Abstract
We use a recently proposed method called Spectral Gap Optimization of Order Parameters (SGOOP) [P. Tiwary and B. J. Berne, Proc. Natl. Acad. Sci. U. S. A. 113, 2839 (2016)], to determine an optimal 1-dimensional reaction coordinate (RC) for the unbinding of a bucky-ball from a pocket in explicit water. This RC is estimated as a linear combination of the multiple available order parameters that collectively can be used to distinguish the various stable states relevant for unbinding. We pay special attention to determining and quantifying the degree to which water molecules should be included in the RC. Using SGOOP with under-sampled biased simulations, we predict that water plays a distinct role in the reaction coordinate for unbinding in the case when the ligand is sterically constrained to move along an axis of symmetry. This prediction is validated through extensive calculations of the unbinding times through metadynamics and by comparison through detailed balance with unbiased molecular dynamics estimate of the binding time. However when the steric constraint is removed, we find that the role of water in the reaction coordinate diminishes. Here instead SGOOP identifies a good one-dimensional RC involving various motional degrees of freedom.
I. INTRODUCTION
The unbinding of ligand-substrate systems is a problem of great theoretical and practical relevance. To take an example from the biological sciences, there is now an emerging view that the pharmacological efficacy of a drug depends not just on its thermodynamic affinity for the host protein, but also, and perhaps even more so, on when and how it unbinds from the protein.1,2 While a variety of experimental techniques can provide unbinding rate constants, gleaning a clear molecular scale understanding from such experiments into the dynamics of unbinding is difficult, and at best indirect. This makes it in principle very attractive to use atomistic molecular dynamics (MD) simulations to study the unbinding process. However, most successful drugs unbind at time scales much longer than milliseconds.1,2 Even with the fastest available supercomputers, this makes it virtually impossible to use MD simulations to obtain statistically reliable insight into unbinding dynamics.
This time scale limitation makes it crucial to complement MD with enhanced sampling techniques. These techniques accelerate the movement between metastable states separated by high (≫kBT) barriers but still allow recovering the unbiased thermodynamics and kinetics. While in principle one could construct Markov State Models (MSM)3 to study the unbinding dynamics from multiple short, unbiased simulations without any enhanced sampling, the associated high barriers typical for unbinding make this extremely difficult. As such, reported applications of MSM to such problems have been indirect, and instead of directly studying unbinding, these studies4,5 have actually looked at the drug binding problem where the barriers tend to be smaller. To directly simulate the unbinding process, it thus becomes unavoidable to use enhanced sampling methods.6
On the other hand, the use of enhanced sampling methods to study high barrier systems has its own caveats. Many such methods involve controlling the probability distribution along a low-dimensional reaction coordinate (RC), which best captures all the relevant slow degrees of freedom. Typically many such order parameters or collective variables (CVs) are available that can distinguish between various metastable states of the system at hand. For ligand unbinding, these CVs could include ligand-host relative displacement, their conformations, and their hydration states. However, often the fluctuations in these CVs can be coupled in a non-trivial manner, and it can be tricky to select a RC without having a prescience of the CVs whose fluctuations matter the most for driving the process of interest.
In this work, we aim to answer the following question: given a certain choice of order parameters (or collective variables) for a ligand-host system, what is the optimal 1-dimensional RC for unbinding that can be expressed as a linear combination of these collective variables? We are especially interested in determining how wet this RC is. Wetness here denotes the weight ascribed to the descriptor of the solvation state of the binding site, relative to other descriptors contributing to the RC. This will indicate how important biasing water density fluctuations in the host binding pocket is to the kinetics of ligand unbinding. While it is well-known through various theoretical, simulation, and experimental studies that collective water motion into/out of binding pockets is correlated with unbinding/binding, respectively,6–12 we wish to have a quantitative measure of the utility of biasing these water fluctuations in the sampling of ligand unbinding.
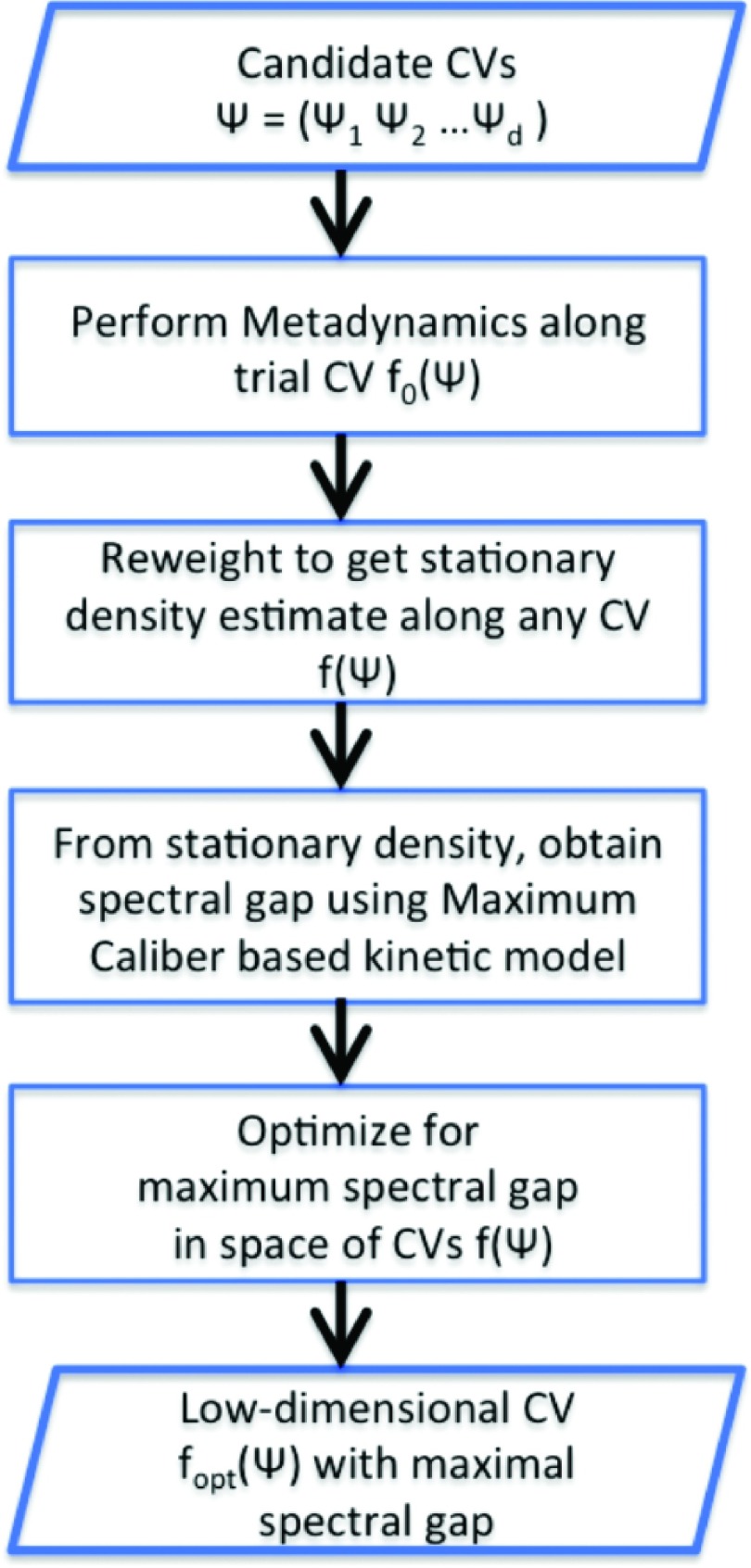
Here, we investigate this question for ligand unbinding in a much studied model hydrophobic ligand-host system (Fig. 1) interacting through Lennard-Jones potential in an aqueous environment made of explicit TIP4P water molecules.9,11,13 Many excellent methods exist for the purpose of RC optimization.14–22 However, the energy barrier for unbinding in this system as reported through previous studies is as high as 30–35 kBT, making it crucial for the purpose of RC optimization to use a method that does not rely on accurate sampling of rare reactive unbinding trajectories. For this reason, we use a recently proposed method SGOOP (spectral gap optimization of order parameters)23 that enables us to determine an optimal RC through relatively short biased simulations performed using a trial RC (see Fig. 2 and Sec. II A for details of SGOOP).
FIG. 1.
Cavity-ligand system in explicit water with axes marked. Red: fullerene shaped ligand atoms. Orange: cavity atoms that interact with the ligand and with water molecules. Blue: wall atoms. The water molecules are not shown for clarity. See Section III and the supplementary material for corresponding interaction potentials and further details.29
FIG. 2.
Flowchart summarizing the various key steps in SGOOP.23 The whole process can in principle be iterated between the second and the last steps to further improve the sampling.
We consider two different scenarios in this work, both of which are expected to arise in the context of ligand unbinding. In the first scenario, we sterically constrain the system so that the ligand can move only along the centro-symmetric axis z (see Fig. 1). In the second, we lift this steric constraint. We find that in the presence of the steric constraint, water density fluctuations in the host cavity must be part of the optimal RC. This is in excellent agreement with the previous work on this and related systems9,11,24–28 where for a sterically constrained setup, there is a bimodal water distribution at a critical ligand-cavity separation, around which the unbinding pathway involves moving from dry to wet states. However we find that when the steric constraint is removed and the ligand is free to move in any direction, the role of water in the optimal RC is minimal to none. In this case, water is less of a driving variable for unbinding, but more of a driven variable that follows the movement of the ligand. Here SGOOP identifies how the optimal RC is distorted from the z-axis (Fig. 1), which turns out to be the minimum free energy pathway for this system as reported in a previous work.11
We validate our results through extensive calculations of unbinding time statistics for the sterically constrained ligand using the infrequent metadynamics approach30 and find that the optimal RC is indeed wet to some extent. In addition, because the analogous barrier for ligand binding is much smaller than for unbinding, we use unbiased MD estimates of the binding time and validate that detailed balance is satisfied between unbinding and binding rates. We perform the infrequent metadynamics calculations using the optimal RC as per SGOOP and two other sub-optimal RCs with no water content and more than optimal water content, respectively. Our findings clearly demonstrate the improvement in the quality and accuracy of the unbinding time statistics by using the optimized RC predicted through SGOOP. With the optimized CV, the unbinding time statistics gives a superior agreement with the binding time statistics obtained through unbiased MD. Furthermore, it also gives a much improved Poisson fit for the cumulative distribution function of unbinding times, as quantified through the Kolmogorov-Smirnov test proposed in Ref. 31. This shows that the optimized RC predicted through SGOOP indeed does a better job of capturing the slow dynamics of the system. Previous applications of SGOOP23 were restricted to using the optimized RC for faster convergence of the free energy. The results reported in this work comprise the first demonstration of improving kinetics calculations using SGOOP and mark a step further towards systematic high-throughput studies of unbinding dynamics.
II. THEORY
In this section, we summarize the key methods23,30,31 used in this work and their underlying principles.
A. Spectral gap optimization of order parameters (SGOOP)
SGOOP23 is a method to optimize low-dimensional order parameters or collective variables for use in enhanced sampling biasing methods like umbrella sampling and metadynamics, when only limited prior information is known about the system (see Fig. 2 for a flowchart summarizing the key steps in SGOOP). This optimization is done from a much larger set of candidate CVs Ψ = (Ψ1, Ψ2, …, Ψd), which are assumed to be known a priori. SGOOP is based on the idea that the best order parameter, which we call the reaction coordinate (RC), is one with the maximum separation of time scales between visible slow and hidden fast processes. This time scale separation is calculated as the spectral gap between the slow and fast eigenvalues of the transition probability matrix on a grid along any CV.23 The transition probability matrix is calculated in SGOOP using an approximate kinetic model that can be derived, for example, through the principle of maximum caliber.23,32,33 Let {λ} denote this set of eigenvalues, with λ0 ≡ 1 > λ1 ≥ λ2…. The spectral gap is then defined as λs − λs+1, where s is the number of barriers apparent from the free energy estimate projected on the CV at hand that are higher than a user-defined threshold (typically ≳kBT). In this case, assuming overdamped dynamics, the eigenvalues beyond the first s + 1 correspond to relaxation times in each of the individual wells,34–36 which for an optimal RC should be much smaller than the escape times from the wells.
The key input to SGOOP as used in this work is an estimate of the stationary probability density (or equivalently the free energy) of the system, accumulated through a biased simulation performed along a sub-optimal trial RC given by some linear or non-linear function f0(Ψ), where Ψ denotes the larger set of candidate CVs. Any type of biased simulation could be used for this purpose, as long as it allows projecting the stationary probability density estimate on generic combinations of CVs without having to repeat the simulation. Metadynamics37 provides this functionality in a straightforward manner and hence we use it here. Given this information, we use the principle of maximum caliber23 to set up an unbiased master equation for the dynamics of various trial CVs f(Ψ). Through a post-processing optimization procedure, we then find the optimal RC as the f(Ψ) which gives the maximal spectral gap of the associated transfer matrix. We refer to Ref. 23 for details of the master equation and the maximum caliber expression that relates the transfer matrix to stationary probabilities and facilitates calculation of the eigenvalues and hence the spectral gap.
As described in the Introduction, for the problem of ligand unbinding in this work, we take this larger set of CVs to be the various components of the separation between the ligand and the host, and the solvation state of the host pocket (Fig. 1). In more complex systems, further members could be added to this set. Since counting the number of barriers in a projected free energy profile could be affected by sampling noise, we smooth the free energy by averaging over bins. To ensure that the calculated spectral gaps are robust with respect to the amount of smoothening, we perform an averaged estimate of the spectral gaps using different amounts of smoothing (see the supplementary material for details).29
Note that the approximate kinetic model used here in SGOOP is equivalent to the Smoluchowski equation whereby (i) the dynamics of any CV is described by a forced diffusion process and (ii) the diffusion constant along this CV is independent of position. This kinetic model is used in SGOOP to improve the choice of the RC that should be biased given limited information starting with a trial RC. The calculation of rates is then done with this improved RC. It is important to note that the infrequent metadynamics method for calculating rate constants30 does not assume Smoluchowski dynamics or constant diffusivity (see Sec. II B for details).
B. Dynamics from infrequent metadynamics
The infrequent metadynamics approach30,31 is a recently proposed method which has been used to obtain rate constants in various molecular systems.11,38 It involves time-dependent biasing of a few selected (typically one to three) order parameters or collective variables (CVs) out of the many available, in order to hasten the escape from metastable free energy basins.39 By periodically adding repulsive bias (typically in the form of Gaussians) in the regions of CV space as they are visited, the system is encouraged to escape stable free energy basins where they would normally be trapped for long periods of time. The central idea in infrequent metadynamics is to deposit bias rarely enough compared to the time spent in the transition state regions so that dynamics in the saddle region is very rarely perturbed. Through this approach, one then increases the likelihood of not corrupting the transition states and preserves the sequence of transitions between stable states. The acceleration of transition rates achieved through biasing can then be calculated by appealing to generalized transition state theory,40 which yields the following simple running average for the acceleration:30
| (1) |
where s is the collective variable being biased, β = 1/kBT is the inverse temperature, V(s, t) is the bias experienced at time t, and the subscript t indicates averaging under the time-dependent potential. This approach is expected to work best in the diffusion controlled regime.41
The infrequent metadynamics method requires a good and small set of slow collective variables demarcating all relevant stable states of interest. Whether this is the case or not can be verified a posteriori by checking if the cumulative distribution function for the transition times out of each stable state is Poissonian,31 as quantified through the Kolmogorov-Smirnov test described in detail in Ref. 31. While metadynamics can still be performed with two, three, or more biasing CVs, the computational gain obtained by compressing the slow dynamics into an optimized 1-dimensional RC is immense, especially given the infrequent nature of biasing (see the supplementary material for detailed simulation parameters such as the frequency of biasing used in this work).29 Using SGOOP (Sec. II A) allows us to select a good 1-dimensional RC as a function of the many available choices of CVs, as we show in this work. This choice increases the probability of passing the test of Ref. 31 once the relatively expensive infrequent metadynamics runs are performed.
III. SYSTEM DETAILS
The model ligand used in this work is a C60 fullerene and the binding pocket is an ellipsoidal cavity carved from a hydrophobic slab, all interacting via Lennard-Jones potentials and enclosed by a periodic box with explicit water and cubic edge length 5.96 nm. This system was introduced previously in works such as Refs. 9 and 11. The pocket sites are fixed and interact with the model ligand with a Lennard-Jones site-site potential having σ = 0.4152 nm, which is kept the same for all interactions. The pocket itself comprises 2 types of atomic species, interacting with the ligand atoms (color red in Fig. 1) as described below. The system comprised a total of 34 296 atoms, with the total number of ligand, cavity, and solvent atoms equaling 60, 9020, and 25 216, respectively.
-
1.
Cavity atoms (color orange in Fig. 1), with Lennard-Jones ϵ = 0.008 kJ/mol.
-
2.
Wall atoms (color blue in Fig. 1), with Lennard-Jones ϵ = 0.0024 kJ/mol.
The solute-solvent interactions are represented by the geometric mean of the respective water and solute parameters, in accordance with the Optimized Potential for Liquid Simulations (OPLS) formalism.42 All simulations are performed in explicit TIP4P water13 with the GROMACS 4.5.4 MD package43 patched with the PLUMED plugin.44 During the equilibration stage, temperature and pressure are controlled with the stochastic velocity rescaling thermostat45 and Berendsen barostat.46 The production runs were NVT (constant number, volume, temperature) with a temperature of 300 K. The PLUMED plugin44 was used to carry out metadynamics calculations. An integration time step of 2 fs was used for all runs. All other relevant simulation details are provided in the supplementary material.29
IV. RESULTS AND DISCUSSION
A. Ligand constrained to move along one direction
In the first investigated case, the system dynamics is sterically constrained so that the ligand can move only along the centro-symmetric axis z (Fig. 1). This system and constraint have already been investigated in studies aimed at understanding hydrophobic interactions.9,11,24–26 Here we consider two descriptors; the z-component of the ligand-cavity separation and the number of water molecules in the host cavity, denoted w. The number of water molecules is computed using a sigmoidal function which makes w continuous and differentiable (see the supplementary material29 for details including precise definition of w) as implemented in the enhanced sampling plugin PLUMED.44 We then seek the best 1-d RC f of the following form:
| (2) |
Throughout this paper mw is a measure of the wetness of the RC, with mw = 0 corresponding to a completely dry RC, and higher values denoting increasingly wetter RCs.
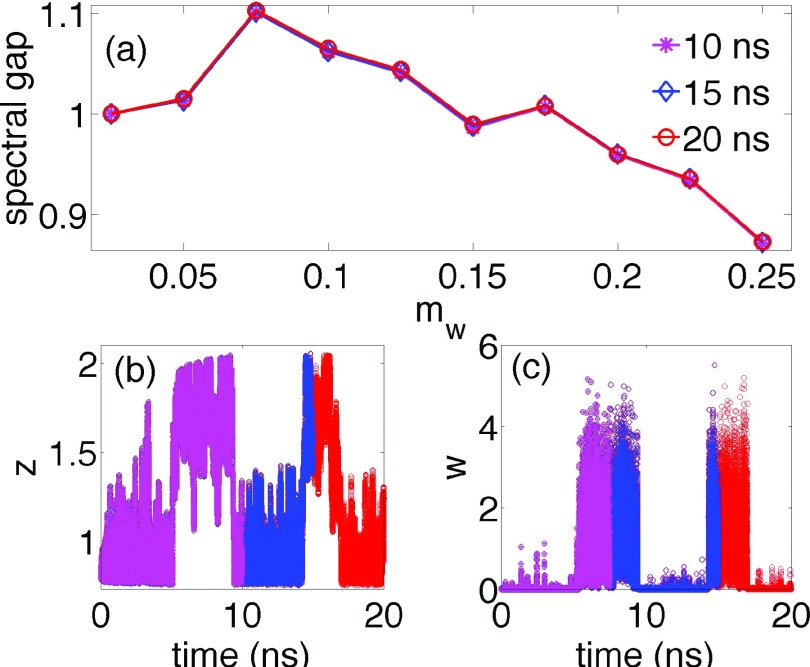
We first perform a short metadynamics simulation by biasing f0 = z. This starting run is performed with frequent biasing since the objective here is to get a sense of the free energy, and not the kinetics (see the supplementary material for various biasing frequencies and other parameters).29 Through this, we can obtain an estimate of the stationary probability density along any f(z, w) by using the reweighting functionality of metadynamics.37 By using SGOOP, we then get an estimate of the optimal mw ≈ 0.075 in Eq. (2) which maximizes the spectral gap. This is shown in Fig. 3(a) where an estimate of the spectral gap versus mw for different lengths of the starting metadynamics trajectory is provided. Other trajectories used in SGOOP shown in Figs. 3(b) and 3(c) are of length 10 ns, 15 ns, and 20 ns, respectively. The results are extremely robust with respect to simulation time and parameters. Fig. 3(a) has 3 different curves calculated which for all practical purposes collapse into one, indicating that the spectral gaps estimated with trajectories of three different simulation times are virtually indistinguishable and well-converged. Furthermore, using an entirely different starting metadynamics trajectory generated using different Gaussian width gives the same optimal wetness of the RC (see the supplementary material).29
FIG. 3.
(a) Spectral gap versus the amount of wetness of the RC, mw (see Eq. (2)) for the case when the ligand constrained to move along a line. The optimal RC can be clearly seen to be at mw ≈ 0.075. Three different profiles are provided, which were calculated by using the starting metadynamics trajectories of different lengths as indicated in legends, performed with biasing CV z. The spectral gap is normalized so that its value for mw = 0 is 1. (b) and (c) are the corresponding trajectories for the distance z and the number of pocket waters w. See Sec. IV A and the supplementary material for precise definition of w.29 In all sub-figures here, magenta stars, blue diamonds, and red circles denote results for trajectories of lengths 10 ns, 15 ns, and 20 ns, respectively.
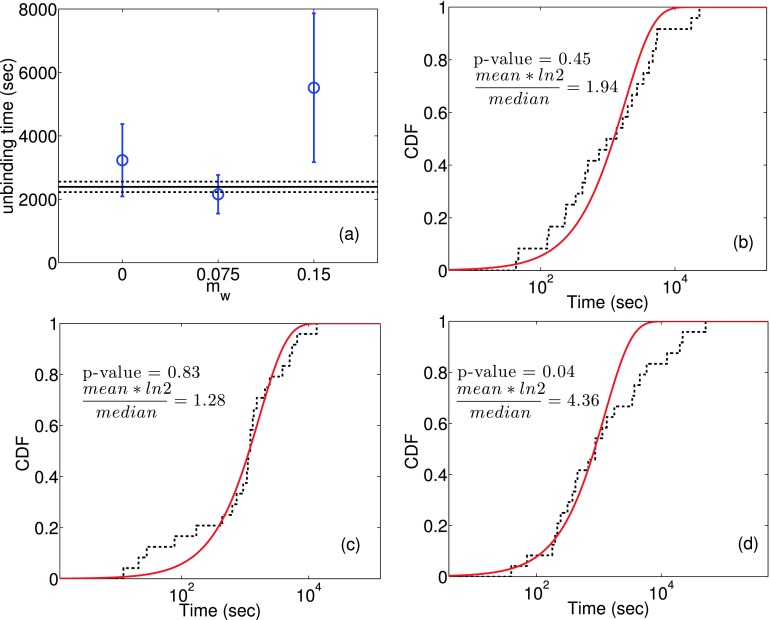
The optimal wetness of the RC in Eq. (2) given by mw ≈ 0.075 is validated by performing extensive multiple independent unbinding simulations using infrequent metadynamics, starting from the bound pose z = 0 (Sec. II B). The unbinding time is calculated as the time taken to reach z = 1.4 nm for the first time.11 We perform three independent sets of 24 simulations (totaling 72 simulations) for (1) mw = 0, a dry RC, (2) mw = 0.075, the RC with optimal wetness found from SGOOP, and (3) mw = 0.15, the RC with more than optimal wetness. The empirical and fitted cumulative distribution functions for the unbinding time statistics using the three different RCs with varying amounts of wetness are shown in Figs. 4(b)-4(d), along with the respective p-values for fits to the ideal Poisson distributions, quantified using the Kolmogorov-Smirnov test from Ref. 31, and mean times log(2) divided by median ratio for each case. An ideal fit to the Poisson distribution would result if both these numbers would be close to 1, and this would suggest that the accelerated time scales found using metadynamics are reliable. The RC with optimal water coefficient mw = 0.075 obtained using SGOOP gives Poisson metrics closest to 1. Fig. 4(a) shows the mean unbinding times obtained using the three RCs with different values of the wetness parameter mw and these are compared with the corresponding estimate provided in the literature9 calculated from accurate free energy calculations together with the principle of detailed balance. While it must be said that the completely dry RC does a reasonable job in terms of the p-value and order of magnitude agreement with unbiased MD, it is very clear from this plot as well that the RC with optimal wetness gives the best performance as per various metrics shown in Fig. 4. Thus to summarize, the optimal RC for this case indeed has a small but distinct amount of wetness.
FIG. 4.
Unbinding times for the sterically constrained ligand using different simulation protocols. In (a) the mean unbinding times as obtained through the three RCs with different water coefficients are plotted along with error bars (blue circles). Also plotted is the corresponding estimate of mean unbinding time (solid black line) with errors (dashed black line) by using the principle of detailed balance with the unbiased estimate of binding time. All error bars correspond to ±standard deviation intervals. (b) to (d) give the empirical (black dashed line) and fitted (solid red line) cumulative distribution functions (CDF) for unbinding time statistics using different RCs with varying amounts of wetness mw (see Eq. (2)). From (b) to (d), respectively, mw is 0, 0.075 and 0.15. Also indicated are respective p-values for fit to the ideal Poisson distribution, quantified using the Kolmogorov-Smirnov test from Ref. 31, and mean times log(2) divided by median ratio for each case. The closer are both these values to 1, the more ideal is the Poisson distribution fit illustrating the reliability of the dynamics generated from metadynamics. As can be seen from these figures, the RC with optimal water coefficient of 0.075 as obtained from SGOOP gives the best Poisson metric as per both these criteria.
B. Ligand free to move in any direction
In this case, we remove the steric constraint forcing the system to move along z and allow the ligand to freely move in any direction (see Fig. 1). Because the system is axially symmetric, we consider 3 order parameters, namely, the z-component of the ligand-cavity separation, , and the number of water molecules in the host cavity, denoted w. We then seek the best 1-d RC f of the following form:
| (3) |
We first perform a short metadynamics simulation by biasing with f0 = z, a purely dry RC. As before, this starting run is performed with frequent biasing since the objective here is to get a sense of the free energy, and not the kinetics. This gives an estimate of the stationary probability density along any f(z, ρ, w) by applying the reweighting functionality of metadynamics.37 We then use SGOOP to obtain an estimate of the optimal values as mρ ≈ 0.6, mw ≈ 0.0 in Eq. (3). These values maximize the spectral gap.
Fig. 5 gives an estimate of the spectral gap versus (mρ, mw) based on an initial metadynamics trajectory of duration 20 ns biasing z. The results are again extremely robust with respect to how long the simulation was run. In this case as well, using an entirely different starting metadynamics trajectory generated using different Gaussian width gives the same optimal value of the RC (see the supplementary material).29
FIG. 5.
Contour plot of spectral gap versus (mρ, mw) with the starting metadynamics trajectory of duration 20 ns used in SGOOP. The optimal RC can be clearly seen to be at (mρ, mw) ≈ (0.6, 0.0). The spectral gap is normalized so that its value for (mρ, mw) = (0, 0) is 1.
As can be seen by comparing Fig. 5 to Fig. 3, the wetness of the optimal RC in the case of unconstrained motion is closer to 0. In a sense, the water fluctuations in the cavity appear to be caused or driven by the unbinding, rather than being a driving variable for unbinding as it is in the constrained case. The primary reaction coordinate depends on z and ρ, the displacement variables of the ligand with respect to the cavity. Indeed SGOOP finds mρ ≈ 0.6, which gives the distortion of the reaction path from the z-axis (see Fig. 1). This is the same as the slope of the minimum free energy pathway in (z, ρ) space reported in the previous work.11 As described in detail in Ref. 11, removing the steric constraint causes the bound ligand to roll over and take a slightly more stable off-center ground state and therefore a different binding pose. This leads to a slight difference in the binding free energies of the two setups considered in this work.
Since the optimal wetness of the RC in this case is close to 0, we do not perform any kinetics calculations. Instead we refer to the results from Ref. 11, where infrequent metadynamics with a similar completely dry RC for this setup gave very good agreement through detailed balance with the unbiased MD estimate of the binding time.
In the supplementary material,29 we also provide illustrative free energy profiles for both setups along a variety of RCs.
V. DISCUSSION AND CONCLUSIONS
In this work, we have applied the recently proposed method SGOOP23 to the problem of determining the reaction coordinate for ligand unbinding in a model system in explicit water. By using short biased metadynamics simulations performed using a sub-optimal reaction coordinate, we find that the true reaction coordinate involves water in the case when the system is sterically constrained to move along an axis of symmetry. In the case when this constraint is lifted, the role of water in the optimal RC is reduced. Our predictions of the optimal RC are validated by extensive calculations of the unbinding rate constant using metadynamics with infrequent biasing30,31 with different RCs. We believe that the application of SGOOP to optimize the choice of RC for ligand unbinding, combined with the approach of Refs. 30 and 31, provides an important step in the quest to invent methods useful for systematic and possibly high throughput calculations of the unbinding rate constant in more complex and realistic protein-ligand systems, a quantity extremely difficult to compute without careful enhanced sampling based approaches.38,47 The hope is that this approach will contribute a step toward the success of computational drug discovery programs that take drug unbinding dynamics into account. It would also be interesting to see how closely the RC and associated unbinding pathways identified through SGOOP correlate with realistic reaction paths obtained from single molecule experiments on dissociation of biomolecular complexes. We also think that the current work is a demonstration of how SGOOP may be used to answer similar questions in systems other than drug unbinding where the role of water density fluctuations in driving the dynamics is believed to play a role but which is hard to quantify.
Using the model system in this work allows us to study an unbinding problem involving solvation and steric related complexities, yet where we can perform extensive simulations of the reverse binding process. Undoubtedly more realistic systems will be harder to tackle than the model system of the current work, possibly involving a much larger set of trial collective variables than used here and requiring more care in coming up with this trial set to begin with. For instance, it might be crucial to incorporate the conformational fluctuations in the receptor or the ligand molecule or both into the trial set of collective variables. In principle, this should be possible with SGOOP and is the subject of current investigations. As long as the system’s intrinsic dynamics displays a time scale separation between few slow and remaining fast processes, and hence possesses an associated spectral gap, we expect SGOOP to be useful in obtaining a sense of fluctuations that matter for driving the dynamics in rare event systems.
We would like to emphasize that the systems considered in this work, in spite of their model nature, are in fact quite challenging test cases. This is due to the enormous barrier height involved (around 30–35 kBT), and the relative insignificance of the barrier in the dewetting related bimodal distribution (around 1–2 kBT)9 relative to the main barrier. As such, even the trial RC that excludes wetness entirely, considered in this work and in Ref. 11, does a remarkably decent job when used with metadynamics.30,31 Yet SGOOP does very well in picking up signals in the right directions for improving the RC towards ideal. This demonstration makes us optimistic that in more complex systems where the barrier associated to movement of water is expected to be higher,38,48–50 the algorithm will be even more useful. Some such studies are already underway and will be the subject of future publications.
Acknowledgments
This work was supported by grants from the National Institutes of Health (Grant No. NIH-GM4330) and the Extreme Science and Engineering Discovery Environment (XSEDE) (Grant No. TG-MCA08X002).
REFERENCES
- 1.Copeland R. A., Pompliano D. L., and Meek T. D., Nat. Rev. Drug Discovery , 730 (2006). 10.1038/nrd2082 [DOI] [PubMed] [Google Scholar]
- 2.Copeland R. A., Nat. Rev. Drug Discovery , 87 (2015). 10.1038/nrd.2015.18 [DOI] [PubMed] [Google Scholar]
- 3.Bowman G. R., Beauchamp K. A., Boxer G., and Pande V. S., J. Chem. Phys. , 124101 (2009). 10.1063/1.3216567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buch I., Giorgino T., and De Fabritiis G., Proc. Natl. Acad. Sci. U. S. A. , 10184 (2011). 10.1073/pnas.1103547108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plattner N. and Noé F., Nat. Commun. , 7653 (2015). 10.1038/ncomms8653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan A. C., Borhani D. W., Dror R. O., and Shaw D. E., Drug Discovery Today , 667 (2013). 10.1016/j.drudis.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 7.Baron R. and McCammon J. A., Annu. Rev. Phys. Chem. , 151 (2013). 10.1146/annurev-physchem-040412-110047 [DOI] [PubMed] [Google Scholar]
- 8.Setny P., Baron R., Kekenes-Huskey P. M., McCammon J. A., and Dzubiella J., Proc. Natl. Acad. Sci. U. S. A. , 1197 (2013). 10.1073/pnas.1221231110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mondal J., Morrone J. A., and Berne B. J., Proc. Natl. Acad. Sci. U. S. A. , 13277 (2013). 10.1073/pnas.1312529110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mondal J., Friesner R. A., and Berne B., J. Chem. Theory Comput. , 5696 (2014). 10.1021/ct500584n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiwary P., Mondal J., Morrone J. A., and Berne B. J., Proc. Natl. Acad. Sci. U. S. A. , 12015 (2015). 10.1073/pnas.1516652112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiß R. G., Setny P., and Dzubiella J., “Solvent fluctuations induce non-Markovian kinetics in hydrophobic pocket-ligand binding,” J. Phys. Chem. B (to be published). 10.1021/acs.jpcb.6b01219 [DOI] [PubMed] [Google Scholar]
- 13.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L., J. Chem. Phys. , 926 (1983). 10.1063/1.445869 [DOI] [Google Scholar]
- 14.Best R. B. and Hummer G., Proc. Natl. Acad. Sci. U. S. A. , 6732 (2005). 10.1073/pnas.0408098102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coifman R. R., Lafon S., Lee A. B., Maggioni M., Nadler B., Warner F., and Zucker S. W., Proc. Natl. Acad. Sci. U. S. A. , 7426 (2005). 10.1073/pnas.0500334102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters B. and Trout B. L., J. Chem. Phys. , 054108 (2006). 10.1063/1.2234477 [DOI] [PubMed] [Google Scholar]
- 17.Ma A. and Dinner A. R., J. Phys. Chem. B , 6769 (2005). 10.1021/jp045546c [DOI] [PubMed] [Google Scholar]
- 18.Rohrdanz M. A., Zheng W., Maggioni M., and Clementi C., J. Chem. Phys. , 124116 (2011). 10.1063/1.3569857 [DOI] [PubMed] [Google Scholar]
- 19.Pérez-Hernández G., Paul F., Giorgino T., De Fabritiis G., and Noé F., J. Chem. Phys. , 015102 (2013). 10.1063/1.4811489 [DOI] [PubMed] [Google Scholar]
- 20.Ceriotti M., Tribello G. A., and Parrinello M., Proc. Natl. Acad. Sci. U. S. A. , 13023 (2011). 10.1073/pnas.1108486108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M., Yu T.-Q., and Tuckerman M. E., Proc. Natl. Acad. Sci. U. S. A. , 3235 (2015). 10.1073/pnas.1418241112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hummer G. and Szabo A., J. Phys. Chem. B , 9029 (2014). 10.1021/jp508375q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiwary P. and Berne B. J., Proc. Natl. Acad. Sci. U. S. A. , 2839 (2016). 10.1073/pnas.1600917113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrone J. A., Li J., and Berne B. J., J. Phys. Chem. B , 378 (2012). 10.1021/jp209568n [DOI] [PubMed] [Google Scholar]
- 25.Li J., Morrone J. A., and Berne B., J. Phys. Chem. B , 11537 (2012). 10.1021/jp307466r [DOI] [PubMed] [Google Scholar]
- 26.Bolhuis P. G. and Chandler D., J. Chem. Phys. , 8154 (2000). 10.1063/1.1315997 [DOI] [Google Scholar]
- 27.Patel A. J., Varilly P., and Chandler D., J. Phys. Chem. B , 1632 (2010). 10.1021/jp909048f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hummer G. and Garde S., Phys. Rev. Lett. , 4193 (1998). 10.1103/PhysRevLett.80.4193 [DOI] [Google Scholar]
- 29.See supplementary material at http://dx.doi.org/10.1063/1.4959969 E-JCPSA6-145-049629 for further simulation details.
- 30.Tiwary P. and Parrinello M., Phys. Rev. Lett. , 230602 (2013). 10.1103/PhysRevLett.111.230602 [DOI] [PubMed] [Google Scholar]
- 31.Salvalaglio M., Tiwary P., and Parrinello M., J. Chem. Theory Comput. , 1420 (2014). 10.1021/ct500040r [DOI] [PubMed] [Google Scholar]
- 32.Pressé S., Ghosh K., Lee J., and Dill K. A., Rev. Mod. Phys. , 1115 (2013). 10.1103/RevModPhys.85.1115 [DOI] [Google Scholar]
- 33.Dixit P. D., Jain A., Stock G., and Dill K. A., J. Chem. Theory Comput. , 5464 (2015). 10.1021/acs.jctc.5b00537 [DOI] [PubMed] [Google Scholar]
- 34.Coifman R. R., Kevrekidis I. G., Lafon S., Maggioni M., and Nadler B., Multiscale Model. Simul. , 842 (2008). 10.1137/070696325 [DOI] [Google Scholar]
- 35.Matkowsky B. and Schuss Z., SIAM J. Appl. Math. , 242 (1981). 10.1137/0140020 [DOI] [Google Scholar]
- 36.Risken H., Fokker-Planck Equation (Springer, 1984). [Google Scholar]
- 37.Tiwary P. and Parrinello M., J. Phys. Chem. B , 736 (2014). 10.1021/jp504920s [DOI] [PubMed] [Google Scholar]
- 38.Tiwary P., Limongelli V., Salvalaglio M., and Parrinello M., Proc. Natl. Acad. Sci. U. S. A. , E386 (2015). 10.1073/pnas.1424461112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valsson O., Tiwary P., and Parrinello M., Annu. Rev. Phys. Chem. , 159 (2016). 10.1146/annurev-physchem-040215-112229 [DOI] [PubMed] [Google Scholar]
- 40.Berne B. J., Borkovec M., and Straub J. E., J. Phys. Chem. , 3711 (1988). 10.1021/j100324a007 [DOI] [Google Scholar]
- 41.Tiwary P. and Berne B. J., J. Chem. Phys. , 134103 (2016). 10.1063/1.4944577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jorgensen W. L., Maxwell D. S., and Tirado-Rives J., J. Am. Chem. Soc. , 11225 (1996). 10.1021/ja9621760 [DOI] [Google Scholar]
- 43.Hess B., Kutzner C., Van Der Spoel D., and Lindahl E., J. Chem. Theory Comput. , 435 (2008). 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- 44.Tribello G. A., Bonomi M., Branduardi D., Camilloni C., and Bussi G., Comput. Phys. Commun. , 604 (2014). 10.1016/j.cpc.2013.09.018 [DOI] [Google Scholar]
- 45.Bussi G., Donadio D., and Parrinello M., J. Chem. Phys. , 014101 (2007). 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- 46.Berendsen H. J., Postma J. P. M., van Gunsteren W. F., DiNola A., and Haak J., J. Chem. Phys. , 3684 (1984). 10.1063/1.448118 [DOI] [Google Scholar]
- 47.Teo I., Mayne C. G., Schulten K., and Lelièvre T., J. Chem. Theory Comput. , 2983 (2016). 10.1021/acs.jctc.6b00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu P., Huang X., Zhou R., and Berne B. J., Nature , 159 (2005). 10.1038/nature03926 [DOI] [PubMed] [Google Scholar]
- 49.Shan Y., Kim E. T., Eastwood M. P., Dror R. O., Seeliger M. A., and Shaw D. E., J. Am. Chem. Soc. , 9181 (2011). 10.1021/ja202726y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jensen M. Ø., Jogini V., Borhani D. W., Leffler A. E., Dror R. O., and Shaw D. E., Science , 229 (2012). 10.1126/science.1216533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4959969 E-JCPSA6-145-049629 for further simulation details.