due to a lack of coverage in the spectra of this particular lysine residue within the TSA-treated experimental group. The reason for a lack of coverage at K109 specifically in the TSA-treated group is unknown at present.
Figure 8. Identification of Ubiquitin-Modified Peptides of PXR Isolated from Hepatocytes using LC-MS/MS.
(A) Primary hepatocytes were isolated from a male 14 week old rat and were cultured overnight in ten separate 15 cM dishes. The adenoviral expression vector encoding the six-histidine-tagged form of PXR [Ad-(His)6-PXR] was added to experimental groups 2 through 5 on the morning of day 2. On day 3, cultures were treated with vehicle (Groups 2 and 3) or 0.5 μM TSA (Groups 4 and 5) for an additional 24 hours. Following IMAC-enrichment under denaturing conditions (1 lane per 15 cM dish), the bands corresponding to PXR and Ub-PXR were excised and in-gel trypsin digestion was performed. (B-F) ESI-CID-MS/MS analysis of in-gel digested PXR resulted in a number of spectra that were assigned to tryptic peptides carrying covalently bound ubiquitin residues of Gly-Gly (ubiquitin di-glycine remnant post-trypsin digestion). The tryptic peptides of ubiquitin-modified PXR were identified with a mass addition of 114 at the lysine residues K109, K160, K170, K198, and K226 based on the assignment of multiple product ions (y and b ions) as indicated in the MS/MS scan.
4. Discussion
The notion that lysine-directed PTMs including SUMOylation, ubiquitylation, and acetylation interact with each other within the context of a given transcriptional regulatory complex is gaining wide acceptance [8]. The class I lysine de-acetylase HDAC3 regulates metabolism through multiple signalling pathways in the liver, and liver-specific deletion of HDAC3 disrupts normal hepatic metabolic homeostasis [36]. A recent article focused on farnesoid x receptor acetylation postulates that targeting acetylation of this receptor and its transcriptional cofactors may provide a new molecular strategy for development of pharmacological agents to treat metabolic disorders [37]. An additional study from the same group indicates that a dysregulated SUMO-acetyl switch on farnesoid x receptor that occurs during morbid obesity may serve as a general mechanism for diminished anti-inflammatory response observed in these patients [20]. Moreover, this group has shown that acetylation of farnesoid x receptor is normally regulated by the acetyl transferase E1A binding protein p300 and the class III NAD-dependent deacetylase Sirtuin-1 [38]. Similarly, the de-acetylation of PXR has been previously suggested to be mediated by both Sirtuin-1 and a TSA-dependent lysine de-acetylase [26].
Previous studies conducted in our laboratory have revealed that SUMOylation of PXR alters its ubiquitination, and likely regulates two distinct facets of PXR biology. The SUMOylation of PXR increases its stability, apparently through protection of this nuclear receptor from the proteasome-mediated degradation pathway [11]. Another likely function of SUMO-modified PXR is the active suppression of the expression of pro-inflammatory cytokines in hepatocytes as well as participation in the resolution of inflammation [27, 39]. While PXR SUMOylation is critical for maintaining cell integrity in response to inflammatory stress, the role of PXR acetylation in this process is not clear. Two studies have independently detected acetylation of PXR by using either biochemical over-expression methods in cultured cell lines [26], or by using a western blotting approach in whole-cell protein extracts from rodent liver [20]. How acetylation of PXR potentially affects its SUMOylation or ubiquitination was not addressed in these studies. However, it was suggested that in addition to Sirtuin 1, other as yet to be identified lysine de-acetylase enzymes likely play an important role in enhancing or assisting with de-acetylation upon PXR activation [26], and our current study suggests that HDAC3 is intimately involved in PXR de-acetylation.
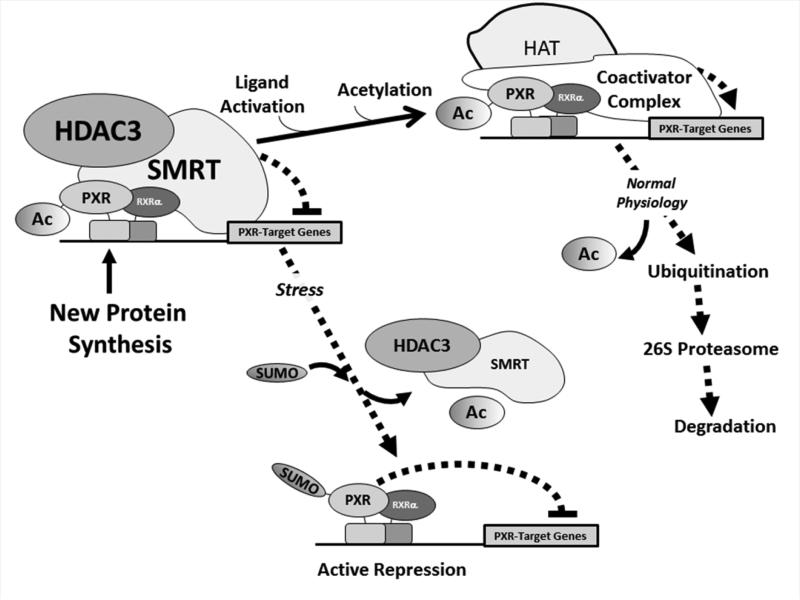
Taken together, the data obtained in our current study put forward a working model of the role of acetylation, SUMOylation, and ubiquitination in regulating PXR biology (Figure 9). We favor the notion that acetylation and SUMOylation are mutually exclusive events, but that acetylation is likely a prerequisite for its subsequent SUMO-modification. Moreover, it appears that ubiquitin migrates to various lysine residues within PXR, likely depending upon the repertoire of other PTMs that co-exist on the protein and its state of ligand activation. The ligand-dependent ubiquitination of PXR likely promotes its degradation through the proteasome mediated pathway during the canonical response, in the absence of concurrent pathological conditions such as inflammatory-related disease states. In the presence of a pathological stimuli, such as inflammation, the acetyl group is removed and a PXR-mediated gene activation program is supplanted by active repression of select PXR-target genes by SUMO-modified and stabilized PXR. Our hypothesis agrees with the previous observation that poly-ubiquitination of transcription factors on a single lysine residue authorizes their transactivation capacity by linking gene activation to their subsequent destruction [40]. Moreover, mono- or multi-monoubiquitination confers distinct biological outcomes when compared with poly-ubiquitination through the formation of ubiquitin chains [41]. Our data show that PXR is heavily multi-monoubiquitinated in a silent state, and that the formation of poly-ubiquitinated PXR at K160 and K170 is stimulated by a more heavily acetylated receptor. We have previously shown that SUMOylation of PXR focuses the formation of K48-linked ubiquitin chains at K170 to support degradation of ligand-activated PXR [11]; and that this event is likely required in order for additional rounds of transcription to proceed [42]. It therefore appears that acetylation of PXR also affects its ubiquitin modification.
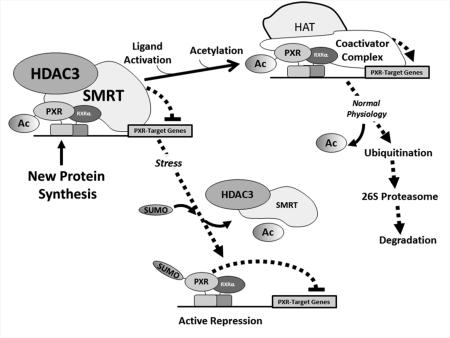
Figure 9. Working model of the Role of the Acetyl-SUMO Switch in PXR Biology.
Newly synthesized PXR protein is acetylated and poised on canonical PXR-target genes in a complex with the HDAC3-SMRT co-repressor multi-protein complex and is transcriptionally silent. Ligand activation promotes hyper-acetylation of the genomic locus, likely through the action of canonical histone/lysine acetyltransferase enzymes belonging to the E1A binding protein p300/CREB-binding protein coactivator family. Following one round of transcription the PXR-associated multi-protein complex is degraded by the 26S proteasome in an ubiquitin-dependent manner, and the promoter is thus cleared and poised to receive another round of transcriptional machinery. In the presence of specific signals, such an inflammatory stress or potentially other extra-cellular stimuli, PXR is de-acetylated and the HDAC3-SMRT co-repressor multi-protein complex is disassociated. The resulting signal-dependent action of a SUMO E3 ligase enzyme, such as PIAS1, promotes PXR-SUMOylation to inhibit PXR-target gene expression in an acetylation-dependent manner.
It is worth noting that while acetyl-lysine modification of PXR was readily detected in this study using an immunological approach, we were not successful in identifying any acetylated lysine residues in PXR using mass spectrometry analysis. This is likely due to the relatively low stoichiometry of the acetyl-modification [43], or perhaps due to the large size of the SUMO- and acetyl-modified target peptide(s), or the inherent limitations of the LC-MS/MS approach. Further refinement of our experimental conditions through the use of proteases other than trypsin, or by the addition of a resolving column pre-LCMS/MS may improve our detection capabilities.
4.1 Conclusions
In conclusion, the acetylation, SUMOylation, and ubiquitination of PXR interface with each other to alter PXR biolgical activity. We show here that PXR is the molecular target of the acetylation signal transduction pathway and that the HDAC3-SMRT multi-protein corepressor complex is a key component of the PXR de-acetylation pathway. The promotion of PXR acetylation enhances its SUMO-modification, while deacetylation of PXR by HDAC3-SMRT tends to inhibit its SUMOylation in cell-based assays. Ligand-mediated activation with rifampicin fuels de-acetylation of PXR, while inhibition of HDAC3 activity using pharmacological methods mildly suppresses the PXR-mediated gene activation program in hepatocytes. Acetylated PXR interacts with HDAC3 and forced over-expression of SMRT further increases PXR-HDAC3 interactions. The SUMOylation of PXR likely forms the molecular basis of PXR-mediated active gene repression during pathological disease states. However, further research in this area should determine the extent to which these and other signaling pathways contribute to this effect. What is abundantly clear is that the regulation of PXR by various PTMs is highly interactive and in a state of constant fluidity. Giving the fact that PXR is a pivatol regultor of drug metabolism and disposition, and is heavily involoved in the pathogenesis of important human diseases, unrevling the molecular and biochemical details of how PTMs determine PXR biology should remian an important thust of future research efforts. While the studies presented here utilize cell-based and biochemical methods, as well as primary cultures of rodent hepatocytes, future experiments should include the use of transgenic mouse models. Specifically, mouse models that express mutant forms of the PXR protein refractory to modification with SUMO and acetyl groups should greatly aid in furthering the knowledge base regarding the effect these signaling pathways have on PXR biology in vivo.
HIGHLIGHTS.
Acetylation of PXR promotes its SUMOylation
Acetylation and SUMOylation negatively affects PXR transactivation capacity
HDAC3 and SMRT cooperate to affect de-acetylation and SUMOylation of PXR
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health National Institute of Digestive, Diabetic, and Kidney Diseases (NIDDK) [Grant R01DK090558] (to J.S.), University of Kansas, Strategic Initiative Grant [INS0073115], and by an award from the Centers of Biomedical Research Excellence (COBRE) and the National Institutes of Health National Institute of General Medical Sciences [Grant P20 GM103549]. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
ABBREVIATIONS
- PXR
pregnane x receptor
- TSA
trichostatin A
- HDAC3
histone deacetylase 3
- SMRT
silencing mediator of retinoic acid and thyroid hormone receptor
- PTM
post-translational modification
- LC-MS/MS
Liquid Chromatography-Tandem Mass Spectrometry
- PIAS1
protein inhibitor of activated STAT-1
- SUMO
small ubiquitin-related modifier
- Rif
rifampicin
- PCN
pregnenolone 16μ-carbonitrile
- RFP
red fluorescent protein
- GFP
green fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Participated in research design: Staudinger, Cui, Galeva
Conducted experiments: Staudinger, Cui, Shupei, Galeva, Sun, and Shen.
Contributed new reagents or analytic tools: Staudinger, Sun, Cui, and Galeva.
Performed data analysis: Staudinger, Cui, Williams, Galeva.
Wrote or contributed to the writing of the manuscript: Staudinger.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, Perlmann T, Lehmann JM. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. The Journal of clinical investigation. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gotoh S, Negishi M. Statin-activated nuclear receptor PXR promotes SGK2 dephosphorylation by scaffolding PP2C to induce hepatic gluconeogenesis. Scientific reports. 2015;5:14076. doi: 10.1038/srep14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao J, Xie W. Pregnane X receptor and constitutive androstane receptor at the crossroads of drug metabolism and energy metabolism. Drug metabolism and disposition: the biological fate of chemicals. 2010;38:2091–2095. doi: 10.1124/dmd.110.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbins D, Chen T. Tissue-specific regulation of pregnane X receptor in cancer development and therapy. Cell & bioscience. 2014;4:17. doi: 10.1186/2045-3701-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng J, Shah YM, Gonzalez FJ. Pregnane X receptor as a target for treatment of inflammatory bowel disorders. Trends in pharmacological sciences. 2012;33:323–330. doi: 10.1016/j.tips.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, Fleet JC, Kortagere S, Mukherjee P, Fasano A, Le Ven J, Nicholson JK, Dumas ME, Khanna KM, Mani S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Molecular cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smutny T, Mani S, Pavek P. Post-translational and post-transcriptional modifications of pregnane X receptor (PXR) in regulation of the cytochrome P450 superfamily. Current drug metabolism. 2013;14:1059–1069. doi: 10.2174/1389200214666131211153307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staudinger JL, Lichti K. Cell signaling and nuclear receptors: new opportunities for molecular pharmaceuticals in liver disease. Molecular pharmaceutics. 2008;5:17–34. doi: 10.1021/mp700098c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui W, Sun M, Galeva N, Williams TD, Azuma Y, Staudinger JL. SUMOylation and Ubiquitylation Circuitry Controls Pregnane X Receptor Biology in Hepatocytes. Drug metabolism and disposition: the biological fate of chemicals. 2015;43:1316–1325. doi: 10.1124/dmd.115.065201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elias A, High AA, Mishra A, Ong SS, Wu J, Peng J, Chen T. Identification and characterization of phosphorylation sites within the pregnane X receptor protein. Biochemical pharmacology. 2014;87:360–370. doi: 10.1016/j.bcp.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lichti-Kaiser K, Brobst D, Xu C, Staudinger JL. A systematic analysis of predicted phosphorylation sites within the human pregnane X receptor protein. The Journal of pharmacology and experimental therapeutics. 2009;331:65–76. doi: 10.1124/jpet.109.157180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lichti-Kaiser K, Xu C, Staudinger JL. Cyclic AMP-dependent protein kinase signaling modulates pregnane x receptor activity in a species-specific manner. The Journal of biological chemistry. 2009;284:6639–6649. doi: 10.1074/jbc.M807426200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Priyanka D, Kotiya M. Rana, Subbarao N, Puri N, Tyagi RK. Transcription regulation of nuclear receptor PXR: Role of SUMO-1 modification and NDSM in receptor function. Molecular and cellular endocrinology. 2015 doi: 10.1016/j.mce.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Rana R, Coulter S, Kinyamu H, Goldstein JA. RBCK1, an E3 ubiquitin ligase, interacts with and ubiquinates the human pregnane X receptor. Drug metabolism and disposition: the biological fate of chemicals. 2013;41:398–405. doi: 10.1124/dmd.112.048728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugatani J, Hattori Y, Noguchi Y, Yamaguchi M, Yamazaki Y, Ikari A. Threonine-290 regulates nuclear translocation of the human pregnane X receptor through its phosphorylation/dephosphorylation by Ca2+/calmodulin-dependent protein kinase II and protein phosphatase 1. Drug metabolism and disposition: the biological fate of chemicals. 2014;42:1708–1718. doi: 10.1124/dmd.114.059139. [DOI] [PubMed] [Google Scholar]
- 18.Sugatani J, Noguchi Y, Hattori Y, Yamaguchi M, Yamazaki Y, Ikari A. Threonine-408 Regulates the Stability of the Human Pregnane X Receptor Through its Phosphorylation and the CHIP/Chaperone-Autophagy Pathway. Drug metabolism and disposition: the biological fate of chemicals. 2015 doi: 10.1124/dmd.115.066308. [DOI] [PubMed] [Google Scholar]
- 19.Blakeslee WW, Wysoczynski CL, Fritz KS, Nyborg JK, Churchill ME, McKinsey TA. Class I HDAC inhibition stimulates cardiac protein SUMOylation through a post-translational mechanism. Cellular signalling. 2014;26:2912–2920. doi: 10.1016/j.cellsig.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim DH, Xiao Z, Kwon S, Sun XX, Ryerson D, Tkac D, Ma P, Wu SY, Chiang CM, Zhou E, Xu HE, Palvimo JJ, Chen LF, Kemper B, Kemper JK. A dysregulated acetyl/SUMO switch of FXR promotes hepatic inflammation in obesity. Embo J. 2015;34:184–199. doi: 10.15252/embj.201489527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. The Journal of clinical investigation. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson DR, Li CW, Chen LY, Ghosh JC, Chen JD. Regulation and binding of pregnane X receptor by nuclear receptor corepressor silencing mediator of retinoid and thyroid hormone receptors (SMRT) Molecular pharmacology. 2006;69:99–108. doi: 10.1124/mol.105.013375. [DOI] [PubMed] [Google Scholar]
- 24.Shaw PG, Chaerkady R, Zhang Z, Davidson NE, Pandey A. Monoclonal antibody cocktail as an enrichment tool for acetylome analysis. Analytical chemistry. 2011;83:3623–3626. doi: 10.1021/ac1026176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staudinger JL, Xu C, Biswas A, Mani S. Post-translational modification of pregnane x receptor. Pharmacological research. 2011;64:4–10. doi: 10.1016/j.phrs.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas A, Pasquel D, Tyagi RK, Mani S. Acetylation of pregnane X receptor protein determines selective function independent of ligand activation. Biochemical and biophysical research communications. 2011;406:371–376. doi: 10.1016/j.bbrc.2011.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu G, Xu C, Staudinger JL. Pregnane X receptor is SUMOylated to repress the inflammatory response. The Journal of pharmacology and experimental therapeutics. 2010;335:342–350. doi: 10.1124/jpet.110.171744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Molecular and cellular biology. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You SH, Lim HW, Sun Z, Broache M, Won KJ, Lazar MA. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nature structural & molecular biology. 2013;20:182–187. doi: 10.1038/nsmb.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawana K, Ikuta T, Kobayashi Y, Gotoh O, Takeda K, Kawajiri K. Molecular mechanism of nuclear translocation of an orphan nuclear receptor, SXR. Molecular pharmacology. 2003;63:524–531. doi: 10.1124/mol.63.3.524. [DOI] [PubMed] [Google Scholar]
- 31.Timsit YE, Negishi M. CAR and PXR: the xenobiotic-sensing receptors. Steroids. 2007;72:231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Molecular cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 33.Ross S, Best JL, Zon LI, Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Molecular cell. 2002;10:831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- 34.Goodwin B, Hodgson E, Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Molecular pharmacology. 1999;56:1329–1339. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- 35.Mottis A, Mouchiroud L, Auwerx J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes & development. 2013;27:819–835. doi: 10.1101/gad.214023.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. Embo J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kemper JK. Regulation of FXR transcriptional activity in health and disease: Emerging roles of FXR cofactors and post-translational modifications. Bba-Mol Basis Dis. 2011;1812:842–850. doi: 10.1016/j.bbadis.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD. FXR Acetylation Is Normally Dynamically Regulated by p300 and SIRT1 but Constitutively Elevated in Metabolic Disease States. Cell Metab. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun M, Cui W, Woody SK, Staudinger JL. Pregnane X receptor modulates the inflammatory response in primary cultures of hepatocytes. Drug metabolism and disposition: the biological fate of chemicals. 2015;43:335–343. doi: 10.1124/dmd.114.062307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- 41.Sadowski M, Suryadinata R, Tan AR, Roesley SN, Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB life. 2012;64:136–142. doi: 10.1002/iub.589. [DOI] [PubMed] [Google Scholar]
- 42.Dennis AP, O'Malley BW. Rush hour at the promoter: how the ubiquitin proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. The Journal of steroid biochemistry and molecular biology. 2005;93:139–151. doi: 10.1016/j.jsbmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 43.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nature reviews. Molecular cell biology. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]