Abstract
Pregnane X receptor (PXR) is a major transcriptional regulator of xenobiotic metabolism and transport pathways in the liver and intestines, which are critical for protecting organisms against potentially harmful xenobiotic and endobiotic compounds. Inadvertent activation of drug metabolism pathways through PXR is known to contribute to drug resistance, adverse drug–drug interactions, and drug toxicity in humans. In both humans and rodents, PXR has been implicated in non-alcoholic fatty liver disease, diabetes, obesity, inflammatory bowel disease, and cancer. Because of PXR's important functions, it has been a therapeutic target of interest for a long time. More recent mechanistic studies have shown that PXR is modulated by multiple PTMs. Herein we provide the first investigation of the role of acetylation in modulating PXR activity. Through LC–MS/MS analysis, we identified lysine 109 (K109) in the hinge as PXR's major acetylation site. Using various biochemical and cell-based assays, we show that PXR's acetylation status and transcriptional activity are modulated by E1A binding protein (p300) and sirtuin 1 (SIRT1). Based on analysis of acetylation site mutants, we found that acetylation at K109 represses PXR transcriptional activity. The mechanism involves loss of RXRα dimerization and reduced binding to cognate DNA response elements. This mechanism may represent a promising therapeutic target using modulators of PXR acetylation levels.
Keywords: Pregnane X receptor (PXR), Acetylation, Drug metabolism, Post-translational modification (PTM), Transcription regulation, Nuclear receptor, E1A binding protein p300 (p300)
1. Introduction
Pregnane X receptor (PXR, NR1I2) is a member of the nuclear receptor (NR) superfamily of ligand-activated transcription factors. It is a major transcriptional regulator of xenobiotic metabolism and transport pathways in the liver and intestines, which are critical for protecting organisms against potentially harmful xenobiotic and endobiotic compounds [1–3]. PXR directly binds xenobiotic compounds via its ligand binding domain (LBD) and coordinately regulates the transcription of drug metabolism enzymes (Phases I & II) and transporters (Phase III), which generally results in detoxification and elimination of drugs [4, 5]. PXR's uniquely large, flexible and promiscuous ligand binding pocket, accommodates binding a vast array of chemically and structurally diverse compounds (e.g., medicinal drugs, environmental pollutants, bile acids). In humans, inadvertent activation of drug metabolism pathways through PXR is known to contribute to drug resistance, adverse drug–drug interactions, and drug toxicity in humans [3, 6–11]. More recent data implicates PXR in hepatic steatosis, diabetes, obesity, inflammation and cancer [4, 12–18]. Surprising roles for PXR have emerged in metabolism and steatosis, underscoring contextual signaling of the receptor [14, 15, 19, 20].
More recently, our lab has shown that PXR regulates intestinal barrier function in vivo through sensing luminal endobiotics (e.g. bacterial metabolites) [21]. Understanding the diverse functions and molecular mechanisms governing PXR action may provide avenues towards pharmacological intervention of relevant diseases [22, 23]. Upon ligand binding, PXR dissociates from sequestering proteins—heat shock protein 90 (HSP90) and cytoplasmic constitutive androstane receptor retention protein (CCRP)—thus allowing translocation to the nucleus [24]. In the nucleus, ligand-bound PXR forms an obligate heterodimer with the retinoid X receptor-α (RXRα), which results in binding cognate xenobiotic response elements (e.g. DR3, ER6) located within the promoters of PXR target genes [25, 26]. In addition to ligand activation, PXR's transcriptional output is modulated by transcriptional coregulator proteins, which include steroid receptor coactivator-1 (SRC-1), peroxisome proliferator-activated receptor-γ coactivator-1 α (PGC-1α), hepatocyte nuclear factor 4α (HNF4α), and E1A binding protein (p300), and corepressors nuclear receptor corepressor (NCoR), silencing mediator for retinoid and thyroid hormone receptor (SMRT), and receptor-interacting protein 140 (RIP140) [5, 27, 28]. PXR target genes include a battery of phase I enzymes (e.g. several cytochrome P450s (CYPs)), phase II enzymes (e.g. glutathione-S-transferase, sulfotransferases), and phase III transporters (e.g. multidrug resistance protein 1 (MDR1)). However, post-transcriptional (e.g., miRNA) [29] and post-translational changes (post- translational modifications or PTMs— phosphorylation, ubiquitination, SUMOylation, methylation, and acetylation) of PXR have recently been characterized [30–34]. PTMs act as signals that collectively fine-tune PXR protein activity producing a context-dependent adaptive cellular response. These PTMs expand PXR function and thus may underlie PXR's pleiotropic roles in normal physiology and pathophysiology [30, 31, 35–37].
Of the myriad of PTMs affecting NRs, dynamic acetylation/deacetylation has been shown to regulate many adopted orphan NRs, (e.g., the farnesoid X receptor or FXR) [38–40]. Overall, acetylation may positively or negatively regulate various NR functions in a site-and context- specific manner, which must be empirically determined. For example, DNA binding and transcriptional activity of the progesterone receptor (PR) is enhanced by p300-mediated acetylation, whereas the opposite effect is true in the case of the FXR [40, 41].
In a previous study, we demonstrated that PXR is acetylated in cells [42]. Treatment with human PXR ligand rifampicin (RIF) reduced relative acetylation levels. This implied that PXR is acetylated in its basal state and deacetylated upon ligand activation. We also showed that deacetylation is at least partially mediated by sirtuin 1 (SIRT1). SIRT1-mediated deacetylation can activate PXR's lipogenic function in vivo independent of a ligand. In the present study, we show that recombinant human PXR protein (hPXR) is acetylated in vitro by p300, but not PCAF. Using LC-MS/MS analysis, we identified lysine 109 (K109) as PXR's major acetylation site and confirmed the acetylation of this site in HepG2 cells. Using various biochemical and cell-based assays, we show that acetylation of PXR at K109 by p300 down-regulates PXR transcriptional activity. The mechanism involves loss of RXRα dimerization and reduced binding to cognate DNA response elements. Implications of our findings are discussed in the context of known roles for PXR in transcription, health, and disease.
2. Experimental procedures
2.1. Cell culture and plasmids
HEK293T (293T), HepG2, and HeLa cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA) and maintained according to ATCC's recommendations in a humidified atmosphere of 5% CO2 at 37 °C. 293T and HeLa cells were maintained in DMEM (10% FBS), while HepG2 cells were maintained in EMEM (10% FBS), with pen-strep. The pSG5-human PXR mammalian expression plasmid (pSG5-PXR) was provided by Dr. S. Kliewer (University of Texas, Dallas, TX), and pSG5-RXRα encoding human RXRα was a generous gift from Dr. C. Carlberg (University of Kuopio, Finland). The chimeric CYP3A4-Luc reporter construct containing the basal promoter (−362/+53) with proximal PXR response element (ER6) and the distal xenobiotic-responsive enhancer module (−7836/−7208) of the CYP3A4 gene 5′-flanking region inserted to pGL3-Basic reporter vector (Promega, Madison, WI) was described previously [43]. The MDR1-Luc reporter construct containing ~8.3 kb of the natural MDR1 promoter (−8055/+261) was a kind gift from Dr. Oliver Burk and described previously [44]. pCDNA3.1-p300 (Addgene #23252) and pCDNA3.1-p300ΔHAT (Addgene #23254) were gifts from Warner Greene [45]. pECE-FLAG- SIRT1 (Addgene #1791) and pECE-FLAG-SIRT1 H363Y (Addgene #1792) plasmids were gifts from Michael Greenberg [46].
2.2. Bacterial expression and purification of PXR protein
Expression and purification of full-length His-MBP-tagged human PXR protein were performed as described [47] with slight modifications. Escherichia coli BL21 (DE3) AI cells were transformed with MBP-LIC pMCSG vector containing a codon-optimized PXR ORF (kind gift from Redinbo Lab, UNC) [47]. Terrific broth was inoculated with a saturated culture of the transformed cells and allowed to shake at 37°C until an OD600 of ~1.5 was reached. To induce protein expression, L-arabinose was added to the culture at a final concentration of 1% and IPTG to a final concentration of 1 mM and allowed to shake overnight at 15 °C. Cells were centrifuged at 4500 ×g for 20 min, and pellets were lysed with Lysis Buffer A (50 mM HEPES pH 7.5, 50 mM imidazole, 500 mM NaCl, 10% glycerol) supplemented with protease inhibitor tablet (Roche) and lysozyme (~1 mg/mL of lysis buffer). Lysates were sonicated to minimize viscosity due to genomic DNA and clarified by high-speed centrifugation at 14,500 ×g for 50 min. Cleared lysates were incubated with Ni Sepharose High Performance affinity beads (GE Healthcare) overnight at 4 °C with agitation. The beads were washed three times with Buffer A (50 mM HEPES pH 7.5, 50 mM imidazole, 500 mM NaCl, 10% glycerol) and eluted three times with Elution Buffer (50 mM HEPES pH 7.5, 500 mM imidazole, 500 mM NaCl, 10% glycerol). Eluate fractions were pooled and further purified using a pre-equilibrated HiLoad™ 16/60 Superdex™ 200 gel filtration column connected to an AKTA FLPC system. Fractions were eluted in elution buffer (20 mM Tris–HCl pH 7.5, 250 mM NaCl, 1 mM TCEP, and 5% glycerol). Pure fractions containing His-MBP-PXR (as assessed by SDS-PAGE/Coomassie staining) were pooled and the His-MBP tag was cleaved using His-tagged ProTEV protease (Promega). The cleaved His-MBP tag and His-tagged proTEV were removed by subtractive purification using Ni Sepharose beads.
2.3. In vitro acetylation assays
In a 40 µL reaction, 200 ng recombinant p300 acetyltransferase (Active Motif, Carlsbad, CA) was incubated with either bacterially purified recombinant PXR or in vitro translated PXR with 6 µg acetyl-CoA in 1× HAT buffer (50 mM Tris, 1 mM DTT, 0.1 mM EDTA, pH 8.0) and incubated for 1–3 h at 30 °C. Relative acetylation levels were assessed by Western blot using an anti-acetyllysine antibody (Cell Signaling #9441S).
2.4. In cell acetylation assays
Cells were seeded 24 h beforehand and transfected using Lipofectamine LTX (Life Technologies) according to the manufacturer's protocol. 5 µg each of pCDNA3.1-FLAG-PXR and HA-p300 was transfected separately or together in 293T cells. Empty pCDNA3.1 vector was used to make total transfected plasmid amounts equal across cell samples. 48 h after transfection, whole cell lysates were harvested and subjected to FLAG-IP. Acetylated PXR was detected by Western blot using the anti-acetyllysine antibody.
2.5. Cell lysis
Adherent cells were washed with cold PBS and resuspended in ice-cold lysis buffer (30 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% NP-40, supplemented with 1× protease inhibitor cocktail (Roche)). Cells were incubated on ice for 20 min then centrifuged for 10 min at 10,000 ×g at 4 °C to pellet insoluble material. The supernatant was then used for immunoprecipitation or directly subjected to Western blotting.
2.6. FLAG immunoprecipitation (FLAG-IP)
Cell lysates were harvested as described above. Lysate from one 10-cm dish of confluent cells was used per co-IP reaction. 30 µL of pre-washed anti-FLAG M2 Affinity Gel (Sigma, #A2220) was added to each co-IP and incubated with agitation at 4 °C for 2 h. Immune complexes were washed three times with 500 µL ice-cold TBS wash buffer (30 mM Tris–HCl, pH 7.4, 150 mM NaCl) and eluted by boiling at 95 °C in Laemmli buffer for 10 min. Eluates were then analyzed by SDS-PAGE/Western blot.
2.7. Western blotting
Western blotting was performed according to standard methods [42]. Briefly, protein samples to be analyzed were boiled at 95 °C for 5 min with 1×Laemmli buffer then allowed to cool. Samples were then loaded into the wells of an 8% SDS-PAGE gel and run at 100 V for 1–3 h. Proteins were then transferred to a nitrocellulose membrane using a wet transfer system run at 350 mA for 2 h. Membranes were blocked in 5% nonfat dry milk for 1 h and then incubated at 4 °C overnight in primary antibody at 1:500 dilutions in 5% BSA. PXR Antibody (H-11) (sc-48340) and pan-acetyllysine antibody (Cell Signaling #9441S) were used throughout the study. The following day the membranes were washed with TBS-T, incubated in appropriate HRP-conjugated secondary antibody (Santa Cruz Biotechnology) at a 1:5000 dilution in 5% BSA, washed again, and visualized using HRP chemiluminescent substrate.
2.8. Luciferase reporter assays
293T cells were seeded at 10,000 cells per well in 96-well plates and transfected 24 h later using Lipofectamine LTX (Life Technologies) with 25 ng pSG5-PXR or K109 mutants, along with 75 ng luciferase reporter plasmid (p3A4-Luc or MDR1-Luc) and 10 ng pCMV-β-galactosidase (β-gal) as a transfection normalization. 24 h after transfection, cells were treated with either DMSO, RIF, or SR12813, at the concentrations indicated. 24 h later, cells were lysed using 60 µL/well 1× Passive Lysis Buffer, and 1/2 lysate was used to assay luciferase activity (Luciferase Assay System, Promega), while the remaining 1/2 was used to measure β-gal activity. To control for differences in transfection efficiencies, luciferase values were normalized to β-gal values, both measured using a SpectraMax M5 instrument (Molecular Devices). Graphing and statistical analysis were performed using GraphPad Prism 6 software.
2.9. Trypsin digestion
Purified recombinant full-length PXR protein (~50 µg) was denatured in 6 M guanidine hydrochloride in 50 mM ammonium bicarbonate solution for 3 h. DTT reducing agent was then added to a final concentration of ~10 mM and incubated for 1 h at 57 °C. Iodoacetamide alkylating agent was then added to a final concentration of ~30 mM and incubated in the dark for 1 h at room temperature. The sample was diluted in 1.5 mL 25 mM ammonium bicarbonate, 5 µg of trypsin was added to digest the protein, and incubated overnight at 37 °C with shaking. Detergent was removed from the digested sample using Pierce Detergent Removal Spin Columns (Thermo Scientific) following the manufacturer's protocol, subjected to SpeedVac, and then diluted to 40 µL with 2% acetonitrile/0.2% trifluoroacetic acid in water.
2.10. Liquid chromatography coupled to mass spectrometry (LC/MS–MS)
Nanospray LC-MS/MS was performed on a LTQ linear ion trap mass spectrometer interfaced with a Rapid Separation LC3000 system (Thermo, San Jose, CA). Nanospray LC–MS/MS was performed on a LTQ linear ion trap mass spectrometer interfaced with a Rapid Separation LC3000 system (Thermo, San Jose, CA). 35 µL of the sample was loaded on an Acclaim PepMap C18 Nanotrap column (5 µm, 100 Å,/100 µm i.d. ×2 cm) from the autosampler with a 50 µL sample loop with the loading buffer (2% Acetonitrile/water + 0.1% trifluoroacetic acid) at a flow rate of 8 µL/min. After 15 min, the trap column was switched in line with the Acclaim PepMap RSLC C18 column (2 µm, 100 Å, 75 µm i.d. × 25 cm) (Dionex Corp). The peptides were eluted with gradient separation using mobile phase A (2% acetonitrile/water + 0.1% formic acid) and mobile phase B (80% acetonitrile/water + 0.1% formic acid). Solvent B was increased from 2 to 35% over 40 min, increased to 90% over a 5-min period and held at 90% for 10 min at a flow rate of 300 nL/min. The 10 most intense ions with charge state from +2 to +4 determined from an initial survey scan from 300 to 1800 m/z, were selected for fragmentation (MS/MS). MS/MS was performed using an isolation width of 2 m/z; normalized collision energy of 35%; activation time of 30 ms and a minimum signal intensity of 5000 counts. The dynamic exclusion option is enabled. Once a certain ion is selected once for MS/MS in 7 s, this ion is excluded from being selected again for a period of 15 s. Mgf files were created from the raw LTQ mass spectrometer LC–MS/MS data using Proteome Discoverer 1.2 (Thermo Scientific). The created mgf files were used to search the NCBI database using the in-house Mascot Protein Search engine (Matrix Science) with the following parameters: trypsin 2 missed cleavages; fixed modification of carbamidomethylation; variable modifications of deamidation (Asn and Gln), pyroglu (Glu and Gln) and oxidation; monoisotopic masses; peptide mass tolerance of 2 Da; product ion mass tolerance of 0.6 Da.
2.11. Homology modeling
The crystal structure of full-length hPXR is currently not available and hence was modeled to understand the role of K109, which resides in the hinge region between the LBD and DBD. The full-length model of hPXR in complex with RXR and DNA was modeled using the crystal structure of PPARγ-RXR-DNA (pdb code: 3DZU) [48] as a template to the homology modeling program Modeler (ver 10) [49]. The full length model consists of the DNA binding domain (DBD) and the ligand binding domain (LBD) connected by a hinge region consisting of 13 amino acid residues. The residue K109 was modeled into an acetylated form using the builder module of MOE. The modeled complexes of PXR-RXR-DNA in its wild type form and with K109 in acetylated form was further refined using energy minimization and 2 ns long molecular dynamics (MD) simulations. Amber force field [50] with 5000 steps of minimization followed by 500 ps of equilibration and 2 ns of production run was performed on the complex which also included an explicit solvent model in an octahedron shell with TIP3P definition [51] and sodium ions were added for neutralizing the system. Electrostatics was calculated using the particle-mesh Ewald algorithm [52] with a cutoff of 10 Ẵ applied to Lennard–Jones interactions.
2.12. Phylogenetic analysis
PXR is in the NR 1I subfamily that also includes the vitamin D receptor (VDR; NR1I1) and the constitutive androstane receptor (CAR; NR1I3). To understand the phylogenetic variation of the position corresponding to human PXR K109, a wide range of vertebrate and invertebrate receptors in the NR1I subfamily were analyzed including amino acid sequences from a total of 50 VDRs, 48 PXRs, and 41 CARs. Sequences were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The Supplemental Data includes accession numbers from either Genbank (http://www.ncbi.nlm.nih.gov/genbank/) or Ensembl (http://www.ensembl.org/index.html), along with the amino acid residue in each receptor that is orthologous to human PXR K109.
2.13. Site-directed mutagenesis
pSG5-PXR plasmid was used as a template to construct K109R and K109Q point mutants using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer's protocol. Primer sequences:
-
K109R forward primer:
5′-GAGCGGCATGAAGAGGGAGATGATCATG-3′.
-
K109R reverse primer:
5′-CATGATCATCTCCCTCTTCATGCCGCTC-3′.
-
K109Q forward primer:
5′-GAGCGGCATGAAGCAGGAGATGATCATG-3′.
-
K109Q reverse primer:
5′-CATGATCATCTCCTGCTTCATGCCGCTC-3′.
2.14. Quantitative polymerase chain reaction (qPCR)
Procedure was performed as previously described [32]. HepG2 cells (106/well) were transiently transfected employing FuGENE HD reagent (Roche) with 1 µg/well of expression plasmid encoding WT PXR or its mutated forms. Cells were seeded in medium supplemented with 10% charcoal/dextran-stripped FBS on 6-well plates and incubated for 16 h prior to the treatments. Cells were treated for 24 h with 10 µM RIF and/or DMSO. After the treatments, total RNA was isolated using TRI Reagent® (Molecular Research Center). cDNA was synthesized from 1000 ng of total RNA using M-MuLV Reverse Transcriptase (F-572, Finnzymes) at 42 °C for 60 min in the presence of random hexamers (3801, Takara). qRT-PCR was carried out on Light Cycler 480 apparatus (Roche Diagnostic Corporation, Prague, Czech Republic). The levels of all mRNAs were determined using primers and Universal Probes Library (UPL; Roche Diagnostic Corporation, Prague, Czech Republic) probes. The following program was used for monitoring the expression of all genes: an activation step at 95 °C for 10 min was followed by 45 cycles of PCR (denaturation at 95 °C for 10 s; annealing with elongation at 60 °C for 30 s). The measurements were performed in triplicates. Gene expression was normalized per glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a housekeeping gene. Data were processed by delta–delta method. Differences in the level of gene expression values were considered statistically significant if p < 0.05.
2.15. Non-radioactive electrophoretic mobility shift assays (EMSA)
EMSA binding assays were performed using the LightShift® Chemiluminescent EMSA Kit (Thermo Scientific, Waltham, MA, USA) as described previously [53]. Briefly, nuclear extracts were harvested from HeLa cells transfected with WT or mutant (K109R or K109Q) PXR or RXRα as described elsewhere [54]. In 10-ul reactions, nuclear extracts and probes were mixed in binding buffer (final: 10 mM Tris, 50 mM KCl, 1 mM DTT, pH 7.5) along with final concentrations of 2.5% glycerol, 0.05% NP-40, ddH2O, and nonspecific competitor poly (dl.dC). PXR-RXR-DNA complexes were allowed to form at room temperature for 30 min before loading in the non-denaturing 4% polyacrylamide gel for electrophoretic separation. The following double-stranded 5′-biotinylated oligonucleotides containing the specific DNA-binding sequence for PXR (DR3 motif from the XREM sequence of CYP3A4 gene promoter) were used as probes: CYP3A4-DR3 wild-type sense 5′-GAATGAACTTGCTGACCCTCT-3′; CYP3A4-DR3 antisense 5′-AGAGGG TCAGCAAGTTCATTC-3′ and their biotinylated forms. The oligonucleotides were synthesized by Generi-Biotech (Hradec Kralove, Czech Republic). For supershift reactions, 1 µg of the anti-RXRα rabbit polyclonal IgG antibody (sc-553X, Santa Cruz Biotechnology, Inc.) was added to the reaction mixture.
2.16. Statistical analysis
Data were expressed as the mean ± standard error of the mean (SEM) and analyzed with Prism 6 software (GraphPad, La Jolla, CA). Group comparisons were performed using the Student t-test or ANOVA as indicated. A value of p < 0.05 was considered statistically significant.
3. Results
3.1. PXR is acetylated by p300 acetyltransferase
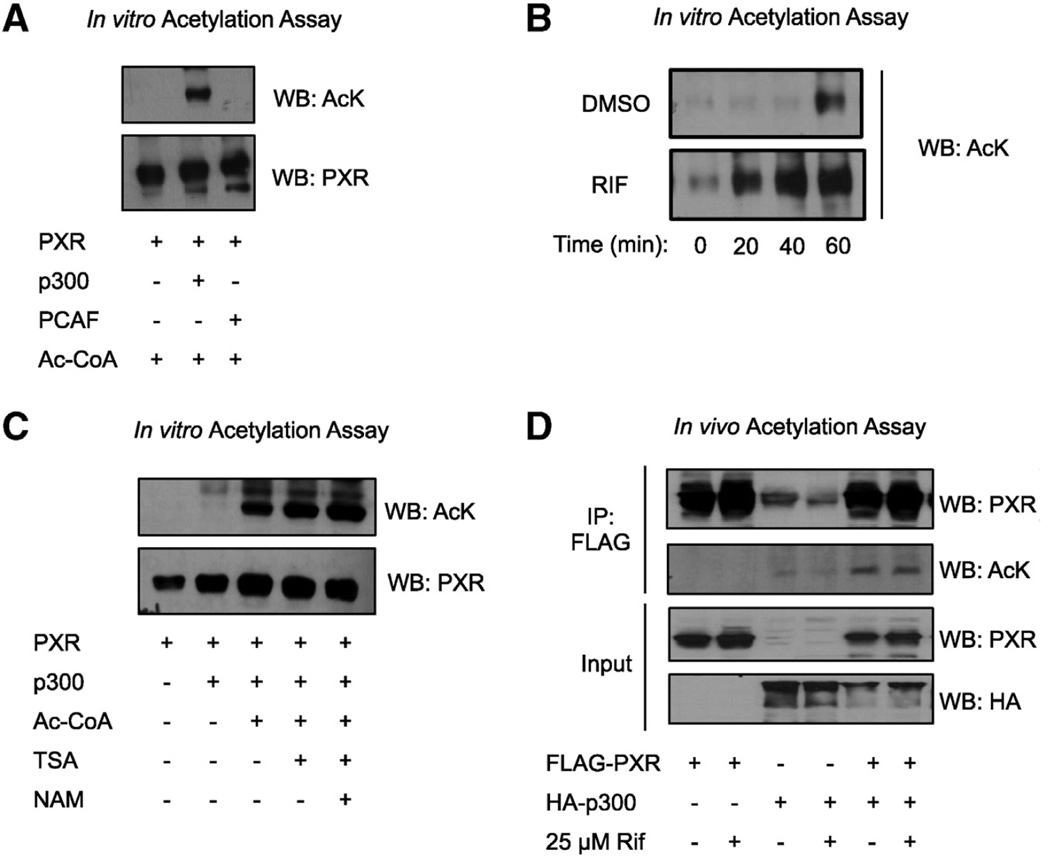
In our previous paper, we detected acetylated PXR in cultured cells but the acetylating enzyme was not identified [42]. Here we tested whether PXR is a substrate for two major acetyltransferases known to target NRs, p300 and p300/CBP-associated factor (PCAF). In Fig. 1A, we show that pure recombinant PXR is efficiently acetylated in vitro by p300 but not PCAF, demonstrating specificity. We also tested the effect of RIF on the rate of p300-mediated acetylation of PXR and show that RIF markedly increased the acetylation rate compared to DMSO (Fig. 1B). This suggests that ligand binding induces a conformational change in PXR that exposes the acetylation site, making it more accessible for p300 and thus enhancing the efficiency of the reaction. We confirmed p300-catalyzed PXR acetylation in a reticulocyte lysate system as well using in vitro translated PXR protein (Fig. 1C). Lysine deacetylase (KDAC) inhibitors Trichostatin A and nicotinamide (NAM, a SIRT1-specific inhibitor) enhance PXR acetylation further (Fig. 1B), suggesting that PXR is dynamically acetylated and deacetylated. We then validated in vivo that FLAG-PXR is acetylated by HA-p300, by ectopically overexpressing both proteins in 293T cells by co-transfection. FLAG-PXR was immunoprecipitated 48 h after transfection and PXR acetylation was visualized by Western blot using a pan-acetyl antibody (Fig. 1D). 24 h treatment with RIF did not affect acetylation levels under the conditions of this assay. These data confirm that p300 acetylates PXR in vitro and in vivo.
Fig. 1.
PXR is acetylated by p300 acetyltransferase. (A) Recombinant PXR was acetylated in vitro by incubating with either recombinant p300 or recombinant PCAF in acetylation buffer containing acetyl-CoA for 1 h at 30 °C. Acetylated PXR was detected by immunoblotting using an acetyl-lysine antibody and the blot was stripped and re-probed using a PXR antibody. (B) PXR was acetylated in vitro by p300 as described in (A), but this time in the presence of DMSO or 1 µM RIF and the reactions were quenched by boiling in Laemmli buffer at 0, 20, 40, or 60 min as indicated. (C) PXR protein was in vitro translated using the cell-free TnT coupled kit and subjected to in vitro acetylation assay using p300 as described in (A), with or without deacetylase inhibitors TSA and NAM. (D) 293T cells were co- transfected with FLAG-PXR and HA-p300 either separately or together. 24 h later, the cells were treated with DMSO or 25 µM RIF for an additional 24 h, nuclear extracts were harvested and subjected to FLAG-IP. To visualize FLAG-PXR acetylation by HA-p300 in vivo, immunoprecipitates were analyzed by immunoblotting using the acetyl-lysine and PXR antibodies. IP = immunoprecipitation, FLAG = FLAG octapeptide, Ac-CoA = acetyl coenzyme A, AcK = acetyllysine, PCAF = p300/CBP associated factor, HA = hemagglutinin, TSA = Trichostatin A, NAM = nicotinamide, Rif = rifampicin.
3.2. p300 enhances PXR transcriptional activity
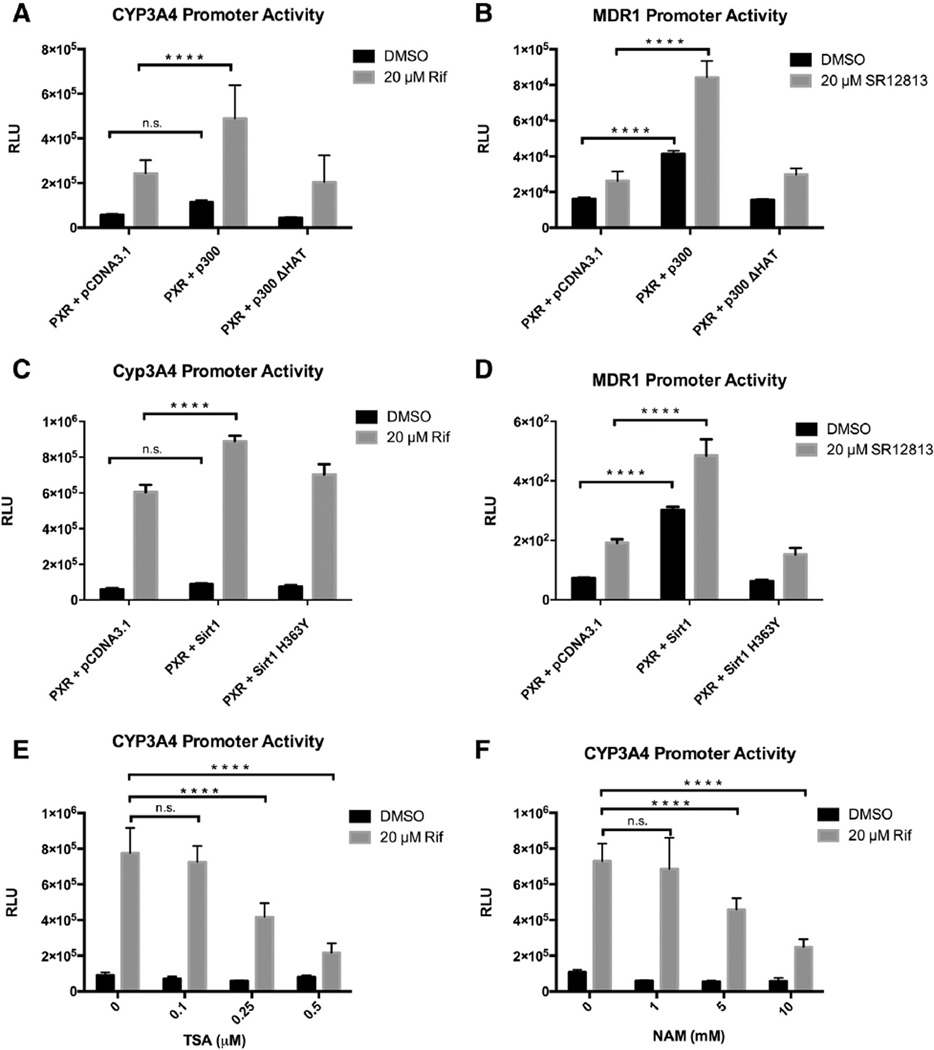
Given that p300 acetylates PXR, we tested the functional effect of p300 on PXR transcriptional activity in cell-based gene reporter assays performed in 293T cells. Co-transfection of pSG5-PXR and CYP3A4 promoter-driven luciferase reporter construct (p3A4-Luc) leads to promoter transactivation upon treatment with RIF. RIF-induced transactivation of the CYP3A4 promoter is enhanced by p300, but not by the catalytically inactive histone acetyltransferase mutant (p300ΔHAT), which lacks acetyltransferase activity (Fig. 2A). The same results were found when the same experiment was performed using a natural MDR1 promoter-driven luciferase reporter construct (MDR1-Luc) activated by a synthetic PXR agonist SR12813 (Fig. 2B). Moreover, p300 significantly enhanced ligand-independent (basal) activity of the MDR1 promoter as well. A similar result was seen using the CYP3A4 promoter but was not statistically significant. Together, these results suggest that p300 co-activates PXR and that p300 acetyl-transferase activity is required.
Fig. 2.
p300 and SIRT1 modulate PXR's transcriptional activity. 293T cells were co-transfected with pSG5-PXR, CYP3A4-Luc (A, C, E, F) or MDR1-Luc reporter (B, D), and pCMV-β-galactosidase control plasmids. 24 h after transfection, cells were treated with DMSO, Rif, or SR12813 as indicated for an additional 24 h. (A, B) Cells were co-transfected with pCDNA3.1. empty vector control, pCDNA3.1-p300, or pCDNA3.1-p300 ΔHAT mutant as well. (C, D) Cells were co-transfected with pCDNA3.1. empty vector control, pECE-SIRT1, or pECE-SIRT1 H363Y. (E) Transfected cells were treated with increasing concentrations of TSA (E), or NAM (F) at the same time with DMSO or RIF as indicated. Cells were then lysed and luciferase and β-galactosidase activities were measured. Luciferase values were normalized to β-galactosidase activities. The data represents the mean and SEM of at least 3 independent experiments performed in triplicate. ****p < 0.05, n.s., not significant 2-way ANOVA. Rif = rifampicin, TSA = Trichostatin A, NAM = nicotinamide.
3.3. SIRT1 enhances PXR transcriptional activity
SIRT1 is established as an important modulator of NR signaling in the liver through its deacetylase function that targets histones and transcription factors and coregulators, such as AR, FXR, LXR, and PGC1α [40, 54–60]. We previously reported that SIRT1 deacetylates PXR and positively regulates PXR's lipogenic function in vivo [42]. Here we test the functional effect of SIRT1 on PXR transcriptional activity similarly as we did with p300. Using either the p3A4-Luc or MDR1-Luc reporter, co-transfection of FLAG-SIRT1 enhanced PXR transactivation upon ligand treatment, while the catalytically inactive mutant FLAG-SIRT1- H363Y had no effect (Figs. 2C, D). Moreover, SIRT1 significantly enhanced ligand-independent (basal) activity of the MDR1 promoter. This was seen using the CYP3A4 promoter but was not statistically significant (Figs. 2C, D). Further, RIF-induced activation of the p3A4-Luc reporter was inhibited when cells were co-treated with TSA and NAM in a dose-dependent manner (Figs. 2E, F). Our previous data confirm that these two inhibitors enhance PXR acetylation levels [42]. These data suggest that SIRT1-catalyzed deacetylation of PXR enhances its transcriptional activity. In corollary, inhibiting deacetylases reduces PXR transcriptional activity. SIRT1 acts as a positive regulator of PXR function, at least partially via deacetylation.
3.4. PXR is acetylated at a highly conserved lysine residue (K109) in the hinge
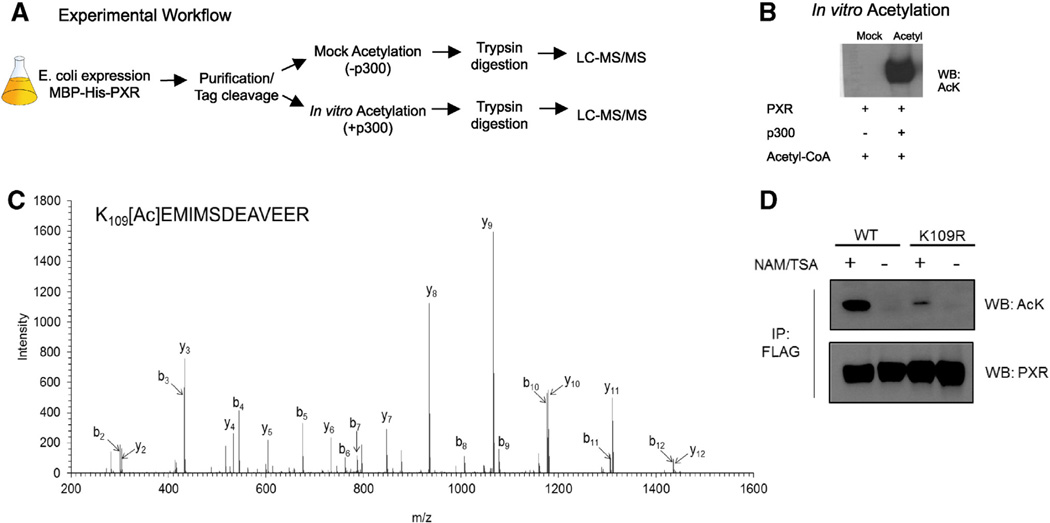
We next determined the specific p300-catalyzed acetylation site of PXR. Recombinant His-MBP-tagged human PXR protein was expressed and purified from E. coli. The pure protein was acetylated, or mock acetylated in vitro with or without recombinant p300, respectively (Fig 3B), and digested by trypsin under denaturing conditions. The tryptic peptides were then subjected to LC–MS/MS analysis (Fig. 3A). A doubly charged precursor ion (m/z = 805.7, retention time = 33.3 min) detected in the acetylated, but not the mock acetylated sample, was found to harbor an acetyl modification based on the presence of a + 42.01 Da mass shift compared to the corresponding unmodified peptide with the same sequence (109KEMIMSDEAVEER121). The MS/MS spectra and fragment ion assignments of the acetylated peptide are shown in Fig. 3C. The b- and y-ions were assigned using Mascot and confirmed by manual analysis (Mascot Peptide View in Fig 3C). The experiment outlined in Fig. 3A was performed three independent times, each time resulting in the identification of the same acetylated peptide indicating acetylated K109.
Fig. 3.
PXR is acetylated at K109) in the hinge. (A) Schematic depicting experimental workflow used to map PXR's acetylation site. (B) Immunoblot confirming the acetylation state of the mock acetylated control and acetylated PXR protein samples that were subjected to LC–MS/MS. (C) Fragment ion spectrum of the acetylated peptide detected in the in vitro acetylated protein sample. (D) Confirmation of K109 acetylation in vivo. HepG2 cells stably expressing the WT or K109R form of FLAG-tagged PXR were treated with or without 10 mM nicotinamide (NAM) and 0.5 µM trichostatin A (TSA) for 2 h. Cells were lysed and lysates were then subjected to FLAG-IP. Eluates were probed for acetylated PXR by Western blot.
To validate that K109 is PXR's major acetylation site in vivo, we measured PXR acetylation in HepG2 cell lines that were stably transfected with either the WT or K109R mutant form of FLAG-tagged PXR (Fig. 3D). Mutation of lysine to arginine blocks acetylation at that residue. After 2 h of treatment with deacetylase inhibitors NAM (10 mM) and TSA (0.5 µM), the cells were lysed and the WT or K109R form of FLAG-PXR was immunoprecipitated using M2 agarose. Eluted protein was probed by Western blot using the acetyllysine antibody to measure relative acetylation levels between the WT and K109R protein. The blot was stripped and re-probed with PXR antibody to confirm equal levels of total PXR protein. As indicated by the results, the K109R mutant protein displays reduced acetylation levels compared to the WT, confirming that K109 is the major site acetylated in living cells (HepG2). This corroborates our MS/MS results in Fig. 3C.
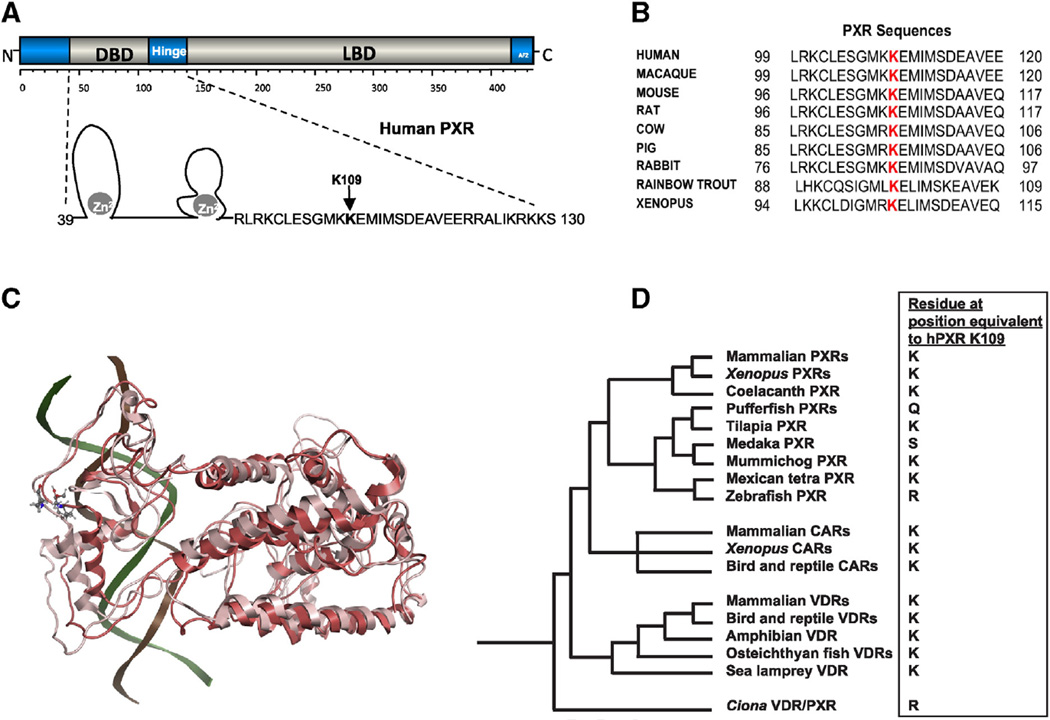
K109 is a critical residue within the hinge domain, immediately adjacent to what is defined as the C-terminal extension (CTE) of the DBD (Fig. 4A) [61]. While there are several crystal structures of PXR in complex with various xenobiotics [62–66] and most recently the heterodimeric complex of the LBDs of PXR and RXR [47] there is no crystal structure of the full length hPXR with the DBD, hinge and LBD. However, the full-length hPXR shares 42% homology with the crystal structure of PPAR-γ and hence was suitable for homology modeling. In the crystal structure complex of PPAR-γ with RXR and DNA, the K154/K155 is positioned in the hinge region and is known to be acetylated, and contributing the binding of SIRT1 [67]. However, sequence alignment of PPAR-γ with hPXR (supplementary Fig 1) suggests that K109 of PXR does not align with the K155 but is present proximal and in the same hinge region that connects the DBD and LBD. In addition, the LBD domain of PXR of the model superimposes with a rmsd of 0.8 Ẵ onto the crystal structure of the LBD of PXR in the PXR-RXR LBD heterodimers (supplementary Fig 2) suggesting that the full length model can be utilized for understanding the role of K109. Results from the MD simulations suggest that this hinge region is flexible and K109 in its non-acetylated form can interact with p300 and other additional cofactor proteins that aid in transcription. However, when K109 is in its acetylated form, the loop region containing K109 and the rest of the DBD undergo a conformational change leading to steric interactions with the DNA bases (Fig 4C). Structural superpositioning of the non-acetylated and acetylated models of PXR show significant deviations in the structure in DBD and hinge regions with an overall rmsd of 2.8 Ẵ (Fig. 4C). These conformational changes suggest that the PTM of PXR at K109 and the resulting conformational changes may lead to the disruption of the PXR-DNA complex described in step-3 of the model of PXR regulation (Fig 7).
Fig. 4.
PXR K109 is a highly conserved amino acid in the hinge. (A) Schematic representation of PXR's modular protein structure with domains labeled. A closer view of the secondary structure of the DNA-binding domain (DBD) and hinge is included. (B) Alignment of PXR sequences from several species depicting the conserved lysine residue 109. (C) Structural superpositioning of PXR wt. (light pink) and K109 acetylated form (red) are represented in the cartoon model with the K109 in both acetylated and non-acetylated represented as licorice sticks and colored atom type (C-cyan, N-blue, O-red). Significant deviations are noted in the hinge and DBD between the two forms. The position of DNA (in cartoon model) in the wt. is shown for reference (see Supplementary Fig 3 for the structural superposition of the entire complex). (D) Phylogeny of PXRs, VDRs, and CARs [117, 118] overlaid with the variation of the amino acid residue orthologous to hPXR K109. Species and accession numbers for all sequences are found in Supplemental Data. Note that mummichog PXR is shown in the phylogeny (with lysine) but that the PXRs from Amazon molly, and platyfish, two other fish species from the order Cyprinodontiformes, also have lysine at this position. The assignment of the chicken CAR (originally indeterminately classified as the chicken X receptor [119]) follows from a recent phylogenetic analysis incorporating amphibian and lizard CARs [118].
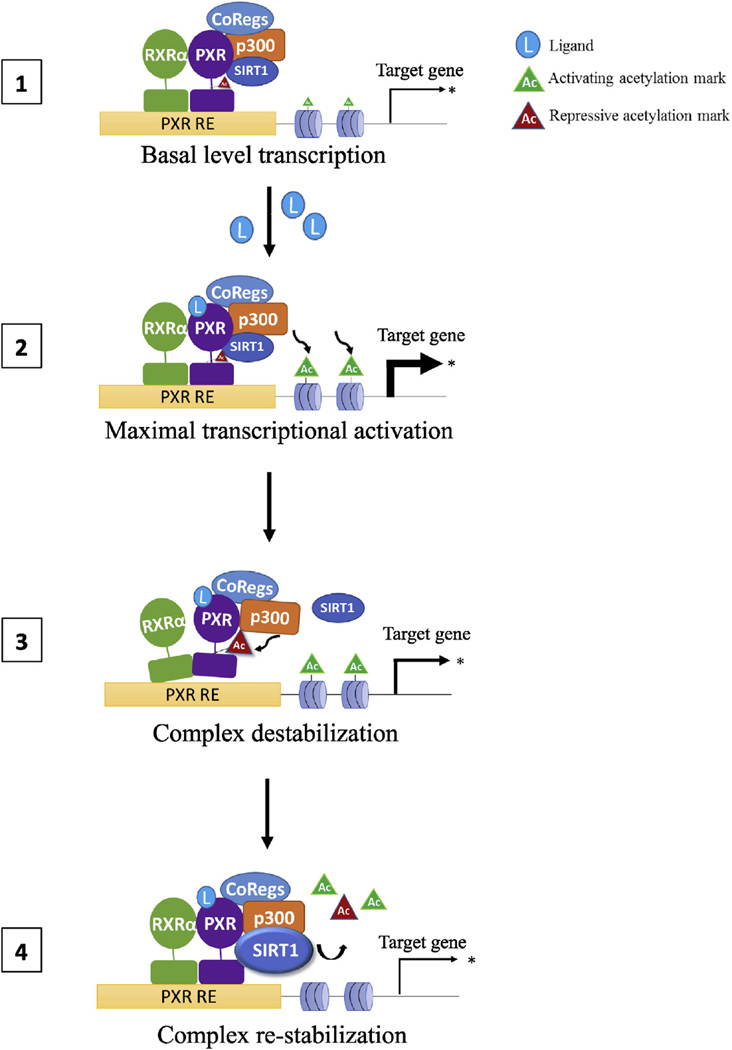
Fig. 7.
Proposed model of PXR regulation by acetylation/deacetylation. Acetylation/deacetylation fine-tunes PXR transcriptional activity depending on cellular signals. 1) At the promoter, unliganded PXR and histones are maintained in a hypoacetylated state by SIRT1 maintaining basal levels of gene transcription. 2) Ligand binding induces a conformational change in PXR that promotes p300-mediated histone acetylation, which facilitates local chromatin relaxing and maximal transcriptional activation ramp-up (rate). 3) After some amount of time, complex rearrangement occurs allowing p300 to acetylate PXR. This induces a conformational change in PXR that reduces its affinity for RXRα and DNA, resulting in destabilization of the transcriptional complex, a transcriptional breaking mechanism, thus reducing the transcription rate. 4) SIRT1 deacetylates histones and PXR, leading to complex re-stabilization, suppression of the stimulated transcriptional response, maintaining transcription until loss of ligand stimulation results in a return to basal transcriptional activity.
K109 is highly conserved across PXR orthologs (Fig. 4B) and across the two most closely related receptors within the NR1I subfamily (CAR and VDR) and their respective orthologs (Supplemental Data). Sequence alignment demonstrates that this position is generally conserved throughout the vertebrate NRI1I subfamily (VDRs, PXRs, CARs) as illustrated in the phylogeny in Fig. 4D (complete list of receptors, accession numbers, and amino acid residue orthologous to human PXR K109 are in Supplemental Data). This position is a lysine residue in all mammalian PXRs, amphibian PXRs, coelacanth PXR, mammalian CARs, amphibian CARs, chicken CAR, coelacanth CAR, and all vertebrate VDRs. The only exceptions are four teleost fish PXRs, which have a different amino acid residue at this position: glutamine, serine, and arginine. Because this residue is highly conserved and important for PXR function, modification by acetylation, which effectively neutralizes lysines, would be expected to alter receptor behavior.
3.5. Lysine 109 acetylation downregulates PXR's transcriptional activity
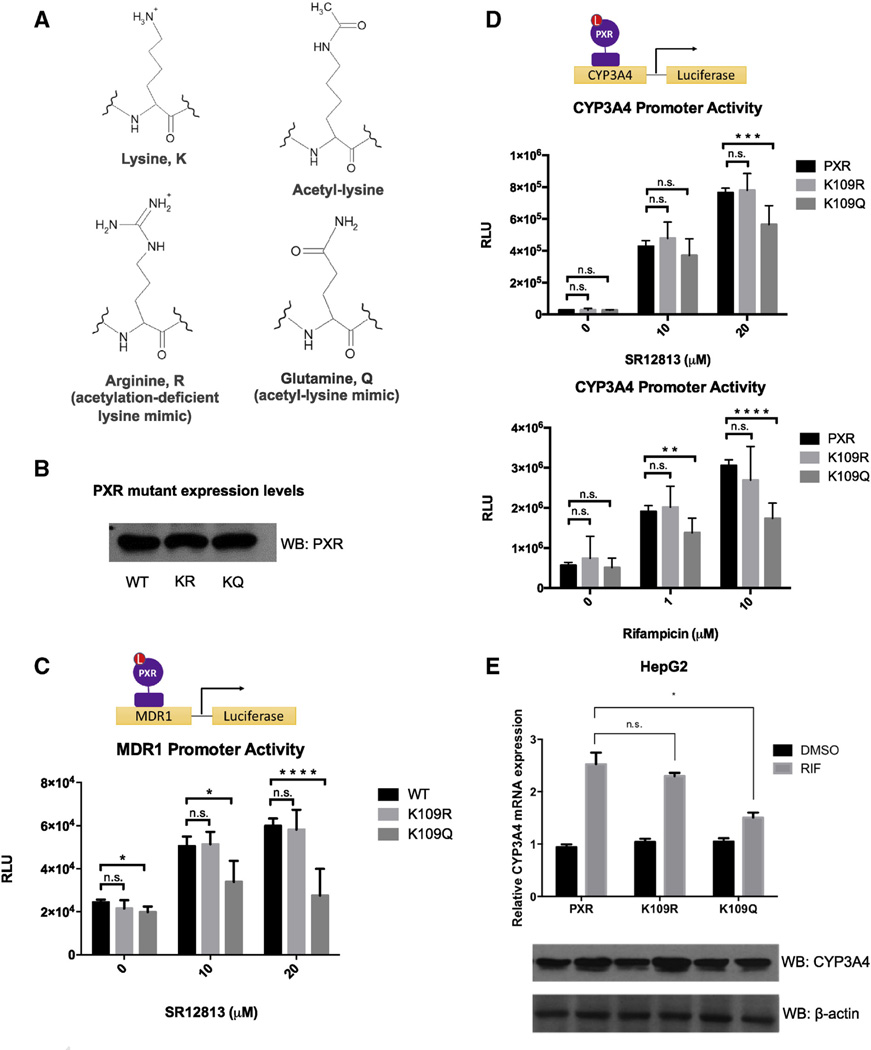
In order to interrogate the function of PXR acetylation specifically at K109, we generated acetylation mutants by site-directed mutagenesis. The lysine to arginine mutant (K109R) mimics the constitutively deacetylated state, while the lysine to glutamine (K109Q) mutant mimics the constitutively acetylated state [68]. These mutational mimics are based on the positive charge of lysine and the neutral charge of an acetyllysine residue as shown (Fig 5A). By Western blot, we show that expression levels of the ectopically expressed WT and mutant proteins are equal to confirm that any changes in activity observed are not due to differences in protein expression or stability (Fig 5B). In luciferase reporter assays using either MDR1-Luc or p3A4-Luc reporter constructs, ligand-activated transcriptional activity of the K109Q acetylation mimic is repressed compared to WT and K109R (Fig 5C and D). Similarly, the K109Q acetylation mimic also has a decreased capacity to induce CYP3A4 mRNA and protein in response to RIF treatment compared to the WT and K109R (Fig 5E). K109R behaves similarly to WT levels in these assays.
Fig. 5.
Lysine 109 acetylation down-regulates PXR's transcriptional activity. (A) Structures of lysine, acetyl-lysine, and corresponding mimicking residues arginine and glutamine, respectively. Mutational mimics are based on charge of the amino acid R-group. (B) Western blot showing equal expression levels of PXR WT, K109R, and K109Q in transfected 293T cells. (C) 293T cells were co-transfected with the MDR1-Luc reporter, and pCMV-β-galactosidase control plasmid along with pSG5- PXR WT, K109R, or K109Q. Cells were treated with DMSO or various concentrations of SR12813 24 h after transfection for an additional 24 h and luciferase and β-galactosidase activities were measured. Luciferase values were normalized to β-galactosidase activities. The data represents the mean and SEM of at least 3 independent experiments performed in triplicate. (D) The experiment described in (C) was repeated using the p3A4-Luc reporter instead. Cells were treated with RIF or SR12813 as indicated. (E) HepG2 cells were transfected with the plasmids as indicated and treated with DMSO or 10 µM RIF for 24 h. CYP3A4 mRNA levels were measured by qRT-PCR and corresponding protein levels measured by immunoblot. Data were processed by delta–delta method. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s., not significant, Student's t-test. 2-way ANOVA was used for data analysis presented in (E).
3.6. Acetylation of PXR inhibits binding to DNA
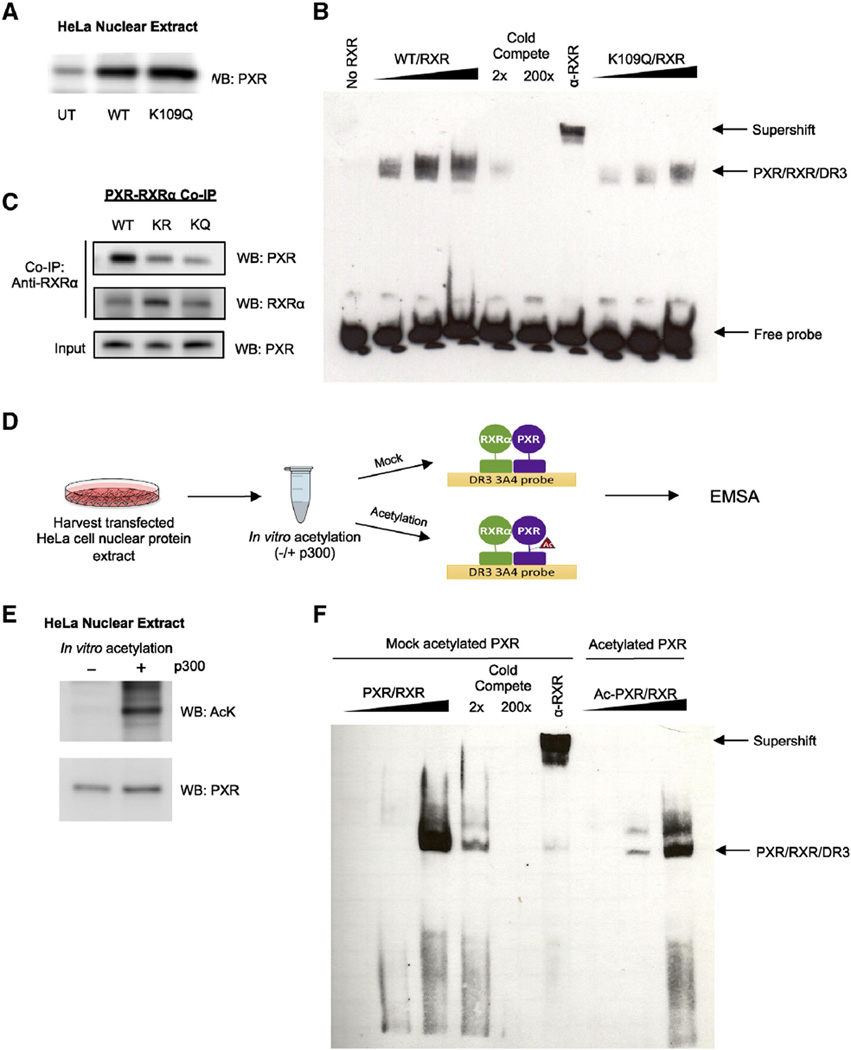
Acetylation can modulate the transcriptional activity of NRs and other transcription factors through altering DNA binding, as was shown for FXR, ERα, PR, p53, and FOXO1 [40, 41, 69, 70]. We tested whether this was the case for PXR since we found that PXR is acetylated in its DBD/hinge. Since acetylation effectively neutralizes positively charged lysine residues, it may disrupt electrostatic interactions that are important for maintaining secondary structure and mediating interactions with the negatively charged phosphate backbone of DNA. We used electrophoretic mobility shift assays (EMSA) to test whether acetylation alters the formation of PXR/RXRα/DNA complexes. HeLa cells were transfected separately with expression vectors encoding either PXR WT or PXR K109Q, then nuclear protein extracts were harvested to use for EMSA binding reactions (Fig. 6A). We show that the K109Q acetylation mimic displays reduced DNA binding compared to the WT at different doses (Fig. 6B). K109Q protein also displays reduced binding to RXRα compared to WT and K109R in co-IP assays, which suggests that reduced RXRα binding due to K109 acetylation contributes to reduced DNA binding and transcriptional activity of PXR (Fig. 6C). Next, as outlined in the schematic (Fig 6D), we tested the effect of direct acetylation of PXR by p300 (Fig 6E). By comparing non-acetylated and acetylated PXR protein, we find that acetylation inhibits DNA binding, which is consistent with our study of the K109Q acetylation mimic (Fig. 6F).
Fig. 6.
Acetylation of PXR inhibits DNA binding and heterodimerization with RXRα. (A) Western blot showing PXR or K109Q ectopic protein expression levels in nuclear extracts harvested from transfected HeLa cells. (B) K109Q mutation reduces DNA binding compared to WT. Increasing amounts of nuclear extract (described in Fig. 5B) enriched with PXR protein (WT vs K109Q) were incubated with nuclear extract enriched with RXRα at a 1:1 ratio along with biotin-labeled oligonucleotide probe containing the DR3 response element. In order to demonstrate specificity, WT reactions were incubated with 2× or 200× unlabeled competing probe or with RXRα antibody. The complexes formed were detected using non-radioactive EMSA. (C) In vitro translated RXRα and WT or mutant PXR were incubated 1:1 at 30 °C for 30 min and then incubated with RXRα antibody-coupled AminoLink Plus Coupling Resin for co-immunoprecipitation. Eluted protein complexes were resolved and analyzed by Western blot to detect amount of PXR that bound to RXRα. (D) Experimental workflow for EMSA using PXR acetylated in vitro. (E) Western blot confirming the acetylation state of the mock acetylated control (−) and acetylated (+) PXR protein samples. (F) In vitro acetylation of PXR reduces DNA binding. PXR-enriched nuclear extracts were mock acetylated or in vitro acetylated (as shown in D) then PXR/RXR/DR3 complex binding was analyzed as described in (B). UT = untransfected, EMSA = electromobility shift assay, IP = immunoprecipitation, WB = Western blot.
4. Discussion
Post-translational modifications (PTMs) have emerged as a major means of NR regulation in normal physiology and disease. In this study, we conducted the first detailed examination of PXR regulation by acetylation. We found that PXR is directly acetylated by p300 at K109 by LC–MS/MS analysis in vitro and in vivo (Fig 3). Note that rifampicin-induced acetylation changes as seen in vitro (Fig 1B) is different that those observed in vivo (Fig 1D) likely because the time course of both experiments is vastly different and we know that acetylation is a time-dependent dynamic process. Furthermore, protein incubations are perfect in stoichiometry; however, transfections to induce proteins in cells are highly variable even across transfection wells. So conditions of the assay per se may have blurred differences in observed acetylation. The K109Q lysine acetyl mimic mutant displayed reduced transcriptional activity (Fig. 5C and D) and reduced ability to induce cyp3A4 target gene mRNA and protein compared to the WT and the K109R lysine acetyl-defective mutant (Fig. 5E). The diminished activity of the K109Q mutant appears to be due to impaired heterodimerization with RXRα (Fig. 6C) and impaired binding of the PXR-RXRα heterodimer to DNA response elements (Fig. 6B and F). Altogether, the data suggests that acetylation at K109 represents an overall “loss of function” effect. As a caution, constitutive mutants likely over-estimate/bias the true dynamic nature of acetylation changes in vivo [71]. Future approaches to investigate dynamic acetylation would require reversible chemical methods for site-selective acetylation/deacetylation in mammalian cells to better appreciate the dynamic nature of this PTM. K109 is located in the C-terminal extension (CTE) of the DBD (within the hinge), which might indicate the structural basis of PXR downregulation by acetylation. The CTE is necessary for high affinity and high specificity DNA binding, as the two DBD zinc fingers alone are insufficient [61]. For the receptors that heterodimerize with RXR, helices in the CTE form significant sequence-specific DNA contacts and additional dimerization surfaces, and are responsible for spacing of the dimer partners, as was demonstrated by the crystal structure of the PPAR-γ-RXRα-DNA ternary complex [48, 61]. The CTE/hinge affords the structural flexibility necessary for simultaneous heterodimerization with RXRα and DNA binding. Hence, we would suspect that any residue modifications in the DBD CTE/hinge, especially those resulting in a change of charge state (i.e. neutralization of positively charged lysine by acetylation or mutation to glutamine), would alter receptor conformation and thus functionality with respect to DNA binding and RXRα heterodimerization. This is what we see when PXR is acetylated at K109—the residue modification induces a conformational change in the receptor, as corroborated by our modeling studies (Fig. 4C), which in turn impairs RXRα dimerization and DBD binding, ultimately repressing receptor function as a whole [72, 73].
Various combinations of recruited coactivator and corepressor proteins modulate NR activities as well, comprising a tightly controlled and complex regulatory program. Many of these coregulator complexes act through catalyzing histone modifications that in turn regulate chromatin structure [74, 75]. p300 is a bona fide transcriptional coactivator for several transcription factors, including NRs. p300 was originally identified based on its histone acetyltransferase activity through which it acetylates specific lysines in histone tails, particularly H3K9/18 and H3K27, promoting chromatin relaxing and active genetranscription [76]. Later it was found that p300 and other HATs (also called KATs or lysine acetyltransferases), target non-histone proteins as well, including the transcription factors and coregulators that they co-activate [77].
Until recently, it was assumed that p300 co- activated PXR, but no one specifically tested it. Smith et al. was the first to confirm their interaction, showing that in primary human hepatocytes exposed to rifampin, p300 is co-recruited to PXR response elements genome-wide [78]. Consistent with this study, we confirmed at the transcriptional level that p300 co-activates basal and ligand-induced PXR (Fig. 2A and B). Initially this result seemed to contradict our hypothesis that p300-mediated acetylation downregulated PXR activity; however, as we see in other receptors such as FXR, recruitment of p300 can have dual functions: enhancing transcription through histone acetylation and repressing transcription through subsequent acetylation of the receptor itself [40]. These opposing effects serve as mechanisms to activate then quickly limit the duration and magnitude of the stimulated transcriptional response, which must be tightly controlled in both directions (Fig. 7).
Like other receptors (e.g. FXR and LXR), PXR is deacetylated and positively regulated by multiple KDACs as well. Pharmacological inhibition of KDACs with TSA (inhibits class I and II KDACs) and NAM (SIRT1-specific inhibitor) leads to increased PXR acetylation levels, suggesting that these various KDACs deacetylate PXR (Fig. 1C). At the transcription-al level, we show that enhancing PXR acetylation levels with TSA and NAM inhibit transactivation in luciferase reporter assays, which suggests that KDACs normally maintain PXR in a deacetylated state allowing it to remain basally active (Fig. 2E and F). Concomitantly, overexpressing SIRT1 promotes PXR transactivation (Fig. 2C and D). These results are consistent with our previous report demonstrating that SIRT1 interacts with and, deacetylates PXR in response to ligand activation which correlates with PXR's lipogenic function in vivo [42].
A study by Sugatani et al. supports our contention that acetylation negatively regulates PXR (while deacetylation positively regulates PXR) and proposes a mechanism involving PTM crosstalk where one PTM affects the function of the other PTM. They propose that phosphorylation of PXR at serine 350 (S350) maintains PXR in an acetylated state by blocking interactions with KDACs. As a result, the dual PTMs synergistically repress PXR activity [36].
Interestingly, Buler et al. reported that pyruvate-activated SIRT1 negatively regulates PXR by physically interfering with PXR's interaction with and co-activation by PGC-1α [79]. There are several reasons why this data might contradict our own. First, they only observe these negative effects of SIRT1 on PXR in a specific context–when cells are both overexpressing PGC-1α and treated with 1 mM pyruvate (simulating fasting conditions). In contrast, we performed our assays without manipulating the metabolic state of the cell. It is possible that SIRT1 may positively regulate PXR under metabolic homeostasis, but then negatively regulate it under fasting conditionsin order to adaptively curb PXR's known effects on lipid and glucose metabolism. And even if SIRT1 can interfere with PXR co-activation when PGC-1α is overexpressed, there are several other important coactivators of PXR (i.e. SRC-1, HNF4α) that could normally compensate since they have redundant functions.
Notably, they also use the mouse isoforms of PXR and SIRT1 in this study, while we used the human isoforms. Furthermore, they used a GAL4-mPXR LBD fusion construct in the mammalian two-hybrid assay used to show that SIRT1 interferes with the PXR-PGC-1α interaction. The GAL4-mPXR LBD fusion protein lacks the DBD and N-terminal half of the hinge, which is the very region we propose to be positively regulated by SIRT1-mediated deacetylation. Therefore, their experiment does not account for this. They also reported that they could not detect acetylated PXR in COS-1 cells by Western blot. This may be because PXR is simply not acetylated efficiently in this cell line and/or the detection method is not sensitive enough. These apparent inconsistencies speak to the complexity and context-dependence (i.e. different cell lines or assay conditions) of these regulatory mechanisms.
PTMs can also antagonize one another and modulate disparate functions independently [80, 81]. In a recent study by Kim et al., they describe an acetyl/SUMO switch that regulates FXR's activation and trans-repression functions depending on nutrient status [82]. Nutrient-excess conditions in the liver (i.e. HFD-induced obese mice) leads to aberrantly elevated acetylation levels of FXR at K217, which then inhibits SUMO2 modification at K277. This promotes pathological inflammation (which is known to occur in obesity) because normally, FXR must undergo SUMOylation in order to trans-repress the NF-κB pro-inflammatory pathway.
It is tempting to hypothesize that PXR may be regulated by an acetyl/SUMO switch like FXR because the mechanisms and functions of the related receptors overlap [83, 84]. First, like FXR, SUMOylation selectively directs PXR towards trans-repressing the NF-κB pro-inflammatory pathway, while not affecting PXR activation of its direct target genes [37, 84]. In this case, SUMOylation acts in a promoter-specific manner. A recent follow-up study demonstrated that PXR is SUMOylated specifically at K108 [85]. It would be interesting to see how SUMOylation of PXR at K108 and adjacent acetylation at K109 influence one another and whether there is functional importance in this interaction. It is easy to imagine that SUMOylation at K108 would perhaps sterically inhibit subsequent acetylation at K109, or vice versa. Second, Kim et al. also showed for comparison, that along with FXR, PXR acetylation levels are also elevated in response to HFD-induced obesity in mouse liver, suggesting that obesity may represent the physiological relevance of PXR acetylation. In this context, HFD in rodents also increases PXR levels, perhaps highlighting the significance for overall PXR acetylation effects in obesity [86]. In fact, it is becoming clear that nutrient excess conditions promote global hepatic acetylation of metabolic regulators in general, including NRs, for metabolic adaptation [87–93]. A counter argument is that this acetylation state/level could simply reflect an adaptation to increased expression of PXR in HFD- fed rodents [86, 94, 95].
Based on our data and emerging mechanisms of NR-mediated transcription, we present a model of PXR regulation by dynamic acetylation and deacetylation in the context of an active promoter (Fig. 7). In the absence of a stimulus, the PXR-RXRα-DNA complex maintains basal gene transcription levels as part of a larger complex of various coregulator proteins and general transcription machinery (Fig. 7.1). In this basal transcriptional state, it is likely that p300 and SIRT1 are present at some levels, whether directly interacting with PXR or not, since they are involved in maintaining local chromatin accessibility. Although SIRT1 and other HDACs are generally associated with corepressor complexes and repressive chromatin marks, they have also been found to bind genes primed for activation [96]. In ChIP-seq analysis of PXR's cistrome, p300 co-localized with PXR independent of ligand stimulation at a proportion of the binding sites, perhaps to help keep promoters primed for rapid ligand-induced activation [78]. Similarly, Navaratnarajah et al. showed in vitro that PXR can bind its coactivator SRC-1 with high affinity with or without a ligand [97]. This study also showed that ligand binding does not reduce PXR's affinity for its corepressor SMRT. These ligand-independent PXR-coregulator interactions challenge the paradigm that ligand binding activates NRs by inducing a conformation that increases affinity for coactivators and decreases affinity for (or displaces) bound corepressors. Instead, the authors suggest that ligand binding may increase transcription by increasing PXR's stability, not its affinity for coactivators [97]. This in turn prolongs the stability and occupancy of the active transcription complex as a whole at the promoter accounting for increased ligand-dependent transcription [98]. This study is corroborated by another report showing that mdr1 (PXR target) gene induction is likely due to increased PXR stability via phosphorylation of its chaperone, heat shock protein (Hsp) 90β [99]. Ligand-induced conformational change of PXR promotes rearrangement of the transcriptional complex into a maximally activated state involving additional p300 recruitment and histone acetylation, thereby accelerating transcription (Fig 7.2). Since the conformational change also exposes PXR's acetylation site, p300 is allowed to then directly acetylate PXR at K109, which decelerates transcription by reducing PXR's affinity for DNA and RXRα, quickly destabilizing the transcription complex (Fig 7.3). Eventually, PXR and histones are deacetylated by SIRT1 and other KDACs, allowing the complex to re-stabilize and return back to a basal state (Fig. 7.4). The complex can then be recycled for a second round of ligand-activated transcription. A likely alternative to complex recycling, which we did not test, is promoter clearance and PXR degradation. Deacetylation of FXR and LXR by SIRT1 was shown to target the receptors for ubiquitin-mediated proteasomal degradation [40, 55]. Since PXR stability is regulated by ubiquitin-mediated proteasomal degradation, this is a likely mechanism, although the role of acetylation and/or deacetylation in this context, thus far, has remained unclear [30, 31].
Throughout the transcription process, protein interactions are in constant flux, as factors continuously cycle on and off the promoter. Recent technological advances in the study of transcriptional processes have revealed that the combination and stoichiometry of factors present at the promoter at any given time are considerably probabilistic and stochastic in nature, although influenced by various signals in the cellular environment [100]. This is in contrast to classical models that describe NR-mediated transcription as a sequence of static, deterministic molecular events that start with ligand binding followed by ordered recruitment of specific factors. Although this is accurate to some levels, it neglects the inherently stochastic interactions that occur in a nucleus packed with an assortment of molecules, including proteins, chromatin structures, and DNA sequences.
NR PTMs have been extensively studied revealing complex, tightly controlled, but highly varied regulatory mechanisms that are in constant flux depending on a combination of external and internal signals [101–103]. Compared to the steroid receptors, in-depth studies on PXR PTMs are relatively under developed, especially for the PTMs other than phosphorylation. In our study, we see that PXR acetylation/deacetylation at K109 is dynamically regulated by p300 and multiple KDACs, including SIRT1. Basal acetylation overall has a repressive effect on PXR but changes in levels of acetylation over time regulate the transcriptional activity of PXR. K109 seems to be a major acetylation site, but we do not rule out the possibility of alternative sites, targeted by other KATs, or present under other physiological situations. Acetylation may also affect PXR interaction with other coregulators (e.g. SRC-1 and SMRT), and may even affect subcellular localization or other PXR functions outside of the active transcription complex. We limited our study to a single promoter therefore it would be interesting to see how acetylation might affect PXR binding genome-wide. Our results are also limited in interpretation because we detected the K109 acetylation modification in vitro. The existence of K109 acetylation in cells and tissues has not been demonstrated in this study. Future studies are aimed towards developing mouse models that express the human PXR transgene while altering K109 via mutations that mimic or loose the acetylation phenotype. This could be coupled to an inducible system (e.g., tamoxifen) to induce transient gene expression. These models could then be used to study effects of transient expression of the PXR acetylation/deacetylation phenotype on PXR target gene expression.
Sirtuin 1 (SirT1/SIRT1) has an important role in both the pathogenesis of hepatic steatosis, inflammation and chemical toxicity. Activation of hepatic SIRT1 is generally thought to inhibit hepatic steatosis [104], inflammation/cancer [105], and neurotoxicity [106]. In our investigations with the PXR wild-type and knockout mice, SIRT1 activation via resveratrol, yielded slightly increased steatosis (~25–30%) when compared with basal levels in wild-type or drug-exposed knockout hepatocytes. Indeed, this contradicts what have been published using rodent models of obesity/diabetes/fat feeding [107]; however, our mice are essentially “normal” and not subject to high-risk features for metabolic syndrome or diabetes. Furthermore, some rodent and clinical studies have provided contradictory findings with respect to SIRT1 activation (resveratrol) in humans [108–111]. Thus, it is potentially possible that the presence of PXR could attenuate the inhibitory properties of resveratrol on hepatic steatosis. SIRT1 mediated deacetylation of PXR could be particularly relevant with environmental contaminants that could enhance fatty liver formation via PXR and other related nuclear receptors [112]. Likewise, dietary changes (e.g., fat feeding) could alter PXR levels as well as influence its acetylation [82, 113], so in these conditions the relative activation of SIRT1 becomes critical in determining the overall effects of PXR in metabolic syndromes. We have shown that SIRT1 is partially responsible for the deacetylation of PXR. However, we have also shown that Trichostatin A (TSA), which inhibits class I (HDACs— 1, 2, 3 and 8) and II (HDACs— 4, 5, 6, 7, 9, and 10) mammalian histone deacetylase enzymes but not class III (e.g., sirtuins) [114, 115] promotes acetylation of PXR in cell based assays. Thus, it is very plausible that other classes of HDAC enzymes could participate in the deacetylation of PXR. Since PXR is known to co-bind several co-regulators like SMRT and NCoR, selective HDACs (e.g., HDAC3) that co-associate with these regulators might also affect PXR acetylation via its deacetylase activity [36, 116]. It is anticipated that several HDACs other than SIRT1 might be involved in the regulation of PXR acetylation. Although many new questions arise, this study provides a basic model of PXR transcriptional regulation by acetylation that may represent a therapeutic approach to modulating PXR function in disease.
Supplementary Material
Acknowledgments
We would like to thank Edward Nieves and Myrasol Callaway of the Einstein Proteomics Facility and Dr. Srinivas Iyer and Timothy Sanchez from Los Alamos National Laboratory for their assistance with the LC– MS/MS procedure, discussions, and data analysis. This work was supported in part by National Institutes of Health Grant CA127231 and CA161879. This work was also supported by a Damon Runyon Foundation Clinical Investigator Award (CI 1502) (to S. Mani). Part of the research was supported by the Czech Science Agency [Grant GACR P303/12/0472].
Abbreviations
- LC–MS/MS
liquid chromatography coupled to mass spectrometry
- PTM
post-translational modification
- DR
direct repeats
- ER
everted repeats
- NR
nuclear receptor
- IP
immunoprecipitation
- FLAG
FLAG octapeptide
- Ac-CoA
acetyl coenzyme A
- AcK
acetyllysine
- PCAF
p300/CBP associated factor
- HA
hemagglutinin
- TSA
Trichostatin A
- NAM
nicotinamide
- Rif
rifampicin
Footnotes
The Transparency document associated with this article can be found, in online version.
Author contributions
DP conceptualized and coordinated the study, performed most of the experiments except where indicated, and wrote the paper. SM conceptualized the study design and advised for all technical and theoretical aspects of the study and edited the paper. AD performed qPCR (Fig. 5E) and experiments in Fig. 6. HL provided technical assistance. SK performed the homology modeling. MDK performed the phylogenetic analysis. AB provided critical advice and comments regarding the study and paper. WGW and MMR provided plasmids and protocols for bacterial protein expression and purification of PXR. ZD contributed general intellectual insights, reagents and nuclear receptor expertise.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagrm.2016.01.006.
References
- 1.Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr. Rev. 2002;23:687–702. doi: 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- 2.Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, Perlmann T, Lehmann JM. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J. Clin. Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kodama S, Negishi M. PXR cross-talks with internal and external signals in physiological and pathophysiological responses. Drug Metab. Rev. 2013;45:300–310. doi: 10.3109/03602532.2013.795585. [DOI] [PubMed] [Google Scholar]
- 6.Morere P, Nouvet G, Stain JP, Paillot B, Metayer J, Hemet J. Information obtained by liver biopsy in 100 tuberculous patients. Sem. Hop. 1975;51:2095–2102. [PubMed] [Google Scholar]
- 7.Chen Y, Nie D. Pregnane X receptor and its potential role in drug resistance in cancer treatment. Recent Pat. Anticancer Drug Discov. 2009;4:19–27. doi: 10.2174/157489209787002498. [DOI] [PubMed] [Google Scholar]
- 8.Fichtenbaum CJ, Gerber JG. Interactions between antiretroviral drugs and drugs used for the therapy of the metabolic complications encountered during HIV infection. Clin. Pharmacokinet. 2002;41:1195–1211. doi: 10.2165/00003088-200241140-00004. [DOI] [PubMed] [Google Scholar]
- 9.Niemi M, Backman JT, Fromm MF, Neuvonen PJ, Kivisto KT. Pharmacokinetic interactions with rifampicin: clinical relevance. Clin. Pharmacokinet. 2003;42:819–850. doi: 10.2165/00003088-200342090-00003. [DOI] [PubMed] [Google Scholar]
- 10.Plant NJ, Gibson GG. Evaluation of the toxicological relevance of CYP3A4 induction. Curr. Opin. Drug Discov. Devel. 2003;6:50–56. [PubMed] [Google Scholar]
- 11.Wolf KK, Wood SG, Hunt JA, Walton-Strong BW, Yasuda K, Lan L, Duan SX, Hao Q, Wrighton SA, Jeffery EH, Evans RM, Szakacs JG, von Moltke LL, Greenblatt DJ, Court MH, Schuetz EG, Sinclair PR, Sinclair JF. Role of the nuclear receptor pregnane X receptor in acetaminophen hepatotoxicity. Drug metabolism and disposition: the biological fate of chemicals. 2005;33:1827–1836. doi: 10.1124/dmd.105.005256. [DOI] [PubMed] [Google Scholar]
- 12.Hoekstra M, Lammers B, Out R, Li Z, Van Eck M, Van Berkel TJ. Activation of the nuclear receptor PXR decreases plasma LDL-cholesterol levels and induces hepatic steatosis in LDL receptor knockout mice. Mol. Pharm. 2009;6:182–189. doi: 10.1021/mp800131d. [DOI] [PubMed] [Google Scholar]
- 13.Hukkanen J, Hakkola J, Rysa J. Pregnane X receptor (PXR)—a contributor to the diabetes epidemic? Drug Metabol. Drug Interact. 2014;29:3–15. doi: 10.1515/dmdi-2013-0036. [DOI] [PubMed] [Google Scholar]
- 14.Lee JH, Zhou J, Xie W. PXR and LXR in hepatic steatosis: a new dog and an old dog with new tricks. Mol. Pharm. 2008;5:60–66. doi: 10.1021/mp700121u. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Febbraio M, Wada T, Zhai Y, Kuruba R, He J, Lee JH, Khadem S, Ren S, Li S, Silverstein RL, Xie W. Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARgamma in promoting steatosis. Gastroenterology. 2008;134:556–567. doi: 10.1053/j.gastro.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, Ren S, Evans RM, Xie W. A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem. 2006;281:15013–15020. doi: 10.1074/jbc.M511116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou W, Zhang J, Li H, Kortagere S, Sun K, Ding L, Ren G, Wang Z, Mani S. Plant flavonol isorhamnetin attenuates chemically induced inflammatory bowel disease via a PXR-dependent pathway. J. Nutr. Biochem. 2014;25:923–933. doi: 10.1016/j.jnutbio.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou C, Verma S, Blumberg B. The steroid and xenobiotic receptor (SXR), beyond xenobiotic metabolism. Nucl. Recept. Signal. 2009;7:e001. doi: 10.1621/nrs.07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wada T, Gao J, Xie W. PXR and CAR in energy metabolism. Trends Endocrinol. Metab. 2009;20:273–279. doi: 10.1016/j.tem.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Rysa J, Buler M, Savolainen MJ, Ruskoaho H, Hakkola J, Hukkanen J. Pregnane X receptor agonists impair postprandial glucose tolerance. Clin. Pharmacol. Ther. 2013;93:556–563. doi: 10.1038/clpt.2013.48. [DOI] [PubMed] [Google Scholar]
- 21.Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, Fleet JC, Kortagere S, Mukherjee P, Fasano A, Le Ven J, Nicholson JK, Dumas ME, Khanna KM, Mani S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mani S, Dou W, Redinbo MR. PXR antagonists and implication in drug metabolism. Drug Metab. Rev. 2013;45:60–72. doi: 10.3109/03602532.2012.746363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukherjee S, Mani S. Orphan nuclear receptors as targets for drug development. Pharm. Res. 2010;27:1439–1468. doi: 10.1007/s11095-010-0117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Squires EJ, Sueyoshi T, Negishi M. Cytoplasmic localization of pregnane X receptor and ligand-dependent nuclear translocation in mouse liver. J. Biol. Chem. 2004;279:49307–49314. doi: 10.1074/jbc.M407281200. [DOI] [PubMed] [Google Scholar]
- 25.Masuyama H, Suwaki N, Tateishi Y, Nakatsukasa H, Segawa T, Hiramatsu Y. The pregnane X receptor regulates gene expression in a ligand- and promoter-selective fashion. Mol. Endocrinol. 2005;19:1170–1180. doi: 10.1210/me.2004-0434. [DOI] [PubMed] [Google Scholar]
- 26.Goodwin B, Redinbo MR, Kliewer SA. Regulation of cyp3a gene transcription by the pregnane x receptor. Annu. Rev. Pharmacol. Toxicol. 2002;42:1–23. doi: 10.1146/annurev.pharmtox.42.111901.111051. [DOI] [PubMed] [Google Scholar]
- 27.Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, Schuetz EG, Kim RB. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat. Med. 2003;9:220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 28.Tian Y. Epigenetic regulation of pregnane X receptor activity. Drug Metab. Rev. 2013;45:166–172. doi: 10.3109/03602532.2012.756012. [DOI] [PubMed] [Google Scholar]
- 29.Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor bymicro-RNA affects the expression of cytochrome P450 3A4. J. Biol. Chem. 2008;283:9674–9680. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- 30.Rana R, Coulter S, Kinyamu H, Goldstein JA. RBCK1 an E3 ubiquitin ligase, interacts with and ubiquinates the human pregnane X receptor. Drug Metab. Dispos. 2013;41:398–405. doi: 10.1124/dmd.112.048728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staudinger JL, Xu C, Biswas A, Mani S. Post-translational modification of pregnane x receptor. Pharmacological research: the official journal of the Italian Pharmacological Society. 2011;64:4–10. doi: 10.1016/j.phrs.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doricakova A, Novotna A, Vrzal R, Pavek P, Dvorak Z. The role of residues T248, Y249 and T422 in the function of human pregnane X receptor. Arch. Toxicol. 2013;87:291–301. doi: 10.1007/s00204-012-0937-9. [DOI] [PubMed] [Google Scholar]
- 33.Elias A, High AA, Mishra A, Ong SS, Wu J, Peng J, Chen T. Identification and characterization of phosphorylation sites within the pregnane X receptor protein. Biochem. Pharmacol. 2014;87:360–370. doi: 10.1016/j.bcp.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smutny T, Mani S, Pavek P. Post-translational and post-transcriptional modifications of pregnane X receptor (PXR) in regulation of the cytochrome P450 superfamily. Curr. Drug Metab. 2013;14:1059–1069. doi: 10.2174/1389200214666131211153307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ong SS, Goktug AN, Elias A, Wu J, Saunders D, Chen T. Stability of the human pregnane X receptor is regulated by E3 ligase UBR5 and serine/threonine kinase DYRK2. Biochem. J. 2014;459:193–203. doi: 10.1042/BJ20130558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugatani J, Uchida T, Kurosawa M, Yamaguchi M, Yamazaki Y, Ikari A, Miwa M. Regulation of pregnane X receptor (PXR) function and UGT1A1 gene expression by posttranslational modification of PXR protein. Drug Metab. Dispos. 2012;40:2031–2040. doi: 10.1124/dmd.112.046748. [DOI] [PubMed] [Google Scholar]
- 37.Hu G, Xu C, Staudinger JL. Pregnane X receptor Is SUMOylated to repress the inflammatory response. J. Pharmacol. Exp. Ther. 2010;335:342–350. doi: 10.1124/jpet.110.171744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popov VM, Wang C, Shirley LA, Rosenberg A, Li S, Nevalainen M, Fu M, Pestell RG. The functional significance of nuclear receptor acetylation. Steroids. 2007;72:221–230. doi: 10.1016/j.steroids.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Tian L, Popov VM, Pestell RG. Acetylation and nuclear receptor action. J. Steroid Biochem. Mol. Biol. 2011;123:91–100. doi: 10.1016/j.jsbmb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu S-Y, Chiang C-M, Veenstra TD. FXR Acetylation Is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung HH, Sze SK, Tay AS, Lin VC. Acetylation at lysine 183 of progesterone receptor by p300 accelerates DNA binding kinetics and transactivation of direct target genes. J. Biol. Chem. 2014;289:2180–2194. doi: 10.1074/jbc.M113.517896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biswas A, Pasquel D, Tyagi RK, Mani S. Acetylation of pregnane X receptor protein determines selective function independent of ligand activation. Biochem. Biophys. Res. Commun. 2011;406:371–376. doi: 10.1016/j.bbrc.2011.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodwin B, Hodgson E, Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol. Pharmacol. 1999;56:1329–1339. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- 44.Geick A, Eichelbaum M, Burk O. Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J. Biol. Chem. 2001;276:14581–14587. doi: 10.1074/jbc.M010173200. [DOI] [PubMed] [Google Scholar]
- 45.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 47.Wallace BD, Betts L, Talmage G, Pollet RM, Holman NS, Redinbo MR. Structural and functional analysis of the human nuclear xenobiotic receptor PXR in complex with RXRalpha. J. Mol. Biol. 2013;425:2561–2577. doi: 10.1016/j.jmb.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR-nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 50.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 51.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:10. [Google Scholar]
- 52.Essmann U, Perera L, Berkowitz ML. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:17. [Google Scholar]
- 53.Vavrova A, Vrzal R, Dvorak Z. A nonradioactive electrophoretic mobility shift assay for measurement of pregnane X receptor binding activity to CYP3A4 response element. Electrophoresis. 2013;34:1863–1868. doi: 10.1002/elps.201300079. [DOI] [PubMed] [Google Scholar]
- 54.Kadiyala V, Smith CL. Minireview: the versatile roles of lysine deacetylases in steroid receptor signaling. Molecular Endocrinology (Baltimore, Md.) 2014;28:607–621. doi: 10.1210/me.2014-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore RL, Dai Y, Faller DV. Sirtuin 1 (SIRT1) and steroid hormone receptor activity in cancer. J. Endocrinol. 2012;213:37–48. doi: 10.1530/JOE-11-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodgers J, Lerin C, Gerharthines Z, Puigserver P. Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugden MC, Caton PW, Holness MJ. PPAR control: it's SIRTainly as easy as PGC. J. Endocrinol. 2010;204:93–104. doi: 10.1677/JOE-09-0359. [DOI] [PubMed] [Google Scholar]
- 58.Yang T, Fu M, Pestell R, Sauve AA. SIRT1 and endocrine signaling. Trends Endocrinol. Metab. 2006;17:186–191. doi: 10.1016/j.tem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 59.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 60.Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, Accili D. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helsen C, Kerkhofs S, Clinckemalie L, Spans L, Laurent M, Boonen S, Vanderschueren D, Claessens F. Structural basis for nuclear hormone receptor DNA binding. Mol. Cell. Endocrinol. 2012;348:411–417. doi: 10.1016/j.mce.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 62.Cheng Y, Redinbo MR. Activation of the human nuclear xenobiotic receptor PXR by the reverse transcriptase-targeted anti-HIV drug PNU-142721. Protein Sci. 2011;20:1713–1719. doi: 10.1002/pro.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, Wisely GB, Lambert MH, Kliewer SA, Redinbo MR. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol. Endocrinol. 2005;19:1125–1134. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- 64.Khan JA, Camac DM, Low S, Tebben AJ, Wensel DL, Wright MC, Su J, Jenny V, Gupta RD, Ruzanov M, Russo KA, Bell A, An Y, Bryson JW, Gao M, Gambhire P, Baldwin ET, Gardner D, Cavallaro CL, Duncia JV, Hynes J., Jr Developing adnectins that target SRC co-activator binding to PXR: a structural approach toward understanding promiscuity of PXR. J. Mol. Biol. 2015;427:924–942. doi: 10.1016/j.jmb.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 65.Xue Y, Chao E, Zuercher WJ, Willson TM, Collins JL, Redinbo MR. Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism. Bioorg. Med. Chem. 2007;15:2156–2166. doi: 10.1016/j.bmc.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watkins RE, Maglich JM, Moore LB, Wisely GB, Noble SM, Davis-Searles PR, Lambert MH, Kliewer SA, Redinbo MR. 2.1 A crystal structure of human PXR in complex with the St. John's wort compound hyperforin. Biochemistry. 2003;42:1430–1438. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- 67.Tian L, Wang C, Hagen FK, Gormley M, Addya S, Soccio R, Casimiro MC, Zhou J, Powell MJ, Xu P, Deng H, Sauve AA, Pestell RG. Acetylation-defective mutant of Ppargamma is associated with decreased lipid synthesis in breast cancer cells. Oncotarget. 2014;5:7303–7315. doi: 10.18632/oncotarget.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 69.Arbely E, Natan E, Brandt T, Allen MD, Veprintsev DB, Robinson CV, Chin JW, Joerger AC, Fersht AR. Acetylation of lysine 120 of p53 endows DNA-binding specificity at effective physiological salt concentration. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8251–8256. doi: 10.1073/pnas.1105028108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsuzaki H. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fujimoto H, Higuchi M, Koike M, Ode H, Pinak M, Bunta JK, Nemoto T, Sakudoh T, Honda N, Maekawa H, Saito K, Tsuchida K. A possible overestimation of the effect of acetylation on lysine residues in KQ mutant analysis. J. Comput. Chem. 2012;33:239–246. doi: 10.1002/jcc.21956. [DOI] [PubMed] [Google Scholar]
- 72.Nagy L, Schwabe JW. Mechanism of the nuclear receptor molecular switch. Trends Biochem. Sci. 2004;29:317–324. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 73.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 74.Lonard DM, O'Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol. Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 75.Lonard DM, O'Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat. Rev. Endocrinol. 2012;8:598–604. doi: 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J, Li Q. Life and death of transcriptional co-activator p300. Epigenetics. 2011;6:957–961. doi: 10.4161/epi.6.8.16065. [DOI] [PubMed] [Google Scholar]
- 78.Smith RP, Eckalbar WL, Morrissey KM, Luizon MR, Hoffmann TJ, Sun X, Jones SL, Force Aldred S, Ramamoorthy A, Desta Z, Liu Y, Skaar TC, Trinklein ND, Giacomini KM, Ahituv N. Genome-wide discovery of drug-dependent human liver regulatory elements. PLoS Genet. 2014;10:e1004648. doi: 10.1371/journal.pgen.1004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buler M, Aatsinki SM, Skoumal R, Hakkola J. Energy sensing factors PGC-1alpha and SIRT1 modulate PXR expression and function. Biochem. Pharmacol. 2011;82:2008–2015. doi: 10.1016/j.bcp.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 80.Yang X-J. Multisite protein modification and intramolecular signaling. Oncogene. 2004;24:1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- 81.van Beekum O, Fleskens V, Kalkhoven E. Posttranslational modifications of PPAR-gamma: fine-tuning the metabolic master regulator. Obesity (Silver Spring) 2009;17:213–219. doi: 10.1038/oby.2008.473. [DOI] [PubMed] [Google Scholar]
- 82.Kim DH, Xiao Z, Kwon S, Sun X, Ryerson D, Tkac D, Ma P, Wu SY, Chiang CM, Zhou E, Xu HE, Palvimo JJ, Chen LF, Kemper B, Kemper JK. A dysregulated acetyl/SUMO switch of FXR promotes hepatic inflammation in obesity. Embo J. 2015;34:184–199. doi: 10.15252/embj.201489527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zimber A, Gespach C. Bile acids and derivatives, their nuclear receptors FXR, PXR and ligands: role in health and disease and their therapeutic potential. Anticancer Agents Med Chem. 2008;8:540–563. doi: 10.2174/187152008784533008. [DOI] [PubMed] [Google Scholar]
- 84.Treuter E, Venteclef N. Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim. Biophys. Acta. 2011;1812:909–918. doi: 10.1016/j.bbadis.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 85.Cui W, Sun M, Galeva N, Williams TD, Azuma Y, Staudinger JL. SUMOylation and ubiquitylation circuitry controls pregnane X receptor biology in hepatocytes. Drug Metab. Dispos. 2015 doi: 10.1124/dmd.115.065201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ghoneim RH, Ngo Sock ET, Lavoie JM, Piquette-Miller M. Effect of a high-fat diet on the hepatic expression of nuclear receptors and their target genes: relevance to drug disposition. Br. J. Nutr. 2015:1–10. doi: 10.1017/S0007114514003717. [DOI] [PubMed] [Google Scholar]
- 87.Russell MJ, Martin W. The rocky roots of the acetyl-CoA pathway. Trends Biochem. Sci. 2004;29:358–363. doi: 10.1016/j.tibs.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 88.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scott I. Regulation of cellular homoeostasis by reversible lysine acetylation. Essays Biochem. 2012;52:13–22. doi: 10.1042/bse0520013. [DOI] [PubMed] [Google Scholar]
- 90.Xiong Y, Guan KL. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell Biol. 2012;198:155–164. doi: 10.1083/jcb.201202056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shi L, Tu BP. Protein acetylation as a means to regulate protein function in tune with metabolic state. Biochem. Soc. Trans. 2014;42:1037–1042. doi: 10.1042/BST20140135. [DOI] [PubMed] [Google Scholar]
- 92.Oosterveer MH, Schoonjans K. Hepatic glucose sensing and integrative pathways in the liver. Cell. Mol. Life Sci. 2014;71:1453–1467. doi: 10.1007/s00018-013-1505-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 94.Spruiell K, Jones DZ, Cullen JM, Awumey EM, Gonzalez FJ, Gyamfi MA. Role of human pregnane X receptor in high fat diet-induced obesity in pre-menopausal female mice. Biochem. Pharmacol. 2014;89:399–412. doi: 10.1016/j.bcp.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.He J, Gao J, Xu M, Ren S, Stefanovic-Racic M, O'Doherty RM, Xie W. PXR ablation alleviates diet-induced and genetic obesity and insulin resistance in mice. Diabetes. 2013;62:1876–1887. doi: 10.2337/db12-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Navaratnarajah P, Steele BL, Redinbo MR, Thompson NL. Rifampicin-independent interactions between the pregnane X receptor ligand binding domain and peptide fragments of coactivator and corepressor proteins. Biochemistry. 2012;51:19–31. doi: 10.1021/bi2011674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Watkins RE, Davis-Searles PR, Lambert MH, Redinbo MR. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J. Mol. Biol. 2003;331:815–828. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- 99.Kim SW, Hasanuzzaman M, Cho M, Heo YR, Ryu MJ, Ha NY, Park HJ, Park HY, Shin JG. CK2-mediated phosphorylation of Hsp90beta as a novel mechanism of rifampin-induced MDR1 expression. J. Biol. Chem. 2015 doi: 10.1074/jbc.M114.624106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Berrabah W, Aumercier P, Lefebvre P, Staels B. Control of nuclear receptor activities in metabolism by post-translational modifications. FEBS Lett. 2011;585:1640–1650. doi: 10.1016/j.febslet.2011.03.066. [DOI] [PubMed] [Google Scholar]
- 102.Prabakaran S, Lippens G, Steen H, Gunawardena J. Post-translational modification: nature's escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012;4:565–583. doi: 10.1002/wsbm.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Benayoun BA, Veitia RA. A post-translational modification code for transcription factors: sorting through a sea of signals. Trends Cell Biol. 2009;19:189–197. doi: 10.1016/j.tcb.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 104.Li Y, Wong K, Giles A, Jiang J, Lee JW, Adams AC, Kharitonenkov A, Yang Q, Gao B, Guarente L, Zang M. Hepatic SIRT1 attenuates hepatic steatosis and controls energy balance in mice by inducing fibroblast growth factor 21. Gastroenterology. 2014;146:539–549. e537. doi: 10.1053/j.gastro.2013.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu TF, McCall CE. Deacetylation by SIRT1 reprograms inflammation and cancer. Genes & Cancer. 2013;4:135–147. doi: 10.1177/1947601913476948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Montie HL, Pestell RG, Merry DE. SIRT1 modulates aggregation and toxicity through deacetylation of the androgen receptor in cell models of SBMA. J. Neurosci. Off. J. Soc. Neurosci. 2011;31:17425–17436. doi: 10.1523/JNEUROSCI.3958-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Faghihzadeh F, Hekmatdoost A, Adibi P. Resveratrol and liver: a systematic review. Journal of research in medical sciences: the official journal of Isfahan University of Medical Sciences. 2015;20:797–810. doi: 10.4103/1735-1995.168405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chachay VS, Macdonald GA, Martin JH, Whitehead JP, O'Moore-Sullivan TM, Lee P, Franklin M, Klein K, Taylor PJ, Ferguson M, Coombes JS, Thomas GP, Cowin GJ, Kirkpatrick CM, Prins JB, Hickman IJ. Resveratrol does not benefit patients with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2014;12:2092–2103. e2091–e2096. doi: 10.1016/j.cgh.2014.02.024. [DOI] [PubMed] [Google Scholar]
- 109.Chen S, Zhao X, Ran L, Wan J, Wang X, Qin Y, Shu F, Gao Y, Yuan L, Zhang Q, Mi M. Resveratrol improves insulin resistance, glucose and lipid metabolism in patients with non-alcoholic fatty liver disease: a randomized controlled trial. Dig. Liver Dis. 2015;47:226–232. doi: 10.1016/j.dld.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 110.Heeboll S, Thomsen KL, Clouston A, Sundelin EI, Radko Y, Christensen LP, Ramezani-Moghadam M, Kreutzfeldt M, Pedersen SB, Jessen N, Hebbard L, George J, Gronbaek H. Effect of resveratrol on experimental non-alcoholic steatohepatitis. Pharmacol. Res. 2015;95–96:34–41. doi: 10.1016/j.phrs.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 111.Heeboll S, El-Houri RB, Hellberg YE, Haldrup D, Pedersen SB, Jessen N, Christensen LP, Gronbaek H. The effect of resveratrol on experimental nonalcoholic fatty liver disease depends on severity of pathology and timing of treatment. J. Gastroenterol. Hepatol. 2015 doi: 10.1111/jgh.13151. [DOI] [PubMed] [Google Scholar]
- 112.Wahlang B, Song M, Beier JI, Cameron Falkner K, Al-Eryani L, Clair HB, Prough RA, Osborne TS, Malarkey DE, Christopher States J, Cave MC. Evaluation of Aroclor 1260 exposure in a mouse model of diet-induced obesity and nonalcoholic fatty liver disease. Toxicol. Appl. Pharmacol. 2014;279:380–390. doi: 10.1016/j.taap.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ghoneim RH, Ngo Sock ET, Lavoie JM, Piquette-Miller M. Effect of a high-fat diet on the hepatic expression of nuclear receptors and their target genes: relevance to drug disposition. Br. J. Nutr. 2015;113:507–516. doi: 10.1017/S0007114514003717. [DOI] [PubMed] [Google Scholar]
- 114.Grozinger CM, Schreiber SL. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 115.Yang XJ, Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 2005;25:2873–2884. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karagianni P, Wong J. HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene. 2007;26:5439–5449. doi: 10.1038/sj.onc.1210612. [DOI] [PubMed] [Google Scholar]
- 117.Ekins S, Reschly EJ, Hagey LR, Krasowski MD. Evolution of pharmacologic specificity in the pregnane X receptor. BMC Evol. Biol. 2008;8:103. doi: 10.1186/1471-2148-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mathas M, Burk O, Qiu H, Nusshag C, Godtel-Armbrust U, Baranyai D, Deng S, Romer K, Nem D, Windshugel B, Wojnowski L. Evolutionary history and functional characterization of the amphibian xenosensor CAR. Molecular endocrinology (Baltimore, Md.) 2012;26:14–26. doi: 10.1210/me.2011-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Handschin C, Podvinec M, Meyer UA. CXR, a chicken xenobiotic-sensing orphan nuclear receptor, is related to both mammalian pregnane X receptor (PXR) and constitutive androstane receptor (CAR) Proc. Natl. Acad. Sci. U. S. A. 2000;97:10769–10774. doi: 10.1073/pnas.97.20.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.