Abstract
Nocardia farcinica is the most clinically significant species within the Nocardia asteroides complex. Differentiation of N. farcinica from other members of N. asteroides complex is important because this species characteristically demonstrates resistance to several extended-spectrum antimicrobial agents. Traditional phenotypic characterization of this species is time- and labor-intensive and often leads to misidentification in the clinical microbiology laboratory. We previously observed a 409-bp product for all strains of N. farcinica by using randomly amplified polymorphic DNA analysis with the primer DKU49. In this investigation, the 409-bp fragment was sequenced and then used to design a specific primer pair, Nf1 (16-mer) and Nf2 (16-mer), complementary to the 409-bp fragment. PCR amplification of genomic DNA from 28 N. farcinica isolates with Nf1 and Nf2 generated a single intense 314-bp fragment. The specificity of the assay with these primers was verified, since there were no PCR amplification products observed from heterologous nocardial species (n = 59) or other related bacterial genera (n = 41). Restriction enzyme digestion using CfoI and direct sequencing of the 314-bp fragment further confirmed the specificity of the assay for N. farcinica. This highly sensitive and specific PCR assay provides a rapid (within 1 day of obtaining DNA) method for identification of this medically important emerging pathogen. Rapid diagnosis of N. farcinica infection may allow for earlier initiation of effective therapy, thus improving patient outcome.
Members of the genus Nocardia, which are partially acid-fast, aerobic, branched gram-positive bacilli, are opportunistic pathogens commonly found in patients with acute or chronic suppurative or granulomatous diseases (18). Of the greatest clinical importance within this genus is N. farcinica, a member of the N. asteroides complex composed of N. asteroides complex drug pattern type I (N. abscessus), N. asteroides complex drug pattern type VI (N. cyriacigeorgica), N. nova, and N. farcinica (28). N. farcinica causes localized and disseminated infections, predominantly affecting immunocompromised patients. Differentiation of N. farcinica from other members of N. asteroides complex is important, because N. farcinica has a high degree of resistance to various antibiotics, especially to the extended-spectrum cephalosporins, which may make it difficult to treat (28, 29), and because mouse pathogenicity studies have demonstrated that it may be more virulent than the other N. asteroides complex species (8).
Traditional biochemical identification of N. farcinica is often laborious, difficult to replicate, and time-consuming; species identification usually requires up to 3 weeks. In addition, misidentification of N. farcinica may occur because it shares some phenotypic similarities with Gordonia, Rhodococcus, and rapidly growing Mycobacterium. Commercially available systems in combination with a few traditional tests have shortened the identification time of Nocardia species to 7 days (2, 13, 20); however, phenotypic identification to the species level within this genus remains problematic (2, 13, 20). For example, within the genus Nocardia are N. vaccinii and two recently described species, N. africana and N. veterana, that share similar phenotypic (biochemical and susceptibility profiles) and molecular characteristics to N. nova (6). The use of molecular approaches such as PCR targeting portions of the hsp gene and the 16S rRNA gene coupled with restriction endonuclease digestion of PCR products has been the focus of recent investigations for the separation of mycobacteria from the nocardiae, as well as for the recognition of species within the genera Mycobacterium and Nocardia (6, 7, 14, 15, 25, 31). Such methodology has proven to be sensitive, less time-consuming, and less labor-intensive than traditional biochemical methods. However, accurate identification may still be difficult because it relies upon analysis of a relatively few restriction fragments from a single gene. The molecular weight of fragments of different species may be the same, or approximately the same, and thus they may migrate similarly. Recently, randomly amplified polymorphic DNA (RAPD) analysis has been described as an identification method for the Nocardia species (12). RAPD analysis has also been described as a useful method for intraspecies discrimination in an epidemiological study of N. farcinica (10). Exmelin et al. (10), using RAPD analysis for subtyping, incorporated a single short primer, DKU49, in a low-stringency PCR to amplify genomic DNA. In a previous evaluation of the usefulness of RAPD as a tool for the rapid identification of Nocardia species using DKU49 profiles, our investigators observed an intense, conserved 409-bp band in all isolates of N. farcinica that was not present in related N. asteroides complex species, suggesting this DNA fragment could be used as a potential diagnostic marker (L. A. Lentnek, B. A. Lasker, M. M. McNeil, R. S. Weyant, and J. M. Brown, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. D-75, 1997). The sequencing information of this 409-bp band was utilized in this study to design the N. farcinica PCR primers Nf1 (16-mer) and Nf2 (16-mer), which we evaluated for the rapid and specific identification of clinical and environmental isolates of N. farcinica by using a species-specific PCR assay.
MATERIALS AND METHODS
Bacterial strains.
A list of the type, reference, and clinical strains used in this study is shown in Table 1. Thirty-six type and reference strains were obtained from the American Type Culture Collection (ATCC; Manassas, Va.), and the type strain Tsukamurella tyrosinosolvens DSM 44-234 was obtained from Deutsche Sammlung von Mikroorganismen Zellkulturen (DSM; Braunschweig, Germany). The PCR assay primers were derived from the type strain N. farcinica ATCC 3318. The 73 Nocardia species clinical isolates and 18 non-Nocardia species clinical isolates were obtained from the culture collection maintained by the Actinomycete Reference Laboratory, Meningitis and Special Pathogens Branch at the Centers for Disease Control and Prevention (Table 1). The 28 N. farcinica study group isolates comprised the type strain ATCC 3318, reference strain ATCC 23826, and 26 “CDC clinical” isolates of N. farcinica from a variety of clinical sources, as follows: wound specimens, 10 isolates; respiratory specimens, 9 isolates; blood specimens, 2 isolates; 1 isolate each from body fluid and eye; and 3 for which the sources were unknown. All isolates were identified by conventional physiologic and biochemical methods and susceptibility patterns, as previously described (1, 19, 28, 29).
TABLE 1.
Microorganisms used in this study
| Strain |
|---|
| Deitzia maris ATCC 35013T |
| Gordonia aichiensis ATCC 33611T |
| Gordonia bronchialis ATCC 25592T |
| Gordonia rubropertincta ATCC 14352T |
| Gordonia sputi ATCC 29627T |
| Gordonia terrae ATCC 25594T |
| Gordonia/Rhodococcus complex clinical isolates (12) |
| Mycobacterium fortuitum ATCC 6841T and 1 clinical isolate |
| Mycobacterium peregrinum ATCC 14467T and 1 clinical isolate |
| Nocardia abscessus clinical isolates (3) |
| Nocardia asteroides ATCC 19247T |
| Nocardia asteroides complex clinical isolatea (1) |
| Nocardia asteroides complex drug pattern type IV ATCC 49872 and ATCC 49873 |
| Nocardia brasiliensis ATCC 19296T and 6 clinical isolates |
| Nocardia brevicatena ATCC 15727, ATCC 15333, and 3 clinical isolates |
| Nocardia cyriacigeogica clinical isolates (8) |
| Nocardia farcinica ATCC 3318T, ATCC 23826, and 26 clinical isolates |
| Nocardia nova ATCC 33726T, ATCC 33727b, and 22 N. nova complex clinical isolatesb |
| Nocardia otitidiscaviarum ATCC 14629T and 4 clinical isolates |
| Nocardia pseudobrasiliensis ATCC 51512T and ATCC 51511 |
| Nocardia transvalensis ATCC 6865T |
| Rhodococcus coprophilus ATCC 29080T |
| Rhodococcus equi ATCC 6939T and 4 clinical isolates |
| Rhodococcus erythropolis ATCC 4277T |
| Rhodococcus fascians ATCC 12974T |
| Rhodococcus globerulus ATCC 14898T |
| Rhodococcus marinonascens ATCC 35653T |
| Rhodococcus opacus ATCC 51882 |
| Rhodococcus percolatus ATCC 6348T |
| Rhodococcus rhodnii ATCC 35071T |
| Rhodococcus rhodochrous ATCC 13808T |
| Rhodococcus wratislaviensis ATCC 51786T |
| Tsukamurella inchonensis ATCC 700082T |
| Tsukamurella paurometabola ATCC 8368T |
| Tsukamurella pulmonis ATCC 700081T |
| Tsukamurella tyrosinosolvens DSM 44-234T |
Biochemicals and susceptibility profiles could not identify this isolate further.
No attempt was made to differentiate these isolates further. Molecular methods may identify them as N. africana, N. nova, or N. veterana.
DNA preparation.
Single colonies were inoculated onto Lowenstein-Jensen slants (Remel, Lenexa, Kans.), checked for purity on heart infusion agar with 5% rabbit blood (BBL, Microbiology Systems, Cockeysville, Md.), and incubated at 35°C, generally for at least 16 h. The growth was harvested and suspended in 1.5 ml of 10 mM Tris-1 mM EDTA (pH 8.0) and adjusted to a turbidity of a McFarland 4 standard. Then, 0.15-mm silica beads were added to 0.5 ml of the resultant stationary-phase culture. Samples were then submerged in a boiling water bath for 15 min, followed by an immediate cell disruption in a mini-beadbeater for 5 min. Bacterial lysates were clarified by three successive centrifugations at 19,873 × g for 5 min. Template DNA was stored frozen at −20°C.
RAPD analysis of amplicons.
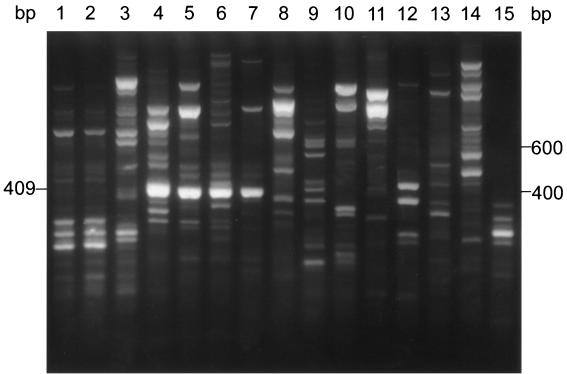
Each RAPD reaction mixture contained 1 μl of template DNA, 2.0 μM primer DKU49 (5′-CCGCCGACCGAG-3′), one Ready-To-Go RAPD analysis bead (Amersham Pharmacia Biotech, Piscataway, N.J.), and 21.5 μl of distilled water. RAPD reaction conditions were previously described by Exmelin et al. (10), except that we used an Applied Biosystems (Foster City, Calif.) Gene Amp PCR System 9700 thermal cycler. Following amplification, a 15-μl sample was electrophoresed through a 2.5% agarose gel (1.5% NuSieve [FMC Bioproducts, Rockland, Maine] and 1% agarose [Life Technologies, Grand Island, N.Y.]). Gels were stained with ethidium bromide (0.5 μg/ml) and then photographed. An intense 409-bp band was observed in the lane for N. farcinica ATCC 3318T. The 409-bp band (Fig. 1, lane 4) was excised with a razor, and DNA from the excised fragment was purified using the buffers and the protocol included in the QIAGEN gel extraction kit (QIAGEN, Chatsworth, Calif.).
FIG. 1.
RAPD patterns of N. farcinica and related species amplified with primer DKU49. Lanes 1 to 3, N. nova isolates W6310 and W6311 and ATCC 33726T; lanes 4 to 7, N. farcinica isolates ATCC 3318T, W6434, W6017, and W6255; lane 8, N. brasiliensis ATCC 19296T; lane 9, M. fortuitum ATCC 6841T; lane 10, N. transvalensis ATCC 6865T; lane 11, N. brasiliensis W6312; lane 12, N. asteroides ATCC 19247T; lanes 13 and 14, N. abscessus W6133 and W6335; lane 15, N. cyriacigeorgica W6344. The molecular size standard consisting of a 100-bp DNA ladder is shown on the right margin. The 409-bp fragment for four strains of N. farcinica (lanes 4 to 7) is designated on the left margin.
DNA sequencing of the 409-bp band.
The purified 409-bp RAPD fragment was cloned into the multiple cloning site of plasmid pCR-2.1 and then used to transform Escherichia coli strain TOP10F′, using the reagents and protocols supplied by the manufacturer for the original TA cloning kit (Invitrogen Corp., San Diego, Calif.). Plasmid DNA containing the RAPD fragment was purified by using the protocol and reagents supplied with the Plasmid Midi protocol (QIAGEN). Using primers M13 reverse and T7 promoter (Invitrogen), both strands of the fragment were sequenced from three independent clones in their entirety by using the ABI PRISM BigDye terminator cycle sequencing ready reaction kit (Perkin-Elmer, Applied Biosystems) with the manufacturer's reagents and recommendations for cycle sequencing. Centrisep columns (Princeton Separations, Adelphia, N.J.) were used to remove unincorporated dye-labeled and unlabeled nucleotides. Sequencing reaction mixtures were resolved and analyzed using an ABI PRISM 310 genetic analyzer (Applied Biosystems). Sequencer version 4.1 software (Gene Codes Corp., Ann Arbor, Mich.) was used to edit and align the sequence data. The sequences of three clones were compared and were subjected to a BLASTN search.
N. farcinica-specific PCR.
Based on the nucleotide sequence of the 409-bp DNA fragment, species-specific PCR primer pairs Nf1 (5′-CCGCAGACCACGCAAC) and Nf2 (5′-ACGAGGTGACGGCTGC) were designed using the OLIGO 4.0 program (National Biosciences, Plymouth, Minn.) and are shown in Fig. 2. Twenty-eight N. farcinica isolates and 100 isolates of related species and genera shown in Table 1 were tested for the expected 314-bp fragment following PCR amplification with primers Nf1 and Nf2. PCR mixtures consisted of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, a 0.2 μM concentration (each) of primers Nf1 and Nf2, 1 to 5 μl of genomic DNA, a 0.2 mM concentration of each deoxynucleotide triphosphate (dATP, dCTP, dGTP, and dTTP), and 2.5 U of Taq DNA polymerase in a volume of 50 μl. PCR was performed in a Gene Amp PCR System 9700 thermal cycler. The amplification profile for the PCR mixture was 25 cycles of 60 s at 94°C, 60 s at 55°C, and 60 s at 72°C. Following amplification, a 15-μl sample was electrophoresed on 1.5% agarose gels, stained with ethidium bromide (0.5 μg/ml), and then photographed.
FIG. 2.
DNA sequence of the 314-bp fragment obtained for N. farcinica isolates ATCC 3318T, W6021, W6032, W6954, and W6889. PCR primers Nf1 and Nf2 are designated by arrows. The two CfoI sites are underlined.
Specificity of PCR assays.
Specificity of the PCRs was confirmed by two methods. Amplicons obtained following PCR of 12 N. farcinica isolates with the primer pair Nf1 and Nf2 were first digested with restriction endonuclease (CfoI; Roche Molecular Biochemicals, Indianapolis, Ind.) at 37°C in the buffer recommended by the manufacturer, and the digestion products were resolved through a 2.5% NuSieve agarose gel (25). Two CfoI fragments of 218 and 78 bp were expected to be observed on ethidium bromide-stained gels, but not the 18-bp fragment located between CfoI sites. The 314-bp amplicons of four N. farcinica clinical isolates, W6934, W6021, W6032, and W6889, and the type strain ATCC 3318 obtained with Nf1 and Nf2 primers were purified using reagents and methods supplied with the QIAquick PCR purification kit (QIAGEN). The specificity of primers Nf1 and Nf2 was confirmed by comparing the direct sequences of four purified amplicons of strains W6934, W6021, W6032, and W6889 with the sequence of N. farcinica ATCC 3318T (Fig. 2). One hundred nanograms of PCR-amplified template was sequenced in both directions using the ABI PRISM BigDye terminator cycle sequencing ready reaction kit (Perkin-Elmer) with Nf1 and Nf2 as the sequencing primers.
A patent application on behalf of the Department of Health and Human Services has been filed.
Nucleotide sequence accession number.
GenBank accession numbers of the 409- and 314-bp species-specific DNA fragments for N. farcinica are CL21561 and CL215612, respectively.
RESULTS
RAPD profiles.
RAPD profiles of two clinical isolates (W6310 and W6311) of N. nova and N. nova ATCC 33726T, N. farcinica ATCC 3318T and three clinical isolates of N. farcinica (W6434, W6017, and W6255), N. brasiliensis ATCC 19296T and one clinical isolate of N. brasiliensis (W6312), Mycobacterium fortuitum ATCC 6841T, N. transvalensis ATCC 6865T, N. asteroides ATCC 19247T, N. asteroides complex drug pattern type I (N. abscessus) isolates (W6133 and W6335), and an N. asteroides complex drug pattern type VI (N. cyriacigeorgica) isolate (W6344) generated with primer DKU49 are shown in Fig. 1. RAPD profiles for four isolates of N. farcinica showed an intense fragment of approximately 409 bp (Fig. 1, lanes 4 to 7). Although the 409-bp band appeared species specific for N. farcinica, bands approximately this size were observed also for closely related species, such as N. brasiliensis ATCC 19296T (Fig. 1, lane 8), M. fortuitum ATCC 6841T (Fig. 1, lane 9), and N. asteroides ATCC 19247T (Fig. 1, lane 12).
Cloning and nucleotide sequencing of the 409-bp RAPD fragment.
Three independent subclones of the 409-bp fragment were sequenced in both directions. The DNA sequences of the subclones were identical, suggesting that the 409-bp fragment was composed of a single homologous DNA element. The nucleotide sequences for primers Nf1 and Nf2 are shown in Fig. 2. The BLASTN search for the 409-bp sequence showed no significant homology to any genes or sequences of other species of bacteria available in the GenBank database.
PCR with primers Nf1 and Nf2.
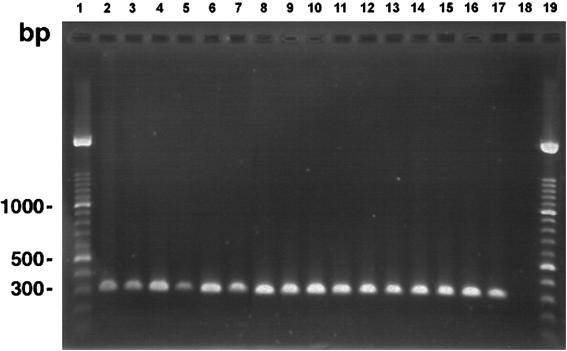
A 314-bp band was obtained from all 28 N. farcinica isolates examined using the Nf1 and Nf2 primers. Figure 3 shows the amplification products obtained for 16 representative N. farcinica isolates. No amplification of the 314-bp band was observed from the genomic DNA of 59 other nocardial species isolates or 41 other phylogenetically related bacterial species isolates (Table 1).
FIG. 3.
Specific amplification of the 314-bp DNA fragment for 16 strains of N. farcinica by using PCR primers Nf1 and Nf2. Lanes 1 and 19, molecular size standard consisting of a 100-bp DNA ladder; lanes 2 to 17, N. farcinica ATCC 3318T, W5185, W5492, W5555, W5871, W5952, W6500, W6544, W6859, W6866, W6889, W6925, W6954, W6993, W7022, and W7028; lane 18, negative control containing no template DNA.
Confirmation of the specificity of primers Nf1 and Nf2.
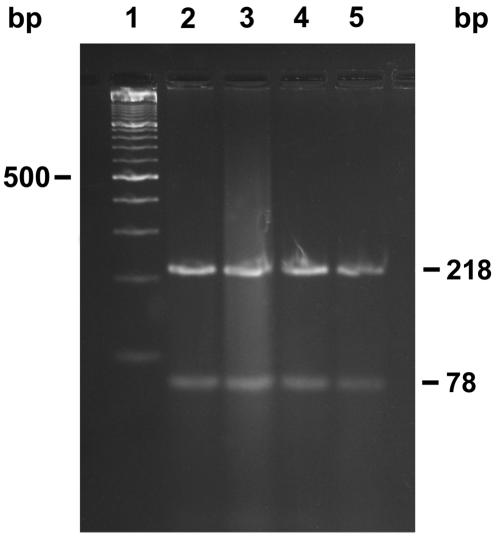
The nucleotide sequence of the 314-bp fragment obtained for N. farcinica ATCC 3318T is shown in Fig. 2. This sequence shows the primer set Nf1 and Nf2. The two CfoI recognition sites are underlined. When we digested Nf1 and Nf2 PCR-amplified fragments of 12 N. farcinica isolates with CfoI, we observed two fragments of 218 and 78 bp; the 18-bp fragment was not visualized as expected. A typical pattern of CfoI restriction fragments for four Nf1 and Nf2 PCR-amplified fragments is shown in Fig. 4. When we sequenced these four Nf1- and Nf2-generated amplicons, they had the identical nucleotide sequence for the 314-bp fragment as the sequence of N. farcinica ATCC 3318T shown in Fig. 2.
FIG. 4.
Restriction endonuclease digestion of the 314-bp fragment from four strains of N. farcinica, using CfoI. Lane 1, molecular size standard consisting of a 100-bp DNA ladder; lanes 2 to 5, N. farcinica W6032, W6021, W6954, and W6889. The 218- and 78-bp CfoI restriction fragments are shown on the right margin.
DISCUSSION
Human and animal clinical infections with N. farcinica may occur more frequently than previously recognized (5, 16, 27). This has been attributed to under diagnosis or, possibly, a change in the spectrum of human nocardiosis in countries such as Germany, where N. farcinica is the prevailing species (22). Other reports from France, Germany, and the United States have implicated N. farcinica as the cause of postoperative wound infections in patients undergoing cardiac and other vascular surgeries (3, 4, 10, 30). From 1987 through 1989, one of the largest known nocardial mastitis epizootics was reported in all 10 Canadian provinces (16). The causative agent of the outbreak was initially reported as Nocardia species but later presumptively identified as N. farcinica (16). Further phenotypic and molecular testing at the Actinomycete Reference Laboratory confirmed the identification. It is important to rapidly identify N. farcinica to enable an earlier diagnosis of infected patients and, as a consequence, optimize their antimicrobial therapy, to enable an improved outcome. These efforts may also likely contribute to more accurate surveillance of the disease to gain a better understanding of the public health impact of this resistant pathogen.
Analyses of restriction fragment length polymorphisms in the hsp and 16S rRNA genes following resolution of PCR-amplified fragments have been reported previously for the identification of Nocardia isolates to the species level (6, 7, 25, 31). Whereas these methods are generally less time-consuming and labor-intensive, discrimination between related species is not always possible, because fragments generated from conserved genes (hsp and 16S rRNA) in different species may result in fragments of similar lengths. The utility of these methods is also hampered because molecular techniques are highly specialized and are not performed in many clinical diagnostic laboratories. The focus on new molecular methods has coincided with an increased number of reports of new Nocardia species. As the number of new nocardial species increases, the interpretation of rapid PCR-based technology devised during the past decade is becoming increasingly complex (6, 7, 25, 31). Conville et al. (6), for example, recommend that care should be used in the interpretation of results of restriction enzyme profiles of the hsp gene alone, as several species, N. africana, N. nova, and N. veterana, have the same enzymatic profile as well as the same antibiotic susceptibility profile. In addition, difficulties have been encountered in the identification of Nocardia isolates solely by comparison of 16S rRNA gene sequences (6, 7, 21, 32). Previously, strains having <3% differences between their 16S rRNA gene sequences were considered the same species (24). However, differences between the 16S rRNA genes for some Nocardia species, such as N. africana and N. veterana, are 1% (6). Yassin et al. (32) have also reported close sequence values (98.3% similarity) between N. puris and N. farcinica. Although these close similarities may reflect improvements in high-quality sequence technologies and analyses, these similarities may lead to misidentification.
In the present study, we have developed a rapid and specific PCR-based assay to improve the identification of N. farcinica. The RAPD method used in this investigation was first described by Exmelin et al. in 1996 (10) and was found to be useful to distinguish among different strains of N. farcinica, though the RAPD profiles were not used for identification of isolates. While relatively easy to perform, identification based on RAPD profiles suffers from the disadvantage of lacking reproducible profiles unless stringent reaction conditions are observed. For instance, profiles were determined to be dependent on reaction parameters such as annealing temperature, magnesium concentration, the type of thermal cycler, source of DNA polymerase, and both primer and template concentrations (9). In addition, except for a pronounced 409-bp band, we observed variation of DKU49 profiles among strains of N. farcinica, making identification based on RAPD profiles alone difficult (Fig. 1). The difficulty of obtaining reproducible RAPD profiles and the changes in the intensities of observed bands may hamper widespread use of this method for clinical identification. RAPD analysis requires adherence to rigorous reaction conditions. Variability of RAPD profiles may be due to annealing of short primers under low-stringency conditions, leading to mismatched hybridization to the non-perfectly complementary target sequences (9), and this suggests the need for a more specific PCR-based test.
A simple PCR assay to rapidly and specifically identify N. farcinica clinical isolates offers an alternative to currently used phenotypic methods and may allow earlier initiation of effective therapy, thus improving patient outcome. Early identification is important for patient management. N. farcinica has been shown to display a high degree of resistance to several antibiotics and requires prompt treatment with appropriate antimicrobial agents (28, 29). A PCR assay to identify N. farcinica has significant advantages beyond phenotypic and other molecular methods for the identification of clinical or environmental isolates. First, a significant reduction in time is required to make an identification. Once DNA is obtained, the assay can be completed in 1 day, in contrast to 1 week for commercially available biochemical identification (2, 13, 20) and 3 weeks for conventional biochemical identification (1). Second, the RAPD-Ready-to-Go beads require less pipetting and therefore decrease the potential for pipetting errors. They also help to standardize reaction conditions. Third, the PCR assay using the Nf1 and Nf2 primer set is performed at high stringency (55°C), allowing only the specific amplification of N. farcinica DNA. No amplification products were observed using the Nf1 and Nf2 primer set for Nocardia spp. other than N. farcinica or for other species of aerobic actinomycetes listed in Table 1. Other advantages include the relatively inexpensive equipment required for amplification of the target DNA and the facts that the PCR assay is easy to perform and the results are easy to interpret.
As the number of new species of Nocardia identified increases, it is becoming evident that taxonomic complexities cause ambiguous interpretation of previously described phenotypic and molecular markers and that earlier identification strategies used over the last decade must be reviewed and revised. The utility and universality of using RAPD profiles to identify a species-specific DNA fragment for the development and testing of PCR assays has been demonstrated in this investigation for the identification of N. farcinica. RAPD analysis has been used to identify species-specific DNA fragments in several different bacterial species, such as Prevotella (11), Bacteroides (26), and Xanthomonas (17). Species-specific DNA fragments have also been obtained by other methods, such as subtractive hybridization (23); however, the latter method is more technically demanding and has the potential to select not only species-specific markers but, more commonly, strain-specific markers.
The PCR assay was able to rapidly and accurately distinguish all isolates of N. farcinica from other species of Nocardia and other closely related aerobic actinomycetes. This PCR assay may simplify identification of this emerging bacterial pathogen to enable an earlier diagnosis and improved outcome for infected patients and provide a useful tool for screening numerous samples rapidly. Such rapid identification may facilitate the availability of N. farcinica strains for further subtyping studies in epidemiologic investigations and enhance the understanding of the role of N. farcinica in the etiology of nocardial infections (16, 30). Future studies will address the sensitivity of the assay as well as evaluate additional epidemiologically linked clinical samples to validate the diagnostic usefulness of Nf1 and Nf2 amplification.
REFERENCES
- 1.Berd, D. 1973. Laboratory identification of clinically important aerobic actinomycetes. Appl. Microbiol. 25:665-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biehle, J. R., S. J. Cavalieri, T. Felland, and B. L. Zimmer. 1996. Novel method for rapid identification of Nocardia species by detection of preformed enzymes. J. Clin. Microbiol. 34:103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumel, J., E. Blumel, A. F. Yassin, H. Schmidt-Rotte, and K. P. Schaal. 1998. Typing of Nocardia farcinica by pulsed-field gel electrophoresis reveals an endemic strain as source of hospital infections. J. Clin. Microbiol. 36:118-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boiron, P., R. Locci, M. Goodfellow, S. A. Gumaa, K. Isik, B. Kim, M. M. McNeil, M. C. Salinas-Carmona, and H. Shojaei. 1998. Nocardia, nocardiosis and mycetoma. Med. Mycol. 36(Suppl. 1):26-37. [PubMed] [Google Scholar]
- 5.Cohen, E., D. Blickstein, E. Inbar, Z. Samra, and M. Weinberger. 2000. Unilateral vocal cord paralysis as a result of a Nocardia farcinica laryngeal abscess. Eur. J. Clin. Microbiol. Infect. Dis. 19:224-227. [DOI] [PubMed] [Google Scholar]
- 6.Conville, P. S., J. M. Brown, A. G. Steigerwalt, J. W. Lee, D. E. Byrer, V. L. Anderson, S. E. Dorman, S. M. Holland, B. Cahill, K. C. Carroll, and F. G. Wistebsky. 2003. Nocardia veterana as a pathogen in North American patients. J. Clin. Microbiol. 41:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conville, P. S., S. H. Fischer, C. P. Cartwright, and F. G. Witebsky. 2000. Identification of Nocardia species by restriction endonuclease analysis of an amplified portion of the 16S rRNA gene. J. Clin. Microbiol. 38:158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmond, E. P., and M. Flores. 1993. Mouse pathogenicity studies of Nocardia asteroides complex species and clinical correlations with human isolates. FEMS Microbiol. Lett. 110:281-284. [DOI] [PubMed] [Google Scholar]
- 9.Ellsworth, D. L., D. Rittenhouse, and R. L. Honetcutt. 1993. Artifactual variation in randomly amplified polymorphic DNA banding patterns. BioTechniques 14:214-217. [PubMed] [Google Scholar]
- 10.Exmelin, L., B. Malbruny, M. Vergnaud, F. Provost, P. Boiron, and C. Morel. 1996. Molecular study of nosocomial nocardiosis outbreak involving heart transplant recipients. J. Clin. Microbiol. 34:1014-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guillot, E., and C. Mouton. 1997. PCR-DNA probe assays for identification and detection of Prevotella intermedia sensu stricto and Prevotella nigrescens. J. Clin. Microbiol. 35:1876-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isik, K., and M. Goodfellow. 2002. Differentiation of Nocardia species by PCR-randomly amplified polymorphic DNA fingerprinting. Syst. Appl. Microbiol. 25:60-67. [DOI] [PubMed] [Google Scholar]
- 13.Kiska, D. L., K. Hicks, and D. J. Pettit. 2002. Identification of medically relevant Nocardia species with an abbreviated battery of tests. J. Clin. Microbiol. 40:1346-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurent, F. J., F. Provost, and P. Boiron. 1999. Rapid identification of clinically relevant Nocardia species to genus level by 16S rRNA gene PCR. J. Clin. Microbiol. 37:99-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lungu, O., P. D. Latta, I. Weitzman, and S. Silverstein. 1994. Differentiation of Nocardia from rapidly growing Mycobacterium species by PCR-RFLP analysis. Diagn. Microbiol. Infect. Dis. 18:13-18. [DOI] [PubMed] [Google Scholar]
- 16.Manninen, K. I., R. A. Smith, and L. O. Kim. 1993. Highly presumptive identification of bacterial isolates associated with the recent Canada-wide mastitis epizootic as Nocardia farcinica. Can. J. Microbiol. 39:635-641. [DOI] [PubMed] [Google Scholar]
- 17.Manulis, S., L. Valinsky, A. Lichter, and D. W. Gabriel. 1994. Sensitive and specific detection of Xanthomonas campestris pv. pelargonii with DNA primers and probes identified by random amplified polymorphic DNA analysis. Appl. Environ. Microbiol. 60:4094-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNeil, M. M., and J. M. Brown. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin. Microbiol. Rev. 7:357-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNeil, M. M., and J. M. Brown. 2002. Nocardia, p. 481-500. In V. L. Yu, R. Weber, and D. Raoult (ed.), Antimicrobial therapy and vaccines. Apple Trees Productions, LLC, New York, N.Y.
- 20.Muir, D. B., and R. C. Pritchard. 1997. Use of the BioMerieux ID 32C yeast identification system for identification of aerobic actinomycetes of medical importance. J. Clin. Microbiol. 35:3240-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth, A., S. Andrees, R. M. Kroppenstedt, D. Harmsen, and H. Mauch. 2003. Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals underspeciation and division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. J. Clin. Microbiol. 41:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaal, K. P., and H. J. Lee. 1992. Actinomycete infections in humans—a review. Gene 15:201-211. [DOI] [PubMed] [Google Scholar]
- 23.Seal, S. E., L. A. Jackson, and M. J. Daniels. 1992. Isolation of a Pseudomonas solanacearum-specific DNA probe by subtraction hybridization and construction of species-specific oligonucleotide primers for sensitive detection by the polymerase chain reaction. Appl. Environ. Microbiol. 58:3751-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 25.Steingrube, V. A., B. A. Brown, J. L. Gibson, R. W. Wilson, J. Brown, Z. Blacklock, K. Jost, S. Locke, R. F. Ulrich, and R. J. Wallace, Jr. 1995. DNA amplification and restriction endonuclease analysis for differentiation of 12 species and taxa of Nocardia, including recognition of four new taxa within the Nocardia asteroides complex. J. Clin. Microbiol. 33:3096-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng, L.-J., P.-R. Hsueh, J.-C. Tsai, F.-L. Chiang, C.-Y. Chen, S.-W. Ho, and K.-T. Luh. 2000. PCR assay for species-specific identification of Bacteroides thetaiotaomicron. J. Clin. Microbiol. 38:1672-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres, O. H., P. Domingo, R. Pericas, P. Boiron, J. A. Montiel, and G. Vazquez. 2000. Infection caused by Nocardia farcinica: case report and review. Eur. J. Clin. Microbiol. Infect. Dis. 19:205-212. [DOI] [PubMed] [Google Scholar]
- 28.Wallace, R. J., Jr., L. C. Steele, G. Sumter, and J. M. Smith. 1988. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob. Agents Chemother. 32:1776-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace, R. J., Jr., M. Tsukamura, B. A. Brown, J. Brown, V. A. Steingrube, Y. S. Zhang, and D. R. Nash. 1990. Cefotaxime-resistant Nocardia asteroides strains are isolates of the controversial species Nocardia farcinica. J. Clin. Microbiol. 28:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wenger, P. N., J. M. Brown, M. M. McNeil, and W. R. Jarvis. 1998. Nocardia farcinica sternotomy site infections in patients following open heart surgery. J. Infect. Dis. 178:1539-1543. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, R. W., V. A. Steingrube, B. A. Brown, and R. J. Wallace, Jr. 1998. Clinical application of PCR-restriction enzyme pattern analysis for rapid identification of aerobic actinomycete isolates. J. Clin. Microbiol. 36:148-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yassin, A. F., B. Straubler, P. Schumann, and K. P. Schaal. 2003. Nocardia puris sp. nov. Int. J. Syst. Evol. Microbiol. 53:1595-1599. [DOI] [PubMed] [Google Scholar]