Abstract
Protein glycation in biological systems occurs predominantly on lysine, arginine and N-terminal residues of proteins. Major quantitative glycation adducts are found at mean extents of modification of 1–5 mol percent of proteins. These are glucose-derived fructosamine on lysine and N-terminal residues of proteins, methylglyoxal-derived hydroimidazolone on arginine residues and Nε-carboxymethyl-lysine residues mainly formed by the oxidative degradation of fructosamine. Total glycation adducts of different types are quantified by stable isotopic dilution analysis liquid chromatography-tandem mass spectrometry (LC-MS/MS) in multiple reaction monitoring mode. Metabolism of glycated proteins is followed by LC-MS/MS of glycation free adducts as minor components of the amino acid metabolome. Glycated proteins and sites of modification within them – amino acid residues modified by the glycating agent moiety - are identified and quantified by label-free and stable isotope labelling with amino acids in cell culture (SILAC) high resolution mass spectrometry. Sites of glycation by glucose and methylglyoxal in selected proteins are listed. Key issues in applying proteomics techniques to analysis of glycated proteins are: (i) avoiding compromise of analysis by formation, loss and relocation of glycation adducts in pre-analytic processing; (ii) specificity of immunoaffinity enrichment procedures, (iii) maximizing protein sequence coverage in mass spectrometric analysis for detection of glycation sites, and (iv) development of bioinformatics tools for prediction of protein glycation sites. Protein glycation studies have important applications in biology, ageing and translational medicine – particularly on studies of obesity, diabetes, cardiovascular disease, renal failure, neurological disorders and cancer. Mass spectrometric analysis of glycated proteins has yet to find widespread use clinically. Future use in health screening, disease diagnosis and therapeutic monitoring, and drug and functional food development is expected. A protocol for high resolution mass spectrometry proteomics of glycated proteins is given.
Keywords: Glycation; Mass spectrometry; Proteomics; Glucose; Methylglyoxal, fructosamine; Hydroimidazolone; Bioinformatics; Orbitrap
Protein glycation in biological systems
Protein glycation is a spontaneous post-translational modification (PTM) of proteins found in biological systems. It involves the non-enzymatic covalent attachment of a reducing sugar or sugar derivative to a protein [1]. It is a PTM that is often thermally and chemically labile when removed from the physiological setting, particularly at high pH and temperature. Analysis of protein glycation is compromised by use of heating and high pH in pre-analytic processing for mass spectrometric analysis [2].
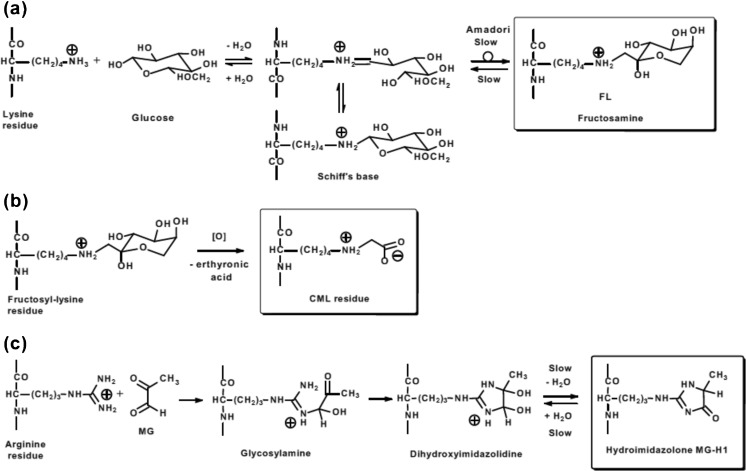
Glycation adducts are classified into two groups: early-stage glycation adducts and advanced glycation endproducts (AGEs). Glucose reacts with amino groups of lysine residue side chains and N-terminal amino acid residues to form sequentially a Schiff’s base and then, via the Amadori rearrangement, Nε-(1-deoxy-D-fructos-1-yl)lysine (FL) and Nα-(1-deoxy-D-fructos-1-yl)amino acid residues – called collectively fructosamines – Fig.1a. FL is also known by synonyms fructosyl-lysine and fructoselysine. These are early-stage glycation adducts. Schiff’s base adducts are usually a minor component of glucose adducts in situ, ca. 10 % of the level of FL residues in the steady-state. They are also relatively rapidly reversed during sample isolation and processing, whereas fructosamines have much slower reversibility of formation; chemical relaxation times for reversal of Schiff’s base and fructosamine formation are ca. 2.5 h and 38 h at pH 7.4 and 37 °C, respectively [6, 7]. Accordingly when adducts of early-stage glycation by glucose are detected and quantified it is typically the fructosamine proteome that is characterised – as the Schiff’s base adduct reverses during protein isolation. Fructosamine modification of proteins is usually low, 5–10 mol% of the protein modified by one fructosamine residue. Examples of proteins found to be susceptible to fructosamine formation are given in Table 1. Collectively these and other proteins susceptible to fructosamine formation constitute the “fructosamine proteome”.
Fig. 1.
Major protein glycation processes in physiological systems. (a) Early glycation. Formation of the Schiff’s base and fructosamine (Amadori product) of lysine residues. (b) Oxidative degradation of fructosyl-lysine to Nε-carboxymethyl-lysine (CML). Similar processes occur on N-terminal amino acid residues. (c) Glycation of arginine residues by methylglyoxal with formation of dihydroxyimidazolidine and hydroimidazolone MG-H1 residues. There are related structural isomers and similar adducts formed from glyoxal and 3-deoxyglucosone [1, 3–5]
Table 1.
Selected components of the fructosamine proteome
| Species | Protein | Hotspot sites | Extent of modification | Functional impairment | Reference |
|---|---|---|---|---|---|
| Human | Apolipoprotein A1 | K239 | 4 % | None | [8] |
| Apolipoprotein E | K93 | Unknown | Impairs heparin binding | [9] | |
| Bisphosphoglycerate mutase | K158 | Unknown | Inactivation | [10] | |
| CD59 | K41 | Unknown | Inactivation | [11] | |
| Complement factor B | K266 | Unknown | [12] | ||
| Gastric inhibitory polypeptide | Y1 | Unknown | Increased insulin release | [13] | |
| Glucagon-like peptide-1 | 1H | Unknown | Decreased insulin release | [14] | |
| Hemoglobin α2β2 | α-K61, β-V1, β-K66 | 5 % (α:β, 0.6:1) | Increased oxygen binding in T state. | [15–17] | |
| Insulin | β-F1 | Decreased activity. | [18] | ||
| Microglobulin, β2- | I1 | Unknown | Aggregation in chronic renal dialysis. | [19] | |
| Serum albumin | D1, K199, K439, K525 | 10 % | Decreased drug binding and leakage through the glomerular filter. | [20, 21] | |
| Superoxide dismutase-1 | K122, K128 | Unknown | Inactivation | [22] | |
| Bovine | Crystallin, αA | K11, K78 | Unknown | [23] | |
| Crystallin, αB | K90, K92 | Unknown | [23] | ||
| Crystallin, γB | G1, K2 | Unknown | [24] | ||
| Glutathione peroxidase-1 | K117 | Unknown | Inactivated. | [25] | |
| Insulin | α-G1, β-F1, β-K29 | Unknown | [26] | ||
| Major intrinsic peptide | K238, K259 | Unknown | Affects membrane permeability. | [27] | |
| Ribonuclease A | K1, K7, K41 | Unknown | [28] | ||
| Serum albumin | K12, K136, K211, K232, K377, K524 | 10 % | [29] | ||
| Rat | Collagen-I | α1-K434 | 50–70 % | Increased susceptibility to cross-linking. | [30] |
| α2-K453 | 27–33 % | ||||
| α2-K479 | 24–29 % | ||||
| α2-K924 | 22–28 % | ||||
| Aldoketo reductase 1 A1 | K67, K84, K140 | 18 % | Inactivation. | [31] |
Extent of modification: data are for extent of modification in vivo of healthy subjects except for rat collagen which are percentage of total fructosamine adducts on each polypeptide chain (α1 and α2) from rat donors 6–36 months of age
In later stage reactions of glycation in physiological systems, Schiff’s base and fructosamine adducts degrade to many stable end-stage adducts or AGEs [1]. Endogenous α-oxoaldehyde metabolites are potent glycating agents and react with proteins to form AGEs directly. An important dicarbonyl glycating agent is methylglyoxal. The well-studied AGE, Nε-carboxymethyl-lysine (CML), is mainly formed by the oxidative degradation of fructosyl-lysine [7, 32] and hence has the same proteome site-specific coverage as the fructosyl-lysine precursor – Fig. 1b. There are also minor contributions to CML from glycation of proteins by glycolaldehyde and glyoxal, which may have a different site-specific distribution [33, 34]. A major AGE quantitatively found in biological systems is the methylglyoxal-derived hydroimidazolone, Nδ-(5-hydro-5-methyl-4-imidazolon-2-yl)-ornithine (MG-H1) [3]. Comparisons of CML and MG-H1 residue contents of proteins, respectively, are (mean ± SD): healthy human subjects - plasma protein 0.038 ± 0.005 mmol/mol lys versus 0.31 ± 0.20 mmol/mol arg, and haemoglobin - 0.075 ± 0.023 mmol/mol lys versus 2.62 ± 0.60 mmol/mol arg (n – 12) [35]; and rats (Sprague Dawley, non-diabetic) – liver 0.100 ± 0.030 mmol/mol lys versus 3.34 ± 0.32 mmol/mol arg, skeletal muscle – 0.188 ± 0.093 mmol/mol lys versus 1.77 ± 0.77 mmol/mol arg, kidney (glomeruli) 0.27 ± 0.15 mmol/mol lys versus 2.42 ± 0.79 mmol/mol arg, brain 0.35 ± 0.07 mmol/mol lys versus 2.73 ± 0.39 mmol/mol arg, and plasma protein 0.042 ± 0.013 mmol/mol lys versus 1.23 ± 0.49 mmol/mol arg (n = 13) [3, 36, 37].
Methylglyoxal reacts predominantly with arginine residues to form sequentially a glycosylamine, dihydroxyimidazolidine and hydroimidazolone MG-H1 residues – Fig. 1c. Other structural isomers are also found: MG-H2 and MG-H3 [4]; isomer MG-H1 is usually dominant in vivo [38]. The half-life for reversal of glycosylamine/ dihydroxyimidazolidine formation is ca. 1.8 days and for reversal of hydroimidazolone is ca. 12 days at pH 7.4 and 37 °C [39]. The stability of the hydroimidazolone decreases with increasing pH; the half-life of MG-H1 is 0.87 days at pH 9.4 [4]. Hence both dihydroxyimidazolidine and hydroimidazolone residues derived from arginine residues may be detected in mass spectrometric analysis of glycated proteins. Glycation of proteins by methylglyoxal is found at levels of 1–5 mol% in most proteins but increases to ca. 50 % in the human lens of elderly subjects where there is limited protein turnover [5, 38]. It often occurs at functional domains of proteins and leads to protein inactivation and dysfunction. This may be because arginine residues have the highest probability (20 %) of any amino acid to be found in a functional domain and there is loss of positive charge on formation of MG-H1 [40, 41]; modification of these arginine residues outside functional domains of proteins is unlikely to lead to protein inactivation, unless involved in a key structural interaction – for example, ion-pair interaction R123 of human apolipoprotein A-1 [42]. Gene knockout of glyoxalase 1 (Glo-1), the enzyme that protects against glycation by methylglyoxal, is embryonically lethal and increased methylglyoxal concentration, or dicarbonyl stress, imposed by Glo-1 deficiency accelerates the ageing process and exacerbates diseases – including cardiovascular disease, diabetes, renal failure and neurological disorders [43]. Proteins susceptible to methylglyoxal glycation are called collectively the “dicarbonyl proteome”. Examples are given in Table 2.
Table 2.
Selected components of the dicarbonyl proteome
| Species | Protein | Hotspot sites | Arginine agent | Extent of modification | Functional impairment | Reference |
|---|---|---|---|---|---|---|
| Human | Apolipoprotein A1 | R27, R123, R149 | Methyl- glyoxal (MG) | ca. 1 % | R27 – increased catabolism; R123 – decreased stability; R149 – impaired functional activity. | [42] |
| Apolipoprotein B100 | R18 | MG | Increased density, proteoglycan binding and atherogenicity | [44] | ||
| Collagen-IV | α1-R390, α2-R889, α2-R1452, α3–1404 | MG | ca. 5 % | Decreased integrin binding. | [45, 46] | |
| Crystallin, αA- | R12, R65, R157, R163 | MG | Unknown | Increased chaperone activity | [47] | |
| ß-Defensin-2 | R22, R23 | Glyoxal /MG | Unknown | Decreased antimicrobial activity. | [48] | |
| Fibrin(ogen) | α-R167, α-R199, α-R491, α-R528, β-R149, β-R304 | MG | Unknown | Abnormal thrombosis and fibrinolysis | [49] | |
| Heat shock protein-27 | R75, R89, R94, R127, R136, R140, R188 | MG | Unknown | Enhanced protection against oxidative stress. | [50] | |
| Hemoglobin α2β2 | α-R31, α-92, α-141, β-R30, β-R 40, β-R104 | MG | ca. 2.6 % | Increased oxygen binding | [35, 51, 52] | |
| HIF1α–co-activator p300 | R354 | MG | Unknown | Decreased hypoxia response | [53] | |
| Insulin | R46 | MG | Unknown | Aggregation | [54] | |
| IgG (monoclonal) | LC-R30 | MG | 5 % | Acidic variant. | [55] | |
| Plasminogen | R504, R530, R561 | MG | Unknown | Likely functional changes to cleavage and lys binding pocket in fibrinolysis. | [56] | |
| Proteasome, 20S subunits | β2-R85, β4-R224, β4–231, β5-123, β5–128 | MG | Unknown | Decreased proteasome activity | [57] | |
| Bovine | Ribonuclease A | R10, R39, R85 | Glyoxal/MG | Unknown | Inhibition | [58, 59] |
| Serum albumin | R114, R186, R218, R257, R410, R428 | MG | ca. 1 % | Inhibition of esterase activity, prostaglandin breakdown and decreased drug binding. | [60, 61] | |
| Mouse | mSin3a co-repressor | R925 | MG | Unknown | Increased angiopoietin-2 activity | [62] |
Extent of modification: data are for extent of modification in vivo of healthy subjects
Glycated proteins undergo proteolysis in physiological systems to release glycated amino acids called “glycation free adducts”. They are trace components of the amino acid metabolome. These are found in plasma and other body fluids. They are excreted from the body in urine. Urinary excretion of glycation free adduct increases from 2 to 15-fold in diabetes and renal failure [35, 63].
Detection of total amounts of glycation adducts in multiplexed assay by liquid chromatography-tandem mass spectrometry
Some of the earliest applications of mass spectrometry to the study of glycated proteins was the detection of chemically-defined glycation adducts by gas chromatography-mass spectrometry. Prior acid hydrolysis and N- and O-acetylation of glycation adducts was required to produce low molecular mass volatile adducts suitable for detection [32]. The use of liquid chromatography-tandem mass spectrometry (LC-MS/MS) with an electrospray ionisation source avoided the requirement of chemical derivatisation of glycation adducts and in multiple reaction monitoring (MRM) data acquisition mode gave the high sensitivity and specificity of detection required to quantify glycation adduct in physiological systems. Use of Hypercarb™ graphitic chromatography retains glycation adducts during the chromatographic step to allow for diversion of non-volatile salts to waste before entry of analyte-containing eluate flow into the mass spectrometer. This minimises ion suppression and maintains a clean electrospray ionisation source for good, stable sample batch-to-batch performance [3]. Alternatively, ion-pair chromatography has been used [64]. Stable isotopic dilution analysis provides for robust quantitation and LC-MS/MS is now the analytical platform that dominates the field for robust, quantitative and multiplexed analysis of glycation adducts [64–66]. A wide range of glycation, oxidation and nitration adducts are routinely analysed by LC-MS/MS multiplexed assay in our laboratory. The protocol, detection conditions and analytical performance have been given elsewhere [65].
Stable isotopic dilution analysis LC-MS/MS may be applied for direct detection of glycation free adducts in ultrafiltrate of physiological fluids. The LC-MS/MS analysis is extended to quantify total glycation adduct contents of purified proteins and protein extracts of cells and extracellular matrix by prior exhaustive enzymatic hydrolysis [3]. The enzymatic hydrolysis method is similar to that employed by Henle et al. [67] with sequential addition of pepsin, pronase E and finally, added together, aminopeptidase and prolidase. We have made multiple modifications for improvement and specific applications: (i) aseptic processing with a sample autoprocessor (CTC-PAL, CTC Analytics, Zwingen, Switzerland) and inclusion of antibiotics after the acidic pepsin step to minimise bacterial contamination; and (ii) a modified procedure for specific proteins - using collagenase instead of pepsin for analysis of collagen [45], omitting pepsin for apolipoprotein B100 as some pepsin fragments are insoluble and resist further digestion [68], and performing the enzymatic hydrolysis under carbon monoxide for hemoglobin or red blood cell lysates to inactivate heme and prevent artefactual heme-catalysed glycoxidation [3, 35]. Analytical recoveries are ca. 90 % or higher for minimally glycated proteins but are lower in highly glycated proteins where there is resistance to proteolysis [3, 69]. An alternative method uses pronase E, aminopeptidase and carboxypeptidase Y and gave lower analytical recovery in the application studied [70]. Conventional acid hydrolysis cannot be used for acid labile AGEs such as hydroimidazolones for which very low analytical recoveries were found – ca. 10 % [4]. Acid hydrolysis may be used for acid-stable AGEs [71].
Acid hydrolysis has also been used for many years in the Nε-(2-furoylmethyl)lysine or furosine-based measurement of FL. The conversion of FL to furosine in acid hydrolysis is 32 %. Furosine is also formed from other Amadori products in food [72]. Recently a LC-MS/MS method has been developed for concurrent quantitation of furosine and acid-stable AGEs, CML and Nε-(1-carboxyethyl)lysine (CEL) [73].
Methodological considerations for application of mass spectrometric proteomics to glycation research – glycated protein detection and quantitation
Proteomics studies provide a powerful approach to identify proteins susceptible to glycation in complex protein mixtures and also identify the lysine and arginine residues within proteins particularly susceptible to glycation. A typical workflow involves: (i) preparation of a protein extracts of samples of interest, (ii) reduction and alkylation of sample protein, (iii) limited proteolysis of proteins – usually by trypsin or lys-C and trypsin sequentially; (iv) partial resolution of tryptic peptides by nanoflow reversed phase liquid chromatography, and (v) detection and sequencing of tryptic peptides by high resolution mass spectrometry. Peptides are sequenced by fragmentation by collision induced dissociation (CID), high-energy collisional dissociation (HCD) or electron transfer dissociation (ETD) and detection and analysis of characteristic fragment ion series.
In all methods for proteomics studies, a critical requirement is unambiguous identification of proteins of interest. For many years a consensus criterion for protein identification was detection and sequencing of a minimum of two tryptic peptides unique in sequence, “unique peptides”, for the protein of interest. With the latest high resolution mass analysers this criterion has now been challenged and one unique peptide with estimation of false discovery rate is gaining acceptance as a criterion for protein identification [74].
Quantitation of tryptic peptides and thereby sample content of related proteins is an on-going challenge in proteomics studies. For analysis of complex protein mixtures quantitation is preferably based on mass spectrometric detection – rather than densitometry of spots of stained gels from 2-dimensional gel electrophoresis. This is because of potential interferences when using gel electrophoretic separation is the only basis for protein resolution. Nano-flow liquid chromatography-high resolution mass spectrometry is the major platform currently used for proteomics studies. A popular operating format is tryptic peptide molecular ion determination by ultrahigh resolution Orbitrap™ mass analyser and rapid consecutive peptide fragmentation for sequencing performed by an on-line ion trap mass analyser. For quantitation of proteins there are label-free and heavy isotopic labelling of sample methods. The most robust method of quantitation is use of stable isotope labelling with amino acids in cell culture (SILAC) and similar stable isotope labelling of mice. Stable isotopic lysine and arginine, [13C6]lysine and [13C6]arginine, are used in cell cultures to label proteins or in animal diets to label mouse tissue protein. Labelled cell culture reagents and mouse tissues are available commercially. Stable isotopic-labelled and normal, natural isotopic abundance, samples are processed identically and cell lysates or tissue extracts mixed prior to tryptic digestion to provide 13C-labelled internal standards for all peptides in subsequent stable isotopic dilution analysis work flow [75]. An alternative method is the introduction of isobaric tags for relative and absolute quantification (iTRAQ), which uses N-hydroxysuccinimide chemistry and N-methyl piperazine reporter group stable isotopic labels [76]. This may pose problems in glycation research for dicarbonyl proteome analysis as previous studies have shown the dicarbonyl moiety of hydroimidazolones migrates between arginine residues during N-hydroxysuccinimide active ester derivatisation conditions – see below. The iTRAQ protocol may require validation for glycation adduct detection and quantitation. A generally available method is label-free quantitation, which requires no additional sample manipulation but rather employs peptide ion responses for quantitation. Initially a normalization procedure for sample total ion current is performed using algorithm-based peak selection and exclusion so that only invariant ion responses are used in the normalization correction. The amount of tryptic peptide is then deduced from the sum of ion intensities of multiply-charged ion series of the peptide. For detection of an unmodified protein, the average, total or 3 most intense molecular ion intensities for unique peptides is used. Several commercial software tools are available for this analysis [77]. We have used Progenesis™ (Nonlinear Dynamics Ltd., Newcastle upon Tyne, U.K.) and Scaffold™ (Proteome Software, Inc., Portland, USA).
For application to detection and quantitation of glycated proteins, the glycated protein and its unglycated counterpart (typically 20–100 fold more abundant) are detected based on the ion intensities of the glycated and related unglycated tryptic peptide. This is only secure if the site of glycation is in a unique peptide; if not, the glycated peptide ion intensity has contributions of unknown proportions from different proteins. Often with glycation, a tryptic cleavage is missed and then the precursor dipeptide is preferably a unique dipeptide for secure identification of the related glycation proteins.
The methods for protein quantitation above provide relative quantitation of analytes. If absolute quantitation is required then the response is compared to that of analyte calibration standards and absolute amounts may be deduced. A further current development is LC-MS/MS with MRM analysis of glycated peptides after trypsinisation for absolute quantitation of particular glycated proteins in clinical and other samples [78, 79]. In this application, it is recommended that quantitation is based on a minimum of three molecular ion > fragment ion MRM transitions [80].
A great challenge for global screening of glycated proteins is to maximize sequence coverage of proteins in mass spectrometric analysis. Leading research teams performing total proteome analysis report a typical median sequence coverage of ca. 20 % [81]. A contributory factor to this is production of short peptides of ambiguous protein origin. This may be improved in some glycated proteins where the glycation adduct causes missed cleavage with trypsin and lys-C with resultant longer peptides. A recent computational approach has indicated that with judicious use of proteases the sequence coverage in proteomics analysis may increase to ca. 90 % [82]. Until this is routinely implemented, only a minor proportion of glycated proteins are likely detected and identified proteomics analysis.
Bioinformatics
Bioinformatics tools for protein glycation are poorly developed. A particularly useful bioinformatics tool for glycation researchers would be a sequence search engine to predict sites susceptible to glycation in proteins. This can be approached empirically – building up a peptide motif glycation site motif on the basis of frequency of occurrence of amino acids on N- terminal and C-terminal sides of the target lysine or arginine residue glycated. For a non-enzymatic process such as glycation, a peptide susceptibility motif may also be predicted from a physicochemical approach where characteristics that make a particular lysine or arginine residue reactive towards glycation are considered. It is not yet possible to predict preferred, hotspot sites of protein glycation with surety. Current empirical data and physicochemical and glycation adduct turnover considerations are now described.
An examination of protein motifs for glucose glycation forming FL residues was made empirically by compiling and combining peptide motifs from published peptide mapping studies. It was found that K and R residues dominate in the N-terminal region and D and E residues dominate in the C-terminal region of FL sites but no clear motif for FL formation was found [83]. In a study of human plasma and red blood cells, detection and filtering for unique peptides with ≥5 spectrum counts gave 361 and 443 unique glycated peptide sequences from native human plasma and red blood cells, respectively. There was only limited evidence to support the hypothesis of N-terminal enrichment of K and R residues and C-terminal enrichment of D and E residues in the sequence motif for hotspot glycation by glucose [84].
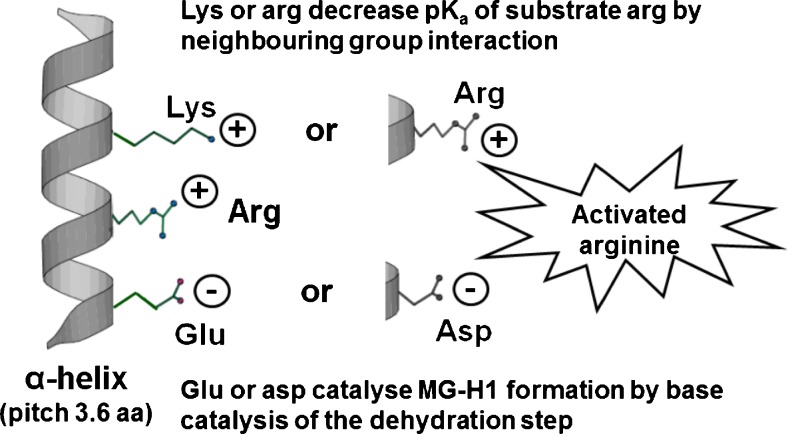
Regarding physiochemical considerations, glycation is a non-enzymatic process and so selectivity for sites of glycation is determined by the reactivity of the lysine, arginine of N-terminal residue under consideration. This is linked to: (i) microscopic pKa of the residue being modified, (ii) surface exposure of the modification site, and (iii) a proximate conjugate base catalyzing the dehydration step involved in FL and MG-H1 residue formation – Fig. 2.
Fig. 2.
Activation of arginine residues in alpha-helix domains of proteins by neighbouring group interactions with basic and acidic amino acid residues. Figure reproduced with permission from [5]
Microscopic pKa values of lysine, N-terminal and arginine residues have a profound influence on the site of glycation by glucose on N-terminal and lysine residues and on glycation by methylglyoxal of arginine residues. Microscopic pKa values may be computed for proteins of known crystal structure – for example, by using the H++ automated system (http://biophysics.cs.vt.edu/H++) [85]. There is marked diversity of pKa values of lys and arg residues in proteins. For example, in human serum albumin (HSA) microscopic pKa values of the 59 lys residues vary from 7.9–14.0 and of the 24 arg residues vary from 12.2–18.6; an expected reactivity range of >106; cf. reactivity of N-terminal serine pKa of 7.9 [86]. The major sites of glycation by glucose in HSA are, in order of reactivity: N-terminal D1, K525, K199 and K439 [20]; cf. their rank order by increasing pKa value of lysine side chain and N-terminal amino groups of first-equal, sixteenth, third and fourteeth. Low pKa values are likely driving glycation of D1 and K199. Activating features of K525 and K439 may be deprotonation catalyzed by proximate E520/R521 and E442, respectively.
In a study of the hotspots sites of glycation of HSA by methylglyoxal, 3 of the 5 sites with MG-H1 residue formation had the lowest microscopic predicted pKa values: R218, pKa = 12.2; and R186 and R410, pKa = 12.5. However, the remaining 2 sites, R114 and R428 with predicted pKa values of 13.6 and 15.1, ranked 8th and 14th of 24 arginine residues in order of increasing microscopic predicted pKa value. R114 has high surface exposure which likely also facilitates methylglyoxal modification. All activated arginine residues have a positively charged R or K residue 3 or 4 residues further along in the sequence that likely decreases the microscopic pKa value and R428 only has a negatively charged residue, E425, preceding in the sequence. A subsequent study confirmed these hotspot sites except for R114 and suggested R257 as a further hotspot modification site, which has a relatively low pKa (= 12.9) [60]. The proximity of a negatively charged D or E residue provides a conjugate base to promote the rate limiting removal of a proton from the protein-glucose Schiff’s base and arginyl-dihydroxyimidazolidine precursors of fructosamine and MG-H1 adducts, respectively. The combination of proximate cationic and anion side chain residues for lysine and arginine residue activation was initially proposed to explain site specificity of lysine residue glycation by glucose [87] and then applied to MG-H1 formation from arginine [5].
The above considerations are features relating to the rate of formation of glycation adducts. FL and MG-H1 residues have half-lives of ca. 25 and 12 days, respectively [4, 7], which exceeds the half-lives of most human proteins (median half-life 1.9 days [88]). Therefore, for many proteins the steady-state extent of protein glycation is also influenced by the half-life of the protein. Hence, early studies found the extent of glycation by glucose of several proteins in vivo was linked to the protein half-life [89]. Glycation leads to protein distortion and misfolding, as indicated by crystallographic studies of HSA glycated by glucose [90] and molecular graphics structural predictions for glycation of HSA, apolipoprotein B100 and apolipoprotein A-1 by methylglyoxal [42, 44, 61]. It is also expected that glycated proteins are targeted for cellular proteolysis and have an unusually decreased half-life. This remains to be determined in robust unfocussed, proteome dynamics studies where half-lives of unique tryptic peptides (which can be unambiguously linked back to particular proteins) and their glycated counterparts are determined in the same cell population. The level of FL and N-terminal fructosamine residues in cellular proteins is also influenced by enzymatic removal and repair by fructosamine-3-phosphokinase (F3PK) [91]. F3PK has different specific activity for FL residues in different sites in proteins. The FL residues detected at different sites in proteins are, therefore, a balance of the intrinsic reactivity for glycation and the reactivity of the FL residue site for repair by FP3K – see glycated haemoglobin, for example [92]. There is no known enzymatic mechanism for repair of MG-H1 residues.
Pre-analytic processing and analytical protocols for mass spectrometric applications in glycation research
We gave updated pre-analytic processing protocols for detection of total amounts of glycation adducts in multiplexed assay by LC-MS/MS recently [65]. We give herein a similar protocol for detection of glycated proteins by high resolution mass spectrometry proteomics – Table 3.
Table 3.
Protocol for high resolution mass spectrometry proteomics of glycated proteins
| Step | Description | Procedure |
|---|---|---|
| 1 | Preparation of biological samples | Prepare fractional proteome cell extract as for analysis of total glycation adduct content e.g. for proteins >10 kDa molecular mass of the cytosolic proteome, cells (ca. 1 x 106) are lysed by sonication in 10 mM sodium phosphate buffer, pH 7.4 and 4 °C, and membranes sedimented by centrifugation (20,000 g, 30 min, 4 °C). The supernatant is removed and washed by 5 cycles of concentration and dilution in water over a 10 kDa microspin ultrafilter. Protein is finally concentrated and assayed by the Bradford method. |
| 2 | Alkylation | To an aliquot of cytosolic protein extract (100 μg, 20 μl), dithiothreitol (6 μl, 6 mM) is added and the sample incubated at 37 °C in the dark for 30 min. Iodoacetamide solution (5.9 μl, 10.8 mM) is then added and the sample incubated at 37 °C in the dark for 30 min. Residual iodoacetamide is quenched by further addition of dithiothreitol (5.9 μl, 6 mM) and incubated at 37 °C in the dark for 30 min. An aliquot of Lys-C protease (1 mg/ml, 5 μl) in 500 mM ammonium bicarbonate, pH 8.0, is added and incubated for 1 h at 37 °C. Then tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (1 mg/ml, 5 μl) in 1 mM calcium chloride/500 mM ammonium bicarbonate, pH 8.0, is added and samples were incubated at 37 °C for 5 h in the dark. The sample is then lyophilised to dryness and re-suspended in an aliquot (100 μl) 0.1 % formic acid in water and analysed by nanoflow liquid-chromatography-Orbitrap mass spectrometry. |
| 3 | Peptide separation, protein identification and quantitation | An aliquot of sample (5 μl) is injected and peptides partially resolved by nanoflow capillary liquid chromatography – see footnote. Peptides were eluted directly (300 nl min−1) via a Triversa Nanomate nanospray source (Advion Biosciences, NY, USA) into a Thermo Orbitrap Fusion (Q-OT-qIT, Thermo Scientific) mass spectrometer. Survey scans of peptide precursors from 350 to 1500 m/z are performed at 120 K resolution (at 200 m/z) with automatic gain control (AGC) 4 × 105. Precursor ions with charge state 2–7 were isolated (isolation at 1.6 Th in the quadrupole) and subjected to HCD fragmentation. HCD was programmed to 35 % and used for rapid scan MS analysis in the ion trap where AGC is set to 1 x 104 and the maximum injection time was 200 ms. Dynamic exclusion duration was set to 45 s with a 10 ppm tolerance around the selected precursor and its isotopes. Monoisotopic precursor selection is turned on. The instrument was run in top speed mode with 2 s cycles. |
| 4 | Data collection | Sequence information from the MS/MS data was managed by converting the raw (.raw) files into a merged file (.mgf) using MSConvert in ProteoWizard Toolkit (version 3.0.5759) [93] The resulting. Mgf files were searched, and the database was searched against protein sequence databases. |
| 5 | Data Analysis | Database search MS2 spectra are searched with Mascot engine (Matrix Science, version 2.5.0) against Homo sapiens database (http://www.uniprot.org/) assuming trypsin digestion. To determine false-positive peptide identifications, spectra are also searched against the corresponding reverse database, common Repository of Adventitious Proteins Database (http://www.thegpm.org/cRAP/index.html). Search parameters for Precursor mass and product ions tolerance are, respectively, ± 5 ppm and ±0.8 Da, allowing for two missed trypsin cleavages, cysteine carbamidomethylation and methionine oxidation. Only fully tryptic peptide matches were allowed. |
| 6 | Validation | Scaffold (version Scaffold_4.3.2, Proteome Software Inc.) is used to validate MS/MS based peptide and protein identifications from MS/MS sequencing results. Peptide identifications are accepted if established at > 95.0 % probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if established at >95.0 % probability and contained at least 3 identified peptides – two of which are unique. Protein probabilities were assigned by the Protein Prophet algorithm [94]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. |
Footnotes: Instrumentation and c hromatography: Reversed phase nanoflow liquid chromatography- mass spectrometry for global protein identification is performed on an Orbitrap Fusion (Thermo) mass spectrometer equipped with a microspray source operating in positive ion mode. The column used is: an Acclaim PepMap μ-pre-column cartridge (trap), 300 μm i.d. × 5 mm, 5 μm particle size, 100 Å pore size, fitted to an Acclaim PepMap RSLC 75 μm i.d. × 50 cm, 2 μm particle size, 100 Å pore size main column (Thermo). It was installed on an Ultimate 3000 RSLC nano system (Tthermo). The peptides are eluted off of the trap onto the analytical column. Mobile phases were: A - 0.1 % formic acid in water, and B - 0.1 % formic acid in acetonitrile. The flow rate was programmed at 0.3 μl/min. Mobile phase B was increased from 3 % to 35 % in 125 to 220 min (depending on the complexity of the sample). Mobile phase B was then increased from 35 % to 80 % in 5 min before being brought back quickly to 3 % in 1 min. The column was equilibrated at 3 % of mobile phase B for 15 min before the next sample
Statistical analysis: The mean, standard deviation, confidence score and ANOVA test for all proteins are determined using datasets of a minimum of 3 independent sample digests using bioinformatics sand statistical analysis by Progenesis QI for proteomics 2.0 (Nonlinear Dynamics). Protein and peptide identification probabilities are performed using Progenesis
Example of application of mass spectrometry in studies of early glycation adducts: fructosamine
Fructosamine residues in peptides and proteins
The mass increment indicating the detection of FL and other fructosamine-modified peptides is +162 Da. Glycation of intact proteins and large peptide chains has been detected by electrospray positive ion mass spectrometry and matrix-assisted laser desorption-time of flight (MALDI-TOF) mass spectrometry. Roberts and co-workers detected and quantified fructosamine modified α- and β-chains of haemoglobin by deconvolution of multiply charged ion series [95], shown in later studies by peptide mapping to reflect fructosamine formation at sites α-K61, β-V1 and β-K66 [15]. Increase in molecular mass of HSA glycated by glucose prepared in vitro was measured by MALDI-TOF. This revealed that preparations of glycated HSA had a large increase in mass due to high extent of glycation, dissimilar from the low increase in mass of glycated HSA in plasma samples in vivo. For example, HSA from human plasma had mean mass increment of +243 Da, whereas model glucose-modified albumin prepared in vitro had a mean mass increment of +6780 Da [96]. This suggested the albumin prepared with very high extent of glycation was a poor model for the albumin with minimal extent of glycation found in vivo.
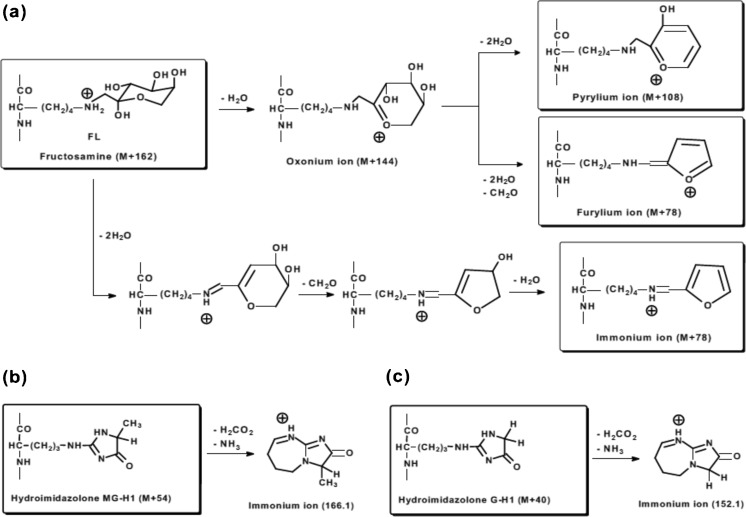
For mass spectrometric analysis of glycated peptides, CID and HCD fragmentation of fructosamine-containing peptides produced characteristic fragment ions of the precursor fructosamines (M + 162): by dehydration to an oxonium ion (M + 144), further dehydration to a pyrylium ion (M + 108), and dehydration and formaldehyde loss immonium ion (M + 78) [29, 97–99] – Fig. 3a. Pyrylium and furylium ions are detected in y ion series providing for fructosamine location [29]. In ETD fragmentation abundant and almost complete series of c- and z-type ions were observed, which greatly facilitated the peptide sequencing and fructosamine site location [100].
Fig. 3.
Fragmentation of fructosamine and hydroimidazolone glycation adducts. (a) Fragmentation of fructosyl-lysine by CID leading to formation of oxonium, pyrylium, furylium and immonium ions. (b) and (c) fragmentation of hydroimidazolones formed by methylglyoxal and glyoxal to immonium ions in CID and HCD [29, 97–99]
The FL degradation product, CML, is detected at the same sites as fructosamines residues in serum albumin, haemoglobin and ribonuclease A [29, 101, 102].
Enrichment strategies of fructosamine-modified proteins
A boronate affinity chromatography enrichment method has been used to facilitate detection of the fructosamine proteome based on the binding of the cis-1,2-diol structure of fructosamine-modified proteins, with subsequent release from the boronate affinity matrix with weak acid. Although some enzymatically glycosylated proteins contain cis-1,2-diol moieties, steric effects, proximate negatively charged groups and acetylation limit the retention and interference in this method by glycoproteins [103]. A similar affinity method is used in the routine separation of hemoglobin in clinical chemistry to quantify glycated hemoglobin HbA1c for assessment of glycaemic control in diabetes [104].
In principle antibodies to fructosamine may be used for immunoaffinity purification and enrichment of proteins glycated by glucose. Fructosamine may be reduced to hexitol-lysine residues prior to enrichment and then immunoaffinity purified with anti-hexitol-lysine antibody [105]. Where immunoaffinity enrichment is employed it is vital to confirm the presence of a glycation adduct residue or hexitol-lysine following reductive pre-analytic processing in the retained proteins by mass spectrometric analysis.
Global analysis of fructosamine-modified protein
For the fructosamine proteome, phenylboronate affinity chromatography was used to enrich glycated proteins and glycated tryptic peptides from both human plasma and erythrocyte membranes. Enriched proteins are processed for limited proteolysis by trypsin or trypsin and lys-C. Trypsinisation cleavage after lysine residues is impaired by glycation by glucose and glycated peptide with missed-cleavage at the glycation site is detected [84].
The rate of fructosamine degradation at 37 °C increases markedly above pH 8 through increased reversal of the Amadori rearrangement and oxidative degradation to CML and related Nα-carboxymethyl-amino acids [106, 107] – Fig. 1b. Use of high pH and high temperatures in pre-analytic processing of samples in glycation studies are avoided to maintain sample glycation analyte integrity. Conventional tryptic digestion in proteomics studies often uses tryptic digestion for 24 h at pH 8.5 with heating at 50 – 60 °C in some protocols [108]. This is preferably avoided in glycation studies where tryptic digestion at pH 7.4 and 37 °C is preferred.
Analysis of tryptic digests by liquid chromatography-tandem mass spectrometry with ETD peptide fragmentation identified 76 and 31 proteins with fructosamine modification from human plasma and erythrocyte membranes, respectively. The ETD fragmentation mode enabled identification of a higher number of glycated peptides (88 % of all identified peptides) compared to CID mode (17 % of all identified peptides) for samples enriched on protein and peptide levels [103]. In other studies with boronate affinity enrichment of proteins in human plasma and red blood cells, 7749 unique glycated peptides corresponding to 3742 unique glycated proteins were identified [84].
Example of application of mass spectrometry in studies of advanced glycation adducts: methylglyoxal-derived hydroimidazolone and dihydroxyimidazolidine
Methylglyoxal-derived hydroimidazolone and dihydroxyimidazolidine in peptides and proteins
Methylglyoxal-derived hydroimidazolone and dihydroxyimidazolidine may be detected in peptides glycated by methylglyoxal and tryptic peptides of proteins glycated in vivo. They have mass increments on arginine residues of +54 Da and +72 Da, respectively. A further minor methylglyoxal-derived and stable AGE, CEL, may be detected as +72 Da on lysine residues [42, 44, 58, 61]. High collision energy fragmentation may dehydrate dihydroxyimidazolidine to hydroimidazolone and so discrimination is provided by detection of the peptide molecular ion [58].
In analysis of methylglyoxal-modified lipoproteins no advantage of ETD over CID fragmentation in the detection of hydroimidazolone and dihydroxyimidazolidine was found [42, 44]. Fragmentation of peptides modified by MG-H1 and related isomers gave complete series of b and y ions with mass increment of 54 Da relative to those of unmodified peptide and no neutral losses [42, 44, 58, 61]. A MG-H1-related, peptide free side chain fragment ion of m/z = 166.1 can be observed in the low-mass region of the MS/MS spectra, with proposed immonium ion structure – Fig. 2b. A similar peptide-free side-chain fragment ion of m/z = 152.1 can be observed for glyoxal-modified peptides [109] – Fig. 2c.
Hydroimidazolone and dihydroxyimidazolidine residues are chemically labile AGEs and conditions of pre-analytic processing for proteomics analysis may influence mass spectrometric analysis outcomes. Tryptic digestion methods with prolonged periods of sample incubation at high pH and/or temperature leads to reversal of hydroimidazolone to dihydroxyimidazolidine and de-glycation. Alternatively, high pH and temperature may also stimulate dicarbonyl formation [110]. In earlier studies using N-hydroxysuccinimidyl active ester derivatisation of MG-H1 in chromatographic analysis, we found that incubation of MG-H1 in the presence of [15N2]arginine at pH 8.8 for 10 min at 55 °C led to migration of the methylglyoxal moiety from MG-H1 to [15N2]arginine [4]. Hence, use of high pH and temperature in pre-analytic processing may induce migration of the methylglyoxal moiety between arginine residues and, potentially, also between proteins. Conventional tryptic digestion techniques require modification to minimise increase of pH and avoid sample heating for peptide mapping and proteomics analysis of methylglyoxal-modified proteins and related PTMs.
Trypsin cleavage after arginine residues is impaired by glycation by methylglyoxal and glycated dipeptides with missed-cleavage at the glycation site are detected [60, 61]. In some cases, cleavage after dicarbonyl glycation of arginine was observed [109].
Enrichment strategies for methylglyoxal-modified proteins
The dihydroxyimidazolidine residues present in proteins glycated by methylglyoxal and glyoxal are also a potential interference in boronate affinity chromatography for enrichment of fructosamine-modified proteins as they contain a side chain with a cis-1,2-diol moiety [111]. This has been exploited to identify proteins with arginine residues activated for reaction with glyoxal derivatives. Reaction of proteins with butan-2,3-dione formed 4,5-dihydroxy-4,5-dimethylimidazolidine residues of proteins containing activated arginine residues. Proteins with such residues on the surface were retained in boronate affinity chromatography [112, 113]. Antibodies to hydroimidazolones may be used for immunoaffinity purification and enrichment of proteins glycated by methylglyoxal. The anti-MG-H1 monoclonal antibody IG7 has been widely used for immunoblotting of MG-H1 and would be suitable for this application [114]. Where employed it is vital to confirm the presence of hydroimidazolone residues in the retained proteins by mass spectrometric analysis [2].
Global analysis of methylglyoxal-modified proteins
In a recent report [109] plasma digests were analysed by nanoflow chromatography-LTQ Orbitrap XL ETD mass spectrometry and tryptic peptides scanned for m/z 152.1 and 166.1 side chain fragment ions indicative of glyoxal- and methylglyoxal-modified peptides – see above and Fig. 2b and c. Forty-four peptides representing 42 proteins were annotated. Arginine modifications were mostly represented by glyoxal-derived hydroimidazolones (34 peptides/39 sites) and methylglyoxal-derived dihydroxyimidazolidine (8 peptides/8 sites) and MG-H1 (14 peptides/14 sites). Use of high temperature and pH processing in this study may have compromised the outcome; many glyoxal modified proteins were detected whereas LC-MS/MS analysis typically shows very low amounts of glyoxal-derived-AGEs, hydroimidazolone and Nω-carboxymethylarginine, in plasma protein [35].
In pilot studies using nanoflow liquid chromatography-Orbitrap Fusion™ mass spectrometry with peptide HCD fragmentation, we analyzed cytosolic protein extracts of primary human periodontal ligament fibroblasts (hPDLFs) cultured in low and high glucose concentration (8 mM and 25 mM glucose, respectively). Cell cytosolic protein had total MG-H1 residue content of ca. 0.42 and 0.72 mmol/mol arg in low and high glucose concentration (P < 0.01), respectively, measured by LC-MS/MS analysis of exhaustive enzymatic digests. In Lys C-tryptic digests, Orbitrap Fusion analysis (see Table 3) detected 1077 proteins in both low and high glucose concentration cultures. Thirty proteins were found modified in hPDLF by MG-H1 residues: 10 in hPDLF in low glucose incubation and 20 proteins were detected in high glucose incubations. As a positive control, cell protein extracts were incubated with methylglyoxal to increased MG-H1 content ca. 20-fold wherein 173 proteins were detected with MG-H1 modification (unpublished observations) – similar to application of mass spectrometric Orbitrap™ analysis of endothelial cell proteins [40].
Example of application of mass spectrometry in studies of advanced glycation adducts: glucosepane
Glucosepane is a major glycation-derived quantitative crosslink in physiological systems [115]. Glycation of ribonuclease A with glucose led to the formation of glucosepane cross-links, which were found at residues K41-R39 and K98-R85. The only other intermolecular cross-link observed was the 3-deoxyglucosone-derived 2-ammonio-6-({2-[(4-ammonio-5-oxido-5-oxopentyl) amino]-4-(2, 3,4-trihydroxybutyl)-4,5-dihydro-1H-imidazol-5-ylidene}amino)hexanoate (DODIC) at residues K1-R39. The identity of cross-linked peptides was confirmed by sequencing with tandem mass spectrometry. Recombinant ribonuclease A mutants R39A, R85A, and K91 A were produced and glycated to confirm importance of these sites for cross-linking [116].
Conclusions/recommendations
Mass spectrometry is often the method of choice for detection and quantitation of glycation adduct content of biological samples where multiplexing for multiple analyte detection is interference-free and addition of further analytes has little incremental cost for analysis. Mass spectrometry proteomics provides for identification of proteins glycated in complex mixtures and concurrent assessment of the effect of glycation on the amounts of all proteins in the sample. Our recommendations for glycation research applications are:
Use of stable isotopic dilution analysis LC-MS/MS for detection and quantitation of early and advanced glycation endproducts;
use of enzymatic hydrolysis for application to protein samples;
immunoassay of glycation adducts be corroborated and referenced to the LC-MS/MS technique where practicable; and
use of nanoflow liquid chromatography-Orbitrap™ mass spectrometry with label-free or SILAC approaches for glycated protein identification and quantification in complex mixtures.
In the future there will likely be introduction of stable isotopic dilution analysis LC-MS/MS based quantitation of glycated proteins in to clinical chemistry laboratories.
Acknowledgments
We thank the Biosciences and Biotechnology Research Council, European Union FP7 programme and British Heart Foundation for support for our glycation research. AA thanks the Ministry of Education, Government of Saudi Arabia, for a PhD studentship.
Abbreviations
- AGC
Automatic gain control
- AGE
Advanced glycation endproduct
- CID
Collision induced dissociation
- CML
Nε-Carboxymethyl-lysine
- CEL
Nε-(1-Carboxyethyl)lysine
- DODIC
2-ammonio-6-({2-[(4-ammonio-5-oxido-5-oxopentyl) amino]-4-(2,3,4-trihydroxybutyl)-4,5-dihydro-1H-imidazol-5-ylidene}amino)hexanoate
- ETD
Electron transfer dissociation
- FDR
False discovery rate
- FL
Nε-(1-Deoxy-D-fructos-1-yl)lysine (fructosyl-lysine or fructoselysine).
- F3PK
Fructosamine-3-phosphokinase
- Glo-1
Glyoxalase 1
- HCD
High-energy collisional dissociation
- hPDLF
Human periodontal ligament fibroblasts
- HSA
Human serum albumin
- iTRAQ
Isobaric tags for relative and absolute quantification
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
- MALDI-TOF
Matrix-assisted laser desorption-time of flight
- MG
Methylglyoxal
- MG-H1
Nδ-(5-hydro-5-methyl-4-imidazolon-2-yl)-ornithine
- MG-H2
2-amino-5-(2-amino-5-hydro-5-methyl-4-imidazolon-1-yl)pentanoic acid
- MG-H3
2-amino-5-(2-amino-4-hydro-4-methyl-5-imidazolon-1-yl)pentanoic acid
- MRM
Multiple reaction monitoring
- PTM
post-translational modification
- SILAC
Stable isotope labelling with amino acids in cell culture
- Th
Thomson unit (m/e = 1)
- TPCK
Tosylphenylalanylchloromethyl ketone
References
- 1.Rabbani N, Thornalley PJ. Glycation research in amino acids: a place to call home. Amino Acids. 2012;42(4):1087–1096. doi: 10.1007/s00726-010-0782-1. [DOI] [PubMed] [Google Scholar]
- 2.Thornalley PJ, Rabbani N. Detection of oxidized and glycated proteins in clinical samples using mass spectrometry - a user's perspective. Biochim Biophys Acta. 2014;1840(2):818–829. doi: 10.1016/j.bbagen.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 3.Thornalley PJ, Battah S, Ahmed N, Karachalias N, Agalou S, Babaei-Jadidi R, Dawnay A. Quantitative screening of advanced glycation endproducts in cellular and extracellular proteins by tandem mass spectrometry. Biochem. J. 2003;375(3):581–592. doi: 10.1042/bj20030763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed N, Argirov OK, Minhas HS, Cordeiro CA, Thornalley PJ. Assay of advanced glycation endproducts (AGEs): surveying AGEs by chromatographic assay with derivatisation by aminoquinolyl-N-hydroxysuccimidyl-carbamate and application to Nε-carboxymethyl-lysine- and Nε-(1-carboxyethyl)lysine-modified albumin. Biochem. J. 2002;364(1):1–14. doi: 10.1042/bj3640001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabbani N, Thornalley PJ. Methylglyoxal, glyoxalase 1 and the dicarbonyl proteome. Amino Acids. 2012;42(4):1133–1142. doi: 10.1007/s00726-010-0783-0. [DOI] [PubMed] [Google Scholar]
- 6.Baynes JW, Thorpe SR, Murtiashaw MH. Nonenzymatic glucosylation of lysine residues in albumin. Methods Enzymol. 1984;106:88–98. doi: 10.1016/0076-6879(84)06010-9. [DOI] [PubMed] [Google Scholar]
- 7.Smith PR, Thornalley PJ. Mechanism of the degradation of non-enzymatically glycated proteins under physiological conditions. (Studies with the model fructosamine, N -(1-deoxy-D-fructose-1-yl)hippuryl-lysine). Eur J Biochem. 1992;210(3):729–739. doi: 10.1111/j.1432-1033.1992.tb17474.x. [DOI] [PubMed] [Google Scholar]
- 8.Calvo C, Ulloa N, Campos M, Verdugo C, Ayrault-Jarrier M. The preferential site of non-enzymatic glycation of human apolipoprotein A-I in vivo. Clin Chim Acta. 1993;217(2):193–198. doi: 10.1016/0009-8981(93)90165-Z. [DOI] [PubMed] [Google Scholar]
- 9.Shuvaev VV, Fujii J, Kawasaki Y, Itoh H, Hamaoka R, Barbier A, Ziegler O, Siest G, Taniguchi N. Glycation of apolipoprotein E impairs its binding to heparin: identification of the major glycation site. Biochim Biophys Acta. 1999;1454(3):296–308. doi: 10.1016/S0925-4439(99)00047-2. [DOI] [PubMed] [Google Scholar]
- 10.Fujita T, Suzuki K, Tada T, Yoshihara Y, Hamaoka R, Uchida K, Matuo Y, Sasaki T, Hanafusa T, Taniguchi N. Human erythrocyte Bisphosphoglycerate mutase: inactivation by glycation in vivo and in vitro. J Biochem. 1998;124(6):1237–1244. doi: 10.1093/oxfordjournals.jbchem.a022243. [DOI] [PubMed] [Google Scholar]
- 11.Acosta J, Hettinga J, Flückiger R, Krumrei N, Goldfine A, Angarita L, Halperin J. Molecular basis for a link between complement and the vascular complications of diabetes. Proc Natl Acad Sci U S A. 2000;97(10):5450–5455. doi: 10.1073/pnas.97.10.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niemann MA, Bhown AS, Miller EJ. The principal site of glycation of human complement factor B. Biochem J. 1991;274(2):473–480. doi: 10.1042/bj2740473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Harte FPM, Abdel-Wahab YHA, Conlon JM, Flatt PR. Amino terminal glycation of gastric inhibitory polypeptide enhances its insulinotropic action on clonal pancreatic B-cells. Biochim Biophys Acta. 1998;1425(2):319–327. doi: 10.1016/S0304-4165(98)00084-1. [DOI] [PubMed] [Google Scholar]
- 14.O'Harte FPM, Abdel-Wahab YHA, Conlon JM, Flatt PR. Glycation of glucagon-like peptide-1(7–36)amide: characterization and impaired action on rat insulin secreting cells. Diabetologia. 1998;41(10):1187–1193. doi: 10.1007/s001250051050. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Medzihradszky KF, Cunningham J, Lee PDK, Rognerud CL, Ou CN, Harmatz P, Witkowska HE. Characterization of glycated hemoglobin in diabetic patients: usefulness of electrospray mass spectrometry in monitoring the extent and distribution of glycation. J ChromatogrB. 2001;759(1):1–15. doi: 10.1016/S0378-4347(01)00196-7. [DOI] [PubMed] [Google Scholar]
- 16.Coletta M, Amiconi G, Bellelli A, Bertollini A, Čarsky J, Castagnola M, Condò S, Brunori M. Alteration of T-state binding properties of naturally glycated hemoglobin, HbA1c. J Mol Biol. 1988;203(1):233–239. doi: 10.1016/0022-2836(88)90104-0. [DOI] [PubMed] [Google Scholar]
- 17.Wang SH, Wang TF, Wu CH, Chen SH. In-depth comparative characterization of hemoglobin glycation in normal and diabetic bloods by LC-MSMS. J Amer Soc Mass Spectrometry. 2014;25(5):758–766. doi: 10.1007/s13361-014-0830-2. [DOI] [PubMed] [Google Scholar]
- 18.O'Harte FPM, Højrup P, Barnett CR, Flatt PR. Identification of the site of glycation of human insulin. Peptides. 1996;17(8):1323–1330. doi: 10.1016/S0196-9781(96)00231-8. [DOI] [PubMed] [Google Scholar]
- 19.Miyata T, Inagi R, Wada Y, Ueda Y, Iida Y, Takahashi M, Taniguchi N, Maeda K. Glycation of human beta 2 -microglobulin in patients with hemodialysis-associated amyloidosis: identification of the glycated sites. Biochemistry. 1994;33(40):12215–12221. doi: 10.1021/bi00206a026. [DOI] [PubMed] [Google Scholar]
- 20.Barnaby OS, Cerny RL, Clarke W, Hage DS. Quantitative analysis of glycation patterns in human serum albumin using 16O/18O-labeling and MALDI–TOF MS. Clin Chim Acta. 2011;412(17–18):1606–1615. doi: 10.1016/j.cca.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabbani N, AntonySunil A, Rossing K, Rossing P, Tarnow L, Parving HH, Thornalley PJ. Effect of Irbesartan treatment on plasma and urinary protein glycation, oxidation and nitration markers in patients with type 2 diabetes and microalbuminuria. Amino Acids. 2011;42(5):1627–1639. doi: 10.1007/s00726-011-0857-7. [DOI] [PubMed] [Google Scholar]
- 22.Arai K, Maguchi S, Fujii S, Ishibashi H, Oikawa K, Taniguchi N. Glycation and inactivation of human Cu-Zn-superoxide dismutase - identification of the invitro glycated sites. J Biol Chem. 1987;262(35):16969–16972. [PubMed] [Google Scholar]
- 23.Abraham EC, Cherian M, Smith JB. Site selectivity in the glycation of αA-crystallin and αB-Crystallins by glucose. Biochem Biophys Res Commun. 1994;201(3):1451–1456. doi: 10.1006/bbrc.1994.1866. [DOI] [PubMed] [Google Scholar]
- 24.Casey EB, Zhao HR, Abraham EC. Role of glycine 1 and lysine 2 in the glycation of bovine g B-crystallin. J.Biol.Chem. 1995;270(35):20781–20786. doi: 10.1074/jbc.270.35.20781. [DOI] [PubMed] [Google Scholar]
- 25.Baldwin JS, Lee L, Leung TK, Muruganandam A, Mutus B. Identification of the site of nonenzymatic glycation of glutathione-peroxidase - rationalization of the glycation-related catalytic alterations on the basis of 3-dimensional protein-structure. Biochim Biophys Acta. 1995;1247(1):60–64. doi: 10.1016/0167-4838(94)00202-R. [DOI] [PubMed] [Google Scholar]
- 26.Guedes S, Vitorino R, Domingues MRM, Amado F, Domingues P. Mass spectrometry characterization of the glycation sites of bovine insulin by tandem mass spectrometry. J Am Soc Mass Spectrom. 2009;20(7):1319–1326. doi: 10.1016/j.jasms.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Swamy Mruthinti S, Schey KL. Mass spectroscopic identification of in vitro glycated sites of MIP. Curr Eye Res. 1997;16(9):936–941. doi: 10.1076/ceyr.16.9.936.5037. [DOI] [PubMed] [Google Scholar]
- 28.Watkins NG, Thorpe SR, Baynes JW. Glycation of amino groups in protein. (Studies on the specificity of modification of RNase by glucose) J Biol Chem. 1985;260(19):10629–10636. [PubMed] [Google Scholar]
- 29.Frolov A, Hoffmann P, Hoffmann R. Fragmentation behavior of glycated peptides derived from D-glucose, D-fructose and D-ribose in tandem mass spectrometry. J Mass Spectrom. 2006;41(11):1459–1469. doi: 10.1002/jms.1117. [DOI] [PubMed] [Google Scholar]
- 30.Reiser KM, Amigable M, Last JA. Nonenzymatic glycation of type I collagen (the effects of aging on preferential glycation sites. J.Biol.Chem. 1992;267(34):24207–24216. [PubMed] [Google Scholar]
- 31.Takahashi M, Lu Y-b, Myint T, Fujii J, Wada Y, Taniguchi N. In vivo glycation of aldehyde reductase, a major 3-deoxyglucosone reducing enzyme: identification of glycation sites. Biochemistry. 1995;34(4):1433–1438. doi: 10.1021/bi00004a038. [DOI] [PubMed] [Google Scholar]
- 32.Ahmed MU, Thorpe SR, Baynes JW. Identification of Nε-carboxymethyl-lysine as a degradation product of fructoselysine in glycated protein. J Biol Chem. 1986;261(11):4889–4894. [PubMed] [Google Scholar]
- 33.Wells-Knecht MC, Thorpe SR, Baynes JW. Pathways of formation of glycoxidation products during glycation of collagen. Biochemistry. 1995;34(46):15134–15141. doi: 10.1021/bi00046a020. [DOI] [PubMed] [Google Scholar]
- 34.Anderson MM, Requena JR, Crowley JR, Thorpe SR, Heinecke JW. The myeloperoxidase system of human phagocytes generates Nε-(carboxymethyl)lysine on proteins: a mechanism for producing advanced glycation end products at sites of inflammation. J Clin Invest. 1999;104(1):103–113. doi: 10.1172/JCI3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed N, Babaei-Jadidi R, Howell SK, Beisswenger PJ, Thornalley PJ. Degradation products of proteins damaged by glycation, oxidation and nitration in clinical type 1 diabetes. Diabetologia. 2005;48(8):1590–1603. doi: 10.1007/s00125-005-1810-7. [DOI] [PubMed] [Google Scholar]
- 36.Babaei-Jadidi R, Karachalias N, Kupich C, Ahmed N, Thornalley PJ. High dose thiamine therapy counters dyslipidaemia in streptozotocin-induced diabetic rats. Diabetologia. 2004;47:2235–2246. doi: 10.1007/s00125-004-1582-5. [DOI] [PubMed] [Google Scholar]
- 37.Karachalias N, Babaei-Jadidi R, Rabbani N, Thornalley PJ. Increased protein damage in renal glomeruli, retina, nerve, plasma and urine and its prevention by thiamine and benfotiamine therapy in a rat model of diabetes. Diabetologia. 2010;53(7):1506–1516. doi: 10.1007/s00125-010-1722-z. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed N, Thornalley PJ, Dawczynski J, Franke S, Strobel J, Stein G, Haik GM., Jr Methylglyoxal-derived hydroimidazolone advanced glycation endproducts of human lens proteins. Investig Ophthalmol Vis Sci. 2003;44(12):5287–5292. doi: 10.1167/iovs.03-0573. [DOI] [PubMed] [Google Scholar]
- 39.Lo TWC, Selwood T, Thornalley PJ. Reaction of methylglyoxal with aminoguanidine under physiological conditions and prevention of methylglyoxal binding to plasma proteins. Biochem Pharmacol. 1994;48(10):1865–1870. doi: 10.1016/0006-2952(94)90584-3. [DOI] [PubMed] [Google Scholar]
- 40.Rabbani N, Thornalley PJ. Dicarbonyl proteome and genome damage in metabolic and vascular disease. Biochem Soc Trans. 2014;42(2):425–432. doi: 10.1042/BST20140018. [DOI] [PubMed] [Google Scholar]
- 41.Gallet X, Charloteaux B, Thomas A, Braseur R. A fast method to predict protein interaction sites from sequences. JMolBiol. 2000;302(4):917–926. doi: 10.1006/jmbi.2000.4092. [DOI] [PubMed] [Google Scholar]
- 42.Godfrey L, Yamada-Fowler N, Smith JA, Thornalley PJ, Rabbani N. Arginine-directed glycation and decreased HDL plasma concentration and functionality. Nutrition and diabetes. 2014;4 doi: 10.1038/nutd.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabbani N, Thornalley PJ. Dicarbonyl stress in cell and tissue dysfunction contributing to ageing and disease. Biochem Biophys Res Commun. 2015;458(2):221–226. doi: 10.1016/j.bbrc.2015.01.140. [DOI] [PubMed] [Google Scholar]
- 44.Rabbani N, Godfrey L, Xue M, Shaheen F, Geoffrion M, Milne R, Thornalley PJ. Conversion of low density lipoprotein to the pro-atherogenic form by methylglyoxal with increased arterial proteoglycan binding and aortal retention. Diabetes. 2011;60(7):1973–1980. doi: 10.2337/db11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobler D, Ahmed N, Song LJ, Eboigbodin KE, Thornalley PJ. Increased dicarbonyl metabolism in endothelial cells in hyperglycemia induces anoikis and impairs angiogenesis by RGD and GFOGER motif modification. Diabetes. 2006;55(7):1961–1969. doi: 10.2337/db05-1634. [DOI] [PubMed] [Google Scholar]
- 46.Pedchenko VK, Chetyrkin SV, Chuang P, Ham AJ, Saleem MA, Mathieson PW, Hudson BG, Voziyan PA. Mechanism of perturbation of integrin-mediated cell-matrix interactions by reactive carbonyl compounds and its implication for pathogenesis of diabetic nephropathy. Diabetes. 2005;54(10):2952–2960. doi: 10.2337/diabetes.54.10.2952. [DOI] [PubMed] [Google Scholar]
- 47.Gangadhariah MH, Wang BL, Linetsky M, Henning C, Spanneberg R, Glomb MA, Nagaraj RH. Hydroimidazolone modification of human alpha A-crystallin: effect on the chaperone function and protein refolding ability. Biochim Biophys Acta. 2010;1802(4):432–441. doi: 10.1016/j.bbadis.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiselar JG, Wang X, Dubyak GR, El Sanadi C, Ghosh SK, Lundberg K, Williams WM. Modification of β-Defensin-2 by Dicarbonyls methylglyoxal and glyoxal inhibits antibacterial and chemotactic function in vitro. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0130533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lund T, Svindland A, Pepaj M, Jensen AB, Berg JP, Kilhovd B, Hanssen KF. Fibrin(ogen) may be an important target for methylglyoxal-derived AGE modification in elastic arteries of humans. Diabetes Vascular Disease Research. 2011;8(4):284–294. doi: 10.1177/1479164111416831. [DOI] [PubMed] [Google Scholar]
- 50.Oya-Ito T, Naito Y, Takagi T, Handa O, Matsui H, Yamada M, Shima K, Yoshikawa T. Heat-shock protein 27 (Hsp27) as a target of methylglyoxal in gastrointestinal cancer. Biochim Biophys Acta. 2011;1812(7):769–781. doi: 10.1016/j.bbadis.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, Ahmed N, Thornalley PJ. Peptide mapping of human hemoglobin modified minimally by methylglyoxal in vitro. AnnNYAcadSci. 2005;1043:905. doi: 10.1196/annals.1333.119. [DOI] [PubMed] [Google Scholar]
- 52.Gao Y, Wang Y. Site-selective modifications of arginine residues in human hemoglobin induced by methylglyoxal. Biochemistry. 2006;45(51):15654–15660. doi: 10.1021/bi061410o. [DOI] [PubMed] [Google Scholar]
- 53.Thangarajah H, Yao DC, Chang EI, Shi YB, Jazayeri L, Vial IN, Galiano RD, Du XL, Grogan R, Galvez MG, Januszyk M, Brownlee M, Gurtner GC. The molecular basis for impaired hypoxia-induced VEGF expression in diabetic tissues. Proc Natl Acad Sci U S A. 2009;106(32):13505–13510. doi: 10.1073/pnas.0906670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliveira L, Lages A, Gomes R, Neves H, Familia C, Coelho A, Quintas A. Insulin glycation by methylglyoxal results in native-like aggregation and inhibition of fibril formation. BMC Biochem. 2011;12(1):41. doi: 10.1186/1471-2091-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chumsae C, Gifford K, Lian W, Liu H, Radziejewski CH, Zhou ZS. Arginine modifications by methylglyoxal: discovery in a recombinant monoclonal antibody and contribution to acidic species. Anal Chem. 2013;85(23):11401–11409. doi: 10.1021/ac402384y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kinsky OR. Dicarbonyl Protein Adduction: Plasminogen as a Target and Metformin as a Scavenging Therapeutic in Type 2 Diabetes. PhD thesis. PhD thesis. Tucson, USA: University of Arizona; 2014. [Google Scholar]
- 57.Queisser MA, Yao D, Geisler S, Hammes HP, Lochnit G, Schleicher ED, Brownlee M, Preissner KT. Hyperglycemia impairs proteasome function by methylglyoxal. Diabetes. 2010;59(3):670–678. doi: 10.2337/db08-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brock JWC, Cotham WE, Thorpe SR, Baynes JW, Ames JM. Detection and identification of arginine modifications on methylglyoxal-modified ribonuclease by mass spectrometric analysis. J Mass Spectrom. 2007;42(1):89–100. doi: 10.1002/jms.1144. [DOI] [PubMed] [Google Scholar]
- 59.Cotham WE, Metz TO, Ferguson PL, Brock JWC, Hinton DJS, Thorpe SR, Baynes JW, Ames JM. Proteomic analysis of arginine adducts on glyoxal-modified ribonuclease. Mol Cell Proteomics. 2004;3(12):1145–1153. doi: 10.1074/mcp.M400002-MCP200. [DOI] [PubMed] [Google Scholar]
- 60.Kimzey, M.J., Yassine, H.N., Riepel, B.M., Tsaprailis, G., Monks, T.J., Lau, S.S.: New site(s) of methylglyoxal-modified human serum albumin, identified by multiple reaction monitoring, alter warfarin binding and prostaglandin metabolism. Chem Biol Interact 192(1–2), 122–128 (2011). [DOI] [PMC free article] [PubMed]
- 61.Ahmed N, Dobler D, Dean M, Thornalley PJ. Peptide mapping identifies hotspot site of modification in human serum albumin by methylglyoxal involved in ligand binding and esterase activity. J. Biol. Chem. 2005;280(7):5724–5732. doi: 10.1074/jbc.M410973200. [DOI] [PubMed] [Google Scholar]
- 62.Yao D, Taguchi T, Matsumura T, Pestell R, Edelstein D, Giardino I, Suske G, Rabbani N, Thornalley PJ, Sarthy VP, Hammes HP, Brownlee M. High glucose increases angiopoietin-2 transcription in microvascular endothelial cells through methylglyoxal modification of mSin3A. J Biol Chem. 2007;282(42):31038–31045. doi: 10.1074/jbc.M704703200. [DOI] [PubMed] [Google Scholar]
- 63.Agalou S, Ahmed N, Babaei-Jadidi R, Dawnay A, Thornalley PJ. Profound mishandling of protein glycation degradation products in uremia and dialysis. J Amer Soc Nephrol. 2005;16(5):1471–1485. doi: 10.1681/ASN.2004080635. [DOI] [PubMed] [Google Scholar]
- 64.Teerlink T, Barto R, ten Brink HJ, Schalkwijk CG. Measurement of Nî-(carboxymethyl)lysine and Nî-(carboxyethyl)lysine in human plasma protein by stable-isotope-dilution tandem mass spectrometry. Clin Chem. 2004;50(7):1222–1228. doi: 10.1373/clinchem.2004.031286. [DOI] [PubMed] [Google Scholar]
- 65.Rabbani N, Shaheen F, Anwar A, Masania J, Thornalley PJ. Assay of methylglyoxal-derived protein and nucleotide AGEs. Biochem Soc Trans. 2014;42(2):511–517. doi: 10.1042/BST20140019. [DOI] [PubMed] [Google Scholar]
- 66.Fan X, Sell DR, Zhang J, Nemet I, Theves M, Lu J, Strauch C, Halushka MK, Monnier VM. Anaerobic vs aerobic pathways of carbonyl and oxidant stress in human lens and skin during aging and in diabetes: a comparative analysis. Free Radic Biol Med. 2010;49(5):847–856. doi: 10.1016/j.freeradbiomed.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henle T, Walter H, Klostermeyer H. Evaluation of the extent of the early Maillard-reaction in milk products by direct measurement of the Amadori-product lactuloselysine. ZLebensmUntersForsch. 1991;193:119–122. doi: 10.1007/BF01193359. [DOI] [PubMed] [Google Scholar]
- 68.Rabbani N, Varma Chittari M, Bodmer CW, Zehnder D, Ceriello A, Thornalley PJ. Increased glycation and oxidative damage to apolipoprotein B100 of LDL in patients with type 2 diabetes and effect of metformin. Diabetes. 2010;59(4):1038–1045. doi: 10.2337/db09-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed N, Thornalley PJ. Chromatographic assay of glycation adducts in human serum albumin glycated in vitro by derivatisation with aminoquinolyl-N-hydroxysuccimidyl-carbamate and intrinsic fluorescence. Biochem J. 2002;364:15–24. doi: 10.1042/bj3640015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Glomb MA, Pfahler C. Amides are novel protein modifications formed by physiological sugars. J Biol Chem. 2001;276(45):41638–41647. doi: 10.1074/jbc.M103557200. [DOI] [PubMed] [Google Scholar]
- 71.Scheffer PG, Bakker SJL, Heine RJ, Teerlink T. Measurement of low-density lipoprotein particle size by high-performance gel-filtration chromatography. Clin Chem. 1997;43(10):1904–1912. [PubMed] [Google Scholar]
- 72.Krause R, Knoll K, Henle T. Studies on the formation of furosine and pyridosine during acid hydrolysis of different Amadori products of lysine. Eur Food Res Technol. 2003;216(4):277–283. [Google Scholar]
- 73.Troise AD, Fiore A, Wiltafsky M, Fogliano V. Quantification of N epsilon-(2-furoylmethyl)-L-lysine (furosine), N epsilon-(carboxymethyl)-L-lysine (CML), N epsilon-(carboxyethyl)-L-lysine (CEL) and total lysine through stable isotope dilution assay and tandem mass spectrometry. Food Chem. 2015;188:357–364. doi: 10.1016/j.foodchem.2015.04.137. [DOI] [PubMed] [Google Scholar]
- 74.Gupta N, Pevzner PA. False discovery rates of protein identifications: a strike against the two-peptide rule. J Proteome Res. 2009;8(9):4173–4181. doi: 10.1021/pr9004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ong S-E, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2007;1(6):2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 76.Noirel J, Evans C, Salim M, Mukherjee J, Ow SY, Pandhal J, Pham TK, Biggs CA, Wright PC. Methods in quantitative proteomics: setting iTRAQ on the right track. Curr Probl Dermatol. 2011;8(1):17–30. [Google Scholar]
- 77.Nahnsen S, Bielow C, Reinert K, Kohlbacher O. Tools for label-free peptide quantification. Mol Cell Proteomics. 2013;12(3):549–556. doi: 10.1074/mcp.R112.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kimzey MJ, Kinsky OR, Yassine HN, Tsaprailis G, Stump CS, Monks TJ, Lau SS. Site specific modification of the human plasma proteome by methylglyoxal. Toxicol Appl Pharmacol. 2015;289(2):155–162. doi: 10.1016/j.taap.2015.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brede C, Hop B, Jørgensen K, Skadberg Ø. Measurement of glycated albumin in serum and plasma by LC-MS/MS. Scand J Clin Lab Invest. 2016;76(3):195–201. doi: 10.3109/00365513.2015.1129671. [DOI] [PubMed] [Google Scholar]
- 80.Liebler DC, Zimmerman LJ. Targeted quantitation of proteins by mass spectrometry. Biochemistry. 2013;52(22):3797–3806. doi: 10.1021/bi400110b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nagaraj N, Kulak NA, Cox J, Neuhauser N, Mayr K, Hoerning O, Vorm O, Mann M. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol Cell Proteomics. 2012;11(3):11. doi: 10.1074/mcp.M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meyer JG. In silico proteome cleavage reveals iterative digestion strategy for high sequence coverage. ISRN Computational Biol. 2014;2014:7. doi: 10.1155/2014/960902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johansen MB, Kiemer L, Brunak S. Analysis and prediction of mammalian protein glycation. Glycobiology. 2006;16(9):844–853. doi: 10.1093/glycob/cwl009. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Q, Monroe ME, Schepmoes AA, Clauss TRW, Gritsenko MA, Meng D, Petyuk VA, Smith RD, Metz TO. Comprehensive identification of glycated peptides and their glycation motifs in plasma and erythrocytes of control and diabetic subjects. J Proteome Res. 2011;10(7):3076–3088. doi: 10.1021/pr200040j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anandakrishnan R, Aguilar B, Onufriev AV. H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012;40(W1):W537–W541. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harris R, Patel SU, Sadler PJ, Viles JH. Observation of albumin resonances in proton nuclear magnetic resonance spectra of human blood plasma: N-terminal assignments aided by use of modified recombinant albumin. Analyst. 1996;121(7):913–922. doi: 10.1039/an9962100913. [DOI] [PubMed] [Google Scholar]
- 87.Venkatraman J, Aggarwal K, Balaram P. Helical peptide models for protein glycation: proximity effects in catalysis of the Amadori rearrangement. Chem Biol. 2001;8(7):611–625. doi: 10.1016/S1074-5521(01)00036-9. [DOI] [PubMed] [Google Scholar]
- 88.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 89.Schleicher E, Wieland OH. Kinetic analysis of glycation as a tool for assesing the half-life of proteins. Biochim Biophys Acta. 1986;884(1):199–205. doi: 10.1016/0304-4165(86)90244-8. [DOI] [PubMed] [Google Scholar]
- 90.Wang Y, Yu H, Shi X, Luo Z, Lin D, Huang M. Structural mechanism of ring-opening reaction of glucose by human serum albumin. J Biol Chem. 2013;288(22):15980–15987. doi: 10.1074/jbc.M113.467027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delpierre G, Rider MH, Collard F, Stroobant V, Vanstapel F, Santos H, Van Schaftingen E. Identification, cloning, and heterologous expression of a mammalian fructosamine-3-kinase. Diabetes. 2000;49(10):1627–1634. doi: 10.2337/diabetes.49.10.1627. [DOI] [PubMed] [Google Scholar]
- 92.Delpierre G, Vertommen D, Communi D, Rider MH, Van Schaftingen E. Identification of fructosamine residues deglycated by fructosamine-3-kinase in human hemoglobin. J. Biol. Chem. 2004;279(26):27613–27620. doi: 10.1074/jbc.M402091200. [DOI] [PubMed] [Google Scholar]
- 93.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24(21):2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 95.Roberts NB, Amara AB, Morris M, Green BN. Long-term evaluation of electrospray ionization mass spectrometric analysis of glycated hemoglobin. Clin Chem. 2001;47(2):316–321. [PubMed] [Google Scholar]
- 96.Thornalley PJ, Argirova M, Ahmed N, Mann VM, Argirov OK, Dawnay A. Mass spectrometric monitoring of albumin in uraemia. Kidney Int. 2000;58(5):2228–2234. doi: 10.1111/j.1523-1755.2000.00398.x. [DOI] [PubMed] [Google Scholar]
- 97.Jerić I, Versluis C, Horvat Š, Heck AJR. Tracing glycoprotein structures: electrospray ionization tandem mass spectrometric analysis of sugar–peptide adducts. J Mass Spectrom. 2002;37(8):803–811. doi: 10.1002/jms.337. [DOI] [PubMed] [Google Scholar]
- 98.Horvat Š, Jakas A. Peptide and amino acid glycation: new insights into the Maillard reaction. J Pept Sci. 2004;10(3):119–137. doi: 10.1002/psc.519. [DOI] [PubMed] [Google Scholar]
- 99.Mennella C, Visciano M, Napolitano A, Del Castillo MD, Fogliano V. Glycation of lysine-containing dipeptides. J Pept Sci. 2006;12(4):291–296. doi: 10.1002/psc.722. [DOI] [PubMed] [Google Scholar]
- 100.Zhang QB, Frolov A, Tang N, Hoffmann R, van de Goor T, Metz TO, Smith RD. Application of electron transfer dissociation mass spectrometry in analyses of non-enzymatically glycated peptides. Rapid Commun Mass Spectrom. 2007;21(5):661–666. doi: 10.1002/rcm.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cai J, Hurst HE. Identification and quantitation of N-(carboxymethyl)valine adducts in hemoglobin by gas chromatography/mass spectrometry. J Mass Spectrom. 1999;34(5):537–543. doi: 10.1002/(SICI)1096-9888(199905)34:5<537::AID-JMS806>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 102.Brock JWC, Hinton DJS, Cotham WE, Metz TO, Thorpe SR, Baynes JW, Ames JM. Proteomic analysis of the site specificity of glycation and Carboxymethylation of ribonuclease. J Proteome Res. 2003;2(5):506–513. doi: 10.1021/pr0340173. [DOI] [PubMed] [Google Scholar]
- 103.Zhang Q, Tang N, Brock JWC, Mottaz HM, Ames JM, Baynes JW, Smith RD, Metz TO. Enrichment and analysis of Nonenzymatically glycated peptides: boronate affinity chromatography coupled with electron-transfer dissociation mass spectrometry. J Proteome Res. 2007;6(6):2323–2330. doi: 10.1021/pr070112q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goodall I.: HbA1c standardisation destination--global IFCC Standardisation. How, why, where and when--a tortuous pathway from kit manufacturers, via inter-laboratory lyophilized and whole blood comparisons to designated national comparison schemes. Clin Biochem Rev. 26(1), (2005) [PMC free article] [PubMed]
- 105.Myint T, Hoshi S, Ookawara T, Miyazawa N, Keiichiro M, Suzuki K, Taniguchi N. Immunological detection of glycated proteins in normal and streptozotocin-induced diabetic rats using anti hexitol-lysine IgG. Biochim Biophys Acta. 1995;1272:73–79. doi: 10.1016/0925-4439(95)00067-E. [DOI] [PubMed] [Google Scholar]
- 106.Thornalley PJ, Langborg A, Minhas HS. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem J. 1999;344(1):109–116. doi: 10.1042/bj3440109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smith PR, Thornalley PJ. Influence of pH and phosphate ions on the kinetics of enolisation and degradation of fructosamines. Studies with the model fructosamine, Nε-1-deoxy-D-fructos-1-yl hippuryllysine. Biochem Internat. 1992;28(3):429–439. [PubMed] [Google Scholar]
- 108.Hustoft HK, Reubsaet L, Greibrokk T, Lundanes E, Malerod H. Critical assessment of accelerating trypsination methods. J Pharm Biomed Anal. 2011;56(5):1069–1078. doi: 10.1016/j.jpba.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 109.Schmidt R, Böhme D, Singer D, Frolov A. Specific tandem mass spectrometric detection of AGE-modified arginine residues in peptides. J Mass Spectrom. 2015;50(3):613–624. doi: 10.1002/jms.3569. [DOI] [PubMed] [Google Scholar]
- 110.Rabbani N, Thornalley PJ. Measurement of methylglyoxal by stable isotopic dilution analysis LC-MS/MS with corroborative prediction in physiological samples. Nat Protoc. 2014;9(8):1969–1979. doi: 10.1038/nprot.2014.129. [DOI] [PubMed] [Google Scholar]
- 111.Lo TWC, Westwood ME, McLellan AC, Selwood T, Thornalley PJ. Binding and modification of proteins by methylglyoxal under physiological conditions. A kinetic and mechanistic study with Nα-acetylarginine, Nα-acetylcysteine, Nα-acetyl-lysine, and bovine serum albumin. J Biol Chem. 1994;269(51):32299–32305. [PubMed] [Google Scholar]
- 112.Leitner A, Lindner W. Functional probing of arginine residues in proteins using mass spectrometry and an arginine-specific covalent tagging concept. Anal Chem. 2005;77(14):4481–4488. doi: 10.1021/ac050217h. [DOI] [PubMed] [Google Scholar]
- 113.Leitner A, Amon S, Rizzi A, Lindner W. Use of the arginine-specific butanedione/phenylboronic acid tag for analysis of peptides and protein digests using matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2007;21(7):1321–1330. doi: 10.1002/rcm.2967. [DOI] [PubMed] [Google Scholar]
- 114.Kilhovd BK, Giardino I, Torjesen PA, Birkeland KI, Berg TJ, Thornalley PJ, Brownlee M, Hanssen KF. Increased serum levels of the specific AGE-compound methylglyoxal-derived hydroimidazolone in patients with type 2 diabetes. Metabolism. 2003;52(2):163–167. doi: 10.1053/meta.2003.50035. [DOI] [PubMed] [Google Scholar]
- 115.Biemel KM, Friedl DA, Lederer MO. Identification and quantification of major Maillard cross-links in human serum albumin and lens protein - evidence for glucosepane as the dominant compound. J Biol Chem. 2002;277(28):24907–24915. doi: 10.1074/jbc.M202681200. [DOI] [PubMed] [Google Scholar]
- 116.Dai Z, Wang B, Sun G, Fan X, Anderson VE, Monnier VM. Identification of glucose-derived cross-linking sites in ribonuclease a. J Proteome Res. 2008;7(7):2756–2768. doi: 10.1021/pr700874a. [DOI] [PMC free article] [PubMed] [Google Scholar]