Abstract
Geographical differences in the genetic diversity of Helicobacter pylori isolates were examined by analyzing rpoB sequences. An extremely high level of allelic diversity among H. pylori strains was found. The rpoB sequences of Asian and non-Asian (North and South American, European, and South African) strains were found to differ. An amino acid polymorphism (alanine and threonine RpoB types) was found at the 497th residue by deduced amino acid analysis. RpoB with a threonine residue (RpoBThr) was uniquely present in East Asian countries, and two-thirds of the H. pylori isolate population in this region was RpoBThr; however, this type was rare or absent in Western countries, where RpoBAla predominated. RpoBThr strains induced a much larger amount of interleukin-8, a chemokine that plays an important role in chronic inflammation, than RpoBAla strains in cultured MKN45 cells.
Helicobacter pylori is disproportionately acquired during childhood and persists in its host for life. H. pylori infection typically leads to chronic inflammation of the gastric mucosa, and this is accompanied by mucosal damage, including the loss of acid-secreting parietal cells and the development of mucous cell metaplasia (30). H. pylori carriers have a higher risk of gastric diseases like gastric cancer, which is the second most common malignancy worldwide, and is particularly common in East Asian countries, such as Korea and Japan (8, 22). However, H. pylori factors involved in gastric carcinogenesis are not well understood.
The presence of the cytotoxin-associated gene A (cagA) of H. pylori has been proposed to be an important risk factor for the development of H. pylori-mediated gastric cancer (7). Recently, it was suggested that Src homology 2-containing tyrosine phosphatase (SHP-2) is an intracellular target of CagA protein (9) and that the prevalent CagA type in East Asian countries binds more strongly to SHP-2, and thus induces more cellular morphological changes, than the CagA type prevalent in Western countries (10). Moreover, it has been suggested that this difference may be correlated with the striking difference in the incidence of gastric cancer in these two geographical areas (10). However, even though nearly 100% of Korean and Japanese isolates possess cagA and express the East Asian type of CagA, relatively few infected individuals develop peptic ulcer or gastric cancer (6). The reason for this remains unresolved (28).
Phylogenetic analysis based on amino acid sequences often provides more significant information than analysis based on the nucleotide sequences of protein-coding genes (8, 20). However, such analyses have not been performed in previous population studies with cagA, oipA, or other housekeeping genes (5, 17, 29, 31). Thus, to test the hypothesis that certain H. pylori strains in Asia are uniquely prone to cause chronic inflammation and metaplastic changes in the gastric mucosa, we studied the population structure of H. pylori isolates from several countries by analyzing rpoB sequences. This allowed us to analyze the H. pylori population by both nucleotide and amino acid sequence analyses. rpoB encodes the β-subunit of RNA polymerase and is a highly conserved housekeeping gene. Comparisons of rpoB sequences have previously been used for phylogenetic analysis and for the differential identification of bacteria (14, 15, 19, 34).
MATERIALS AND METHODS
Bacterial strains.
The DNAs of 535 clinical H. pylori isolates from 12 countries (Table 1) were analyzed; H. cinaedi was used as an outgroup. The strains, which were provided by D. Y. Graham, had been obtained from patients who had signed informed consent forms approved by institutional review boards in the United States.
TABLE 1.
Prevalence of H. pylori types on the basis of the rpoB amino acid, by country
| Geographical region and country | No. (%) of strains (n = 535)
|
|
|---|---|---|
| RpoBAla type | RpoBThr type | |
| East and Southeast Asia | 129 (32.4) | 269 (67.6) |
| Korea | 119 | 241 |
| Japan | 7 | 19 |
| Hong Kong | 1 | 3 |
| Taiwan | 0 | 4 |
| Thailand | 2 | 2 |
| North America | 77 (98.7) | 1 (1.3) |
| United States | 72 | 1a |
| Canada | 5 | 0 |
| South America | 44 (93.6) | 3 (6.4) |
| Colombia | 42 | 3 |
| Brazil | 2 | 0 |
| Europe | 8 (100) | 0 (0.0) |
| France | 4 | 0 |
| Italy | 4 | 0 |
| South Africa | 4 (100) | 0 (0.0) |
| Total | 262 | 273 |
The individual was born in Vietnam and immigrated to the United States.
Preparation of DNA.
H. pylori DNAs were extracted from cultured bacteria and gastric biopsy specimens by the bead beater-phenol extraction method (14). A loopful of a culture of each isolate was suspended in 200 μl of TEN buffer (10 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl [pH 8.0]) and placed in a 2.0-ml screw-cap microcentrifuge tube filled with 100 μl (packed volume) of glass beads (diameter, 0.1 mm; Biospec Products, Bartlesville, Okla.) and 100 μl of phenol-chloroform-isoamyl alcohol (25:24:1; catalog no. P-2069; Sigma Chemical Co.). To disrupt the bacteria, the tube was oscillated on a Mini-Bead Beater (Biospec Products) for 1 min, and to separate the phases the tube was centrifuged (2,300 × g, 15 min). The aqueous phase was then transferred to another clean tube, and 10 μl of 3 M sodium acetate and 250 μl of ice-cold ethanol were added. The mixture was kept at −20°C for 30 min to precipitate the DNA. The DNA pellet obtained was washed with 70% ethanol, dissolved in 60 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl [pH 8.0]), and used as the template for PCR. H. pylori DNAs from countries other than Korea were extracted by use of a commercially available kit (Qiagen, Inc., Hilden, Germany), according to the instructions of the manufacturer.
rpoB DNA amplification.
PCR was performed with forward primer HF (5′-ACTTTAACGCATGAAGATAT-3′) and reverse primer HR (5′-ATATTTTGACCTTCTGGGGT-3′) to amplify rpoB DNA (458 bp) containing the Rifr region (16). Template DNA (50 ng) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer, Daejeon, South Korea) containing 1 U of Taq DNA polymerase, each deoxynucleoside triphosphate at a concentration of 250 μl, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and gel loading dye. The volume was adjusted to 20 μl with distilled water. The reaction mixture was then subjected to 30 cycles of amplification (30 s at 94°C, 45 s at 52°C, and 45 s at 72°C), followed by a 5-min extension at 72°C (model 9600 thermocycler; Perkin-Elmer Cetus). The PCR products were electrophoresed on a 1.2% agarose gel and purified by using a QIAEX II gel extraction kit (Qiagen).
Nucleotide sequencing.
The nucleotide sequences (363 bp) of the purified PCR products were directly determined with forward and reverse primers, using an Applied Biosystems model 373A automatic sequencer and a BigDye Terminator Cycle Sequencing kit (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom). For the sequencing reaction, 60 ng of PCR-amplified DNA, 3.2 pmol of either the forward primer or the reverse primer, and 8 μl of BigDye Terminator RRmix (part no. 430315512114; Perkin-Elmer Applied Biosystems) were mixed and adjusted to a final volume of 20 μl with distilled water. The reaction was run with 5% (vol/vol) dimethyl sulfoxide for 30 cycles of 15 s at 95°C, 10 s at 50°C, and 4 min at 60°C. Both strands were sequenced as a cross-check. PCR and nucleotide sequencing of cagA were also performed as described previously (10, 32).
Sequence alignment and phylogenetic tree.
The partial rpoB sequences (363 bp) were aligned by using the multiple-alignment algorithm in the MegAlign program (Windows version 3.12e; DNASTAR, Madison, Wis.) and the Clustal X program (26), and the amino acids were deduced by using the MegAlign program. On the basis of the aligned sequences, phylogenetic trees were constructed by the neighbor-joining method (21) and the parsimony methods in the PAUP package (25). The rpoB sequence of H. cinaedi was determined simultaneously and was used as an outgroup. Bootstrap values were evaluated from 1,000 replications. Homoplasy test (18) was performed by using the HOMOPLASY program, and split decomposition was analyzed by using the SPLITSTREE program (version 3.1; http://www.mlst.net).
Typing of H. pylori from gastric biopsy specimens.
By using computer-aided analysis (MapDraw package, Windows version 3.12e; DNASTAR) of the rpoB DNA sequences determined as described above, BsmFI was found to distinguish two H. pylori types. Gastric biopsy specimens were obtained from the antrums of the stomachs of 200 Korean patients at gastroscopy. The samples were separately used for genotyping by PCR restriction analysis. H. pylori was primarily detected and identified by the rapid urease test, rpoB PCR restriction analysis, and glmM PCR (16). DNAs were extracted from the biopsy specimens as described above and used for rpoB typing. Ten microliters of the rpoB PCR products was transferred to a fresh microcentrifuge tube and digested with BsmFI (R0572S; New England Biolabs, Beverly, Mass.), according to the supplier's instruction.
IL-8 assay.
Interleukin-8 (IL-8) levels in the culture supernatants were measured as described previously (33). Briefly, MKN45 cells were grown to preconfluent monolayers (5 × 105 cells/ml) and then plated into 24-well plates (1 × 105 cells/well) and cultured for 2 days (∼5 × 105/ml for each well). Twenty strains of each type of H. pylori isolated from Korean patients were cultured on brucella agar (BBL Microbiology Systems, Cockeysville, Md.) plates containing 10% fetal calf serum in a microaerobic atmosphere at 37°C for 48 h and added to the MKN45 cells (ratio of H. pylori cells to MKN45 cells, 100:1), and the mixture was then incubated at 37°C for 20 h in 5% CO2. IL-8 levels were then measured by an enzyme-linked immunosorbent assay (R&D Systems, Minneapolis, Minn.), as instructed by the manufacturer. Values were plotted by using the Box plot program (SigmaPlot 2000 for Windows, version 6.00; SPSS Inc., Chicago, Ill.) to show median values and confidence intervals. Statistical analysis of the IL-8 assay results was performed by the Mann-Whitney rank sum test. A P value of less than 0.05 was accepted as statistically significant.
Nucleotide sequence accession numbers.
The rpoB sequences of H. pylori strains 26695 and J99 were retrieved from GenBank (accession nos. AE000625 and AE001540, respectively).
RESULTS
rpoB genotype.
rpoB DNA (458 bp) containing a highly conserved region was amplified (16) and sequenced from 535 clinical H. pylori isolates obtained from 12 countries. Because many countries were represented by small numbers of isolates and, thus, may not represent the predominant strains of the particular geographical areas, we grouped strains by large geographical regions (Table 1). The rpoB sequences (363 bp) determined were aligned for the Homoplasy test, split decomposition analysis, and phylogenetic study. The Homoplasy index of H. pylori was found to be 0.519, which is higher than those reported for other bacteria (18, 23). This suggests frequent interstrain recombinations among the H. pylori population. Split decomposition analysis of rpoB showed a network topology and star phylogeny (data not shown), which are consistent with a recombinational population structure.
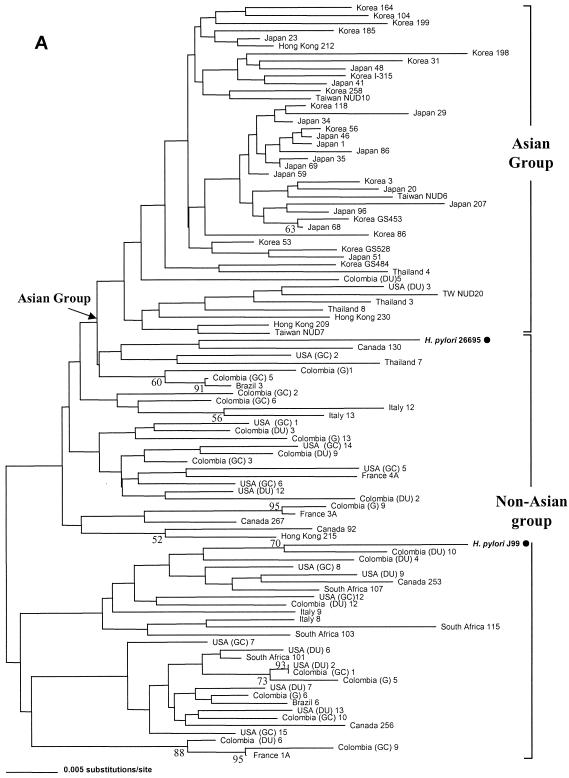
Another interesting finding was that although it was not robustly supported by bootstrap values, the H. pylori population could be separated into two major groups by nucleotide sequence analysis. In accord with previous reports on genotype analysis (1, 5, 8, 12, 13, 29, 32), the geographical distributions of these two groups were in agreement with their phylogenetic relationships. One group was termed the Asian group, and the other was termed the non-Asian group, which was mostly composed of Western H. pylori strains (North and South American, European, and South African strains) and included strains 26695 and J99 (Fig. 1A). Although marked genetic heterogeneity was observed, the clustering of H. pylori strains into different groups by geographical regions by rpoB analysis was also compatible with the findings of other studies (5, 17, 29, 31).
FIG. 1.
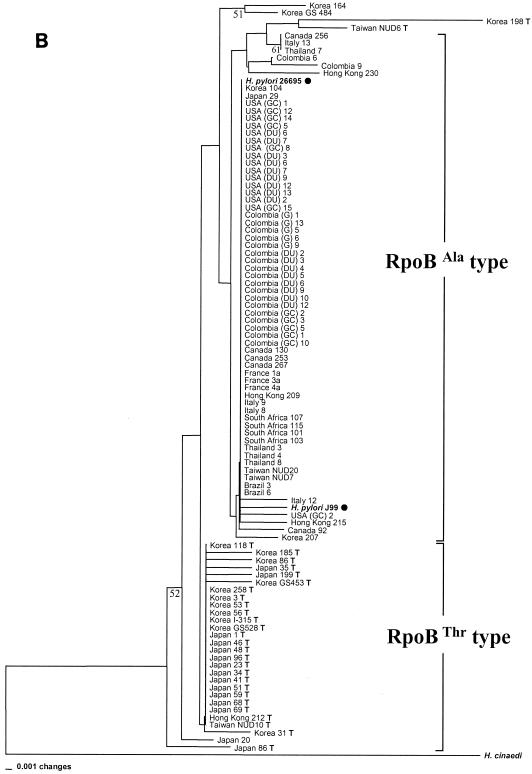
Phylogenetic relationships of 100 H. pylori isolates inferred from partial rpoB DNA sequences (A) and RpoB amino acid sequences (B). The H. pylori population was separated into an Asian group, to which most of the Asian strains belonged, and a non-Asian group, which was mainly composed of Western strains (North and South American, European, and South African strains), including strains H. pylori 26695 and H. pylori J99, by nucleotide sequence analysis (A). Two large groups (RpoBAla and RpoBThr) in the amino acid tree (B) were attributed to the identity of the 497th residue of each strain, which is either alanine or threonine. RpoBThr strains have the suffix T. The tree was constructed by the neighbor-joining method in the PAUP package. The bootstrap values presented at the corresponding branches were evaluated from 1,000 replications, and values less than 50% are not indicated.
Deduced amino acid.
For protein-coding genes, phylogenetic relationships based on amino acid sequences are often more significant than those based on nucleotide sequences (8, 20). Population genetics data based on nucleotide sequences are often inadequate for the study of protein-coding genes because variations are usually found at the third bases of codons (the wobble position), and these variations do not affect the amino acid sequence and thus result in synonymous substitutions (3, 8). Our analysis of the amino acid sequence of rpoB shows that the H. pylori strains could also be separated into another two large groups on the tree based on the amino acid sequences (Fig. 1B), and this was attributed to the identity of the 497th residue of each group, which was either alanine (GCT) or threonine (ACT). These groups were designated types RpoBAla and RpoBThr, respectively. Of the 398 H. pylori isolates from Asian countries, 269 strains (67.6%) were of the RpoBThr type and 129 strains (32.4%) were of the RpoBAla type. Interestingly, almost all H. pylori strains (133 strains [97.1%]) from Western countries, including H. pylori 26695 and J99, whose sequences were retrieved from GenBank, were of the RpoBAla type. Only three Colombian strains and one North American (U.S.) strain (2.9%) were of the RpoBThr type (Table 1). It was of an interest to find that the American individual in the analysis described above was born in Vietnam and had immigrated to the United States at age 7 years. These results indicate that the RpoBThr type H. pylori strains exist almost exclusively in East and Southeast Asia. In other words, the specific amino acid polymorphism at position 497 of RpoB strongly suggests that recombinant strains with different phenotypes may emerge in different human populations and geographical regions.
We also compared the clinical information with the RpoBAla and RpoBThr types of 394 strains from East Asian patients (Table 2). Because many strains were collected retrospectively and the clinical information for many patients was not available, it is not easy to conclude that the two RpoB types correlate with gastrointestinal disease groups. However, the distributions of the two RpoB types were not significantly different among four disease groups (gastric cancer, duodenal ulcer, gastritis, and others, excluding unknown) (P > 0.05 by chi-square test).
TABLE 2.
Distributions of the 394 East Asiana patients from whom the H. pylori strains were isolated
| Clinical status | No (%) of strains
|
||
|---|---|---|---|
| RpoBThr type | RpoBAla type | Subtotal | |
| Gastric cancer | 88 (66.2) | 45 (33.8) | 133 |
| Duodenal ulcer | 24 (75.0) | 8 (25.0) | 32 |
| Gastritis | 59 (66.3) | 30 (33.7) | 89 |
| Benign gastric ulcer | 17 (56.7) | 13 (43.3) | 30 |
| Lymphoid hyperplasia | 2 (100) | 0 (0.0) | 2 |
| Within normal limitb | 3 (75.0) | 1 (25.0) | 4 |
| Unknown | 70 (67.3) | 34 (32.7) | 104 |
| Subtotal | 263 (66.8) | 131 (33.2) | |
Korea, Japan, Hong Kong, and Taiwan.
No pathological finding was observed.
Typing of H. pylori strains in biopsy specimens.
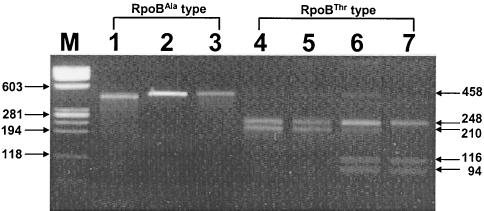
A similar result was obtained when the H. pylori strains in gastric biopsy specimens were typed by restriction fragment length polymorphism analysis. A restriction site, BsmFI, that distinguished between the RpoBAla and RpoBThr types was found on the basis of the rpoB sequences. Only the PCR product of RpoBThr had one or two restriction sites for BsmFI and produced two (248- and 210-bp) or three (248-, 116-, and 94-bp) DNA fragments. DNAs of the RpoBAla type, on the other hand, were not cleaved (458 bp) (Fig. 2). Of the 200 Korean biopsy specimens in which H. pylori was detected and identified, 136 samples (68%) contained the RpoBThr type and the remainder (64 samples [32%]) contained the RpoBAla type.
FIG. 2.
Differentiation of RpoBThr and RpoBAla type H. pylori strains by PCR-restriction fragment length polymorphism analysis (with BsmFI) of rpoB DNA. Amplified rpoB DNAs (458 bp) of H. pylori were digested with BsmFI and electrophoresed in a 3% agarose gel. DNAs from the RpoBThr types were digested (lanes 4 and 5 [248 and 210 bp] and lanes 6 and 7 [248, 116, and 94 bp]), while DNAs from the RpoBAla types were not (lanes 1 to 3 [458 bp]). Lane M, size marker (φX174 replicative-form DNA digested with HaeIII). The numbers next to the gels are in base pairs.
IL-8 production measured in MKN45 cells cocultured with both types of H. pylori.
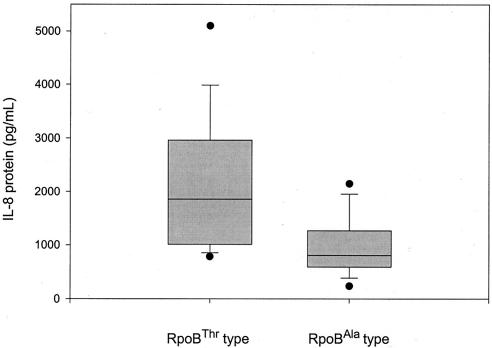
To determine the biological significance of the amino acid polymorphism with respect to the pathogenesis of H. pylori infection, IL-8 production was measured in MKN45 cells cocultured with either type of H. pylori strain, which all turned out to have the East Asian CagA type. IL-8 production reflects a type of H. pylori-epithelial cell interaction. It was noteworthy to find that the amounts of IL-8 secreted by MKN45 cells infected with RpoBThr strains were significantly higher (P < 0.05) than the amounts secreted by cells infected with RpoBAla strains (Fig. 3).
FIG. 3.
Secretion of IL-8 from MKN45 cells cocultured with the two different H. pylori types, RpoBThr (n = 20) and RpoBAla (n = 20). All the H. pylori strains were cagA positive (East Asian-type CagA). The amounts of IL-8 protein excreted by the cells was measured by enzyme-linked immunosorbent assay. The levels of IL-8 secreted by RpoBThr-type infected cells were found to be significantly higher (P < 0.05 by the Mann-Whitney rank sum test) than the levels secreted by RpoBAla-type infected cells.
DISCUSSION
H. pylori infection is a major cause of gastritis and is considered an important risk factor for stomach cancer. However, the presence of H. pylori alone does not sufficiently explain the striking difference in the geographical incidences of gastric cancer. While the association of the cag pathogenicity islands with an increased risk of gastric cancer was proved, it cannot explain the differences in disease presentations caused by the different CagA H. pylori types in different geographical areas, namely, Eastern and Western countries. Furthermore, in Korea and Japan essentially all isolates are positive for the cagA type that encodes the East Asian CagA type. Thus, to test the hypothesis that a more virulent H. pylori population exists in East Asia, we analyzed H. pylori isolates on the basis of the sequence of the protein-coding gene, rpoB, which encodes the β-subunit of DNA-dependent RNA polymerase.
DNA-dependent RNA polymerase is a principal enzyme in the transcriptional process and of many regulatory pathways that control gene expression in living organisms. It is evolutionarily conserved in sequence, structure, and function from bacteria to humans (19, 24, 34). A high level of genetic diversity among H. pylori strains has also been observed by 16S ribosomal DNA sequence analysis (27). However, as a protein-coding gene, rpoB provided several advantages over 16S ribosomal DNA for phylogenetic analysis, which offered only a moderate power to discriminate or distinguish between species and strains (14, 15, 19). With rpoB the analysis can be performed at both the nucleotide and the amino acid sequence levels and the H. pylori strains can be grouped according to both the amino acid and the nucleotide sequences. The amino acid sequence-based grouping of H. pylori led to the discovery of a novel RpoB polymorphism (RpoBAla-RpoBThr) at residue 497. The prevalence of the RpoBThr type was notable only in H. pylori isolates from East and Southeast Asia. However, because the H. pylori strains used for rpoB analysis were collected retrospectively and the clinical information for many patients was unknown, we are very cautious not to conclude that a correlation between an RpoB type with a certain gastrointestinal disease group exists.
Gastric cancer is generally thought to arise through a series of mucosal changes leading to atrophic gastritis caused by chronic H. pylori infection. Chronic H. pylori infection causes abnormal changes in the gastric mucosa, such as severe infiltration of the lamina propria by polymorphonuclear and mononuclear cells, and increases in epithelial cell proliferation, resulting in atrophic gastritis and focal intestinal metaplasia in an animal model (11). The proinflammatory chemokine IL-8 plays an important role in H. pylori-related inflammation by recruiting neutrophils and lymphocytes into the gastric mucosa (2, 4). We measured the IL-8 levels in cultured MKN45 cells after H. pylori infection. Of interest, strains polymorphic at the 497th residue induced different amounts of IL-8 secretion, with strains with the RpoBThr type inducing more IL-8 secretion than those with the RpoBAla type. This difference in levels of IL-8 secretion did not correlate with the recA group (group I and group II), as defined in a previous report (17; data not shown).
These data suggest that additional factors are responsible for enhanced virulence among H. pylori strains and may provide important clues to the question of why the incidence of H. pylori-induced clinical disease differs so markedly between East Asia and the West. While it is clear that the presence of the East Asian-type CagA cannot be solely credited with this difference in IL-8 induction, the specifics of the role of the RpoB polymorphism in IL-8 induction have yet to be explored. The RpoB polymorphism may affect the function of RNA polymerase. Although the presence of the RpoB polymorphisms correlated with the geographical locations of the isolates and with the levels of IL-8 induction in vitro, it did not correlate with clinical presentation. The question remains whether RpoB polymorphisms are directly involved in clinical outcomes or whether they are actually a marker linked to another factor responsible for increased virulence, independent of the polymorphism. Studies are planned to test the effects of the Thr→Ala substitution on IL-8 secretion in vitro to investigate whether the polymorphism is directly related to the induction of enhanced IL-8 secretion.
In conclusion, this H. pylori population study based on rpoB nucleotide sequences, analysis of their deduced amino acid sequences, and the IL-8 assay provides evidence that a polymorphism in RpoB may be related to the pathogenesis of H. pylori-associated gastric diseases. Because of different host-parasite interactions, we suggest that the classification of H. pylori strains according to the RpoB polymorphism should be an integral part of the study of H. pylori-mediated pathogenesis.
Acknowledgments
This study was supported by a grant from the Korea Heath 21 R&D Project, Ministry of Heath and Welfare, Republic of Korea (01-PJ1-PG3-20200-0057) and in part by the BK21 project for Medicine, Dentistry, and Pharmacy; and part of this study was supported by the Office of Research and Development Medical Research Service, U.S. Department of Veterans Affairs, and by Public Health Service grant DK56338, which funds the Texas Gulf Coast Digestive Disease Center.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S., S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Aihara, M., D. Tsuchimoto, H. Takizawa, A. Azuma, H. Wakebe, Y. Ohmoto, K. Imagawa, M. Kikuchi, N. Mukaida, and K. Matsushima. 1997. Mechanisms involved in Helicobacter pylori-induced interleukin-8 production by a gastric cancer cell line, MKN45. Infect. Immun. 65:3218-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 396:176-180. [DOI] [PubMed] [Google Scholar]
- 4.Ando, T., K. Kusugami, M. Ohsuga, K. Ina, M. Shinoda, T. Konagaya, T. Sakai, A. Imada, N. Kasuga, T. Nada, S. Ichiyama, and M. J. Blaser. 1998. Differential normalization of mucosal interleukin-8 and interleukin-6 activity after Helicobacter pylori eradication. Infect. Immun. 66:4742-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ando, T., R. M. Peek, D. Pride, S. M. Levine, T. Takata, Y. C. Lee, K. Kusugami, A. van der Ende, E. J. Kuipers, J. G. Kusters, and M. J. Blaser. 2002. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J. Clin. Microbiol. 40:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 2000. Linking Helicobacter pylori to gastric cancer. Nat. Med. 6:376-377. [DOI] [PubMed] [Google Scholar]
- 7.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A. Nomura. 1995. Infection with. Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111-2115. [PubMed] [Google Scholar]
- 8.Covacci, A., J. L. Telford, G. D. Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 9.Higashi, H., R. Tsutsumi, S. Muto, T. Sugiyama, T. Azuma, M. Asaka, and M. Hatakeyama. 2002. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295:683-686. [DOI] [PubMed] [Google Scholar]
- 10.Higashi, H., R. Tsutsumi, A. Fujita, S. Yamazaki, M. Asaka, T. Azuma, and M. Hatakeyama. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. USA 99:14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda, S., T. Fujioka, M. Tokieda, R. Satoh, A. Nishizono, and M. Nasu. 1998. Development of Helicobacter pylori-induced gastric carcinoma in Mongolian gerbils. Cancer Res. 58:4255-4259. [PubMed] [Google Scholar]
- 12.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatino, W. Su, Z. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcon, M. Lopez-Brea, G. Balakrish Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Wong, S. K. Lam, F. O. Olfat, T. Boren, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 14.Kim, B. J., S. H. Lee, M. A. Lyu, S. J. Kim, G. H. Bai, S. S. Kim, G. T. Chae, E. C. Kim, C. Y. Cha, and Y. H. Kook. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 37:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko, K. S., H. K. Lee, M. Y. Park, M. S. Park, K. H. Lee, S. Y. Woo, Y. J. Yun, and Y. H. Kook. 2002. Population genetic structure of Legionella pneumophila inferred from RNA polymerase gene (rpoB) and DotA gene (dotA) sequences. J. Bacteriol. 184:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim, C. Y., K. H. Lee, M. J. Cho, M. W. Chang, S. Y. Kim, N. H. Myong, W. K. Lee, K. H. Rhee, and Y. H. Kook. 2003. Detection of Helicobacter pylori in gastric mucosa of patients with gastroduodenal diseases by PCR-restriction analysis using the RNA polymerase gene (rpoB). J. Clin. Microbiol. 41:3387-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maggi Solcà, N., M. V. Bernasconi, C. Valsangiacomo, L. J. Van Doorn, and J. C. Piffaretti. 2001. Population genetics of Helicobacter pylori in the southern part of Switzerland analysed by sequencing of four housekeeping genes (atpD, glnA, scoB and recA), and by vacA, cagA, iceA and IS605 genotyping. Microbiology 147:693-707. [DOI] [PubMed] [Google Scholar]
- 18.Maynard Smith, J., and N. H. Smith. 1998. Detecting recombination from gene trees. Mol. Biol. Evol. 15:590-599. [DOI] [PubMed] [Google Scholar]
- 19.Mollet, C., M. Drancourt, and D. Raoult. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26:1005-1011. [DOI] [PubMed] [Google Scholar]
- 20.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 21.Saitou, N., and M. Nei. 1987. The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 22.Shiotani, A., Z. Z. Nurgalieva, Y. Yamaoka, and D. Y. Graham. 2000. Helicobacter pylori. Med. Clin. N. Am. 84:1125-1136. [DOI] [PubMed] [Google Scholar]
- 23.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sweetser, D., M. Nonet, and R. A. Young. 1987. A. prokaryotic and eukaryotic RNA polymerase have homologous core subunits. Proc. Natl. Acad. Sci. USA 84:1192-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swofford, D. L. 1999. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 26.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiveljung, A., K. Borch, J. Jonasson, S. Mardh, F. Petersson, and H. J. Monstein. 1998. Identification of Helicobacter in gastric biopsies by PCR based on 16S rDNA sequences: a matter of little significance for the prediction of H. pylori-associated gastritis? J. Med. Microbiol. 47:695-704. [DOI] [PubMed] [Google Scholar]
- 28.Uemura, N., S. Okamoto, S. Yamamoto, N. Matsumura, S. Yamaguchi, M. Yamakido, K. Taniyama, N. Sasaki, and R. J. Schlemper. 2001. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345:784-789. [DOI] [PubMed] [Google Scholar]
- 29.van der Ende, A., Z. J. Pan, A. Bart, R. W. van der Hulst, M. Feller, S. D. Xiao, G. N. Tytgat, and J. Dankert. 1998. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect. Immun. 66:1822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, T. C., and J. R. Goldenring. 2002. Inflammation intersection: gp130 balances gut irritation and stomach cancer. Nat. Med. 8:1080-1081. [DOI] [PubMed] [Google Scholar]
- 31.Yamaoka, Y., E. Orito, M. Mizokami, O. Gutierrez, N. Saitou, T. Kodama, M. S. Osato, J. G. Kim, F. C. Ramirez, V. Mahachai, and D. Y. Graham. 2002. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 517:180-184. [DOI] [PubMed] [Google Scholar]
- 32.Yamaoka, Y., T. Kodama, K. Kashima, D. Y. Graham, and A. R. Sepulveda. 1998. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J. Clin. Microbiol. 36:2258-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaoka, Y., D. H. Kwon, and D. Y. Graham. 2000. A Mr 34,000 proinflammatory outer membrane protein (OipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 97:7533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zakharova, N., P. S. Hoffman, D. E. Berg, and K. Severinov. 1998. The largest subunits of RNA polymerase from gastric helicobacters are tethered. J. Biol. Chem. 273:19371-19374. [DOI] [PubMed] [Google Scholar]