Abstract
T follicular helper (Tfh) cell is a unique T cell subset specialized in promoting humoral immunity. Bcl6 has been identified as an obligatory transcription factor in Tfh cells; however, the molecular mechanism underlying Bcl6 function remains largely unknown. Here, we defined Bcl6 target genes in Tfh cells by analyzing genome-wide Bcl6 occupancy together with transcriptome profiling. With consensus sequences different from those in Th9, B cells and macrophages, Bcl6 binding in Tfh cell was closely associated with decrease in 5-hydroxymethylcytosine (5hmC). Importantly, Bcl6 promoted Tfh cell differentiation through antagonizing IL-7R (CD127)/signal transducer and activator of transcription (STAT) 5 axis; deletion of the Bcl6 gene in T cells results in enhanced IL-7R-STAT5 signaling and substantial expansion of CD127hi non-Tfh cells. Our study thus systemically examines Bcl6-controlled regulatory networks and provides important insights into its biological functions in Tfh cells.
eTOC Blurb
Liu et al. examine the roles of Bcl6 during Tfh cell programming: Bcl6 binding to chromatin is associated with decreased 5hmC. Bcl6 directs Tfh development, at least in part, through antagonizing the IL-7R/STAT5 axis.
Introduction
The hallmark of T-dependent humoral immunity is the formation of germinal centers (GCs) in secondary lymphoid tissues (MacLennan, 1994; Nieuwenhuis and Opstelten, 1984; Victora and Nussenzweig, 2012). GCs provide a milieu for B cell proliferation, for their antibody affinity maturation and class switching, and for plasma and memory B cell generation (Allen et al., 2007; MacLennan, 1994; Victora and Nussenzweig, 2012). GC generation requires help from Tfh cells (Breitfeld et al., 2000; Crotty, 2011; Dong et al., 2001; Kim et al., 2001; Vinuesa and Cyster, 2011). In the past several years, tremendous efforts have been put on in defining the genetic features of Tfh cells and the molecular mechanisms underlying Tfh differentiation (Crotty, 2011; Liu et al., 2013; Yu and Vinuesa, 2010). Transcriptional factor (TF) Bcl6 expression was first found to be selectively upregulated in Tfh cells; Bcl6-deficient T cells do not give rise to Tfh cells, and constitutive expression of Bcl6 enhances Tfh cell differentiation (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). These findings have recognized Bcl6 as an obligatory transcriptional factor for Tfh development and germinal center reactions (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009), in line with T-bet for Th1, Gata3 for Th2, and RORγt for Th17 (Zhu et al., 2010).
In addition to lineage-specific master transcription factors, T cell differentiation is achieved by a complex network involving multiple transcription factors (Ciofani et al., 2012; Zhu et al., 2010). Recently, Tfh signature gene Cxcr5, initially proposed as a target of Bcl6, is found to be directly regulated by Achaete-Scute homologue 2 (Ascl2) (Liu et al., 2014). In addition, transcription factors STAT3, STAT1, BATF, IRF4, C-Maf, and Ascl2 all have been shown to positively regulate Tfh cell generation (Crotty, 2011; Liu et al., 2014; Liu et al., 2013). In contrast, IL-2-induced STAT5 signaling plays a negative role in Tfh cell development (Ballesteros-Tato et al., 2012; Johnston et al., 2012; Nurieva et al., 2012).
Bcl6 is a member of BTB-Zinc finger family protein, and plays critical roles in various cell types of the innate and adaptive immune system (Chang et al., 1996; Dent et al., 1997; Shaffer et al., 2000; Ye et al., 1997; Ye et al., 1993). Bcl6 was originally identified in diffuse large B cell lymphomas (Ye et al., 1993). The proposed function of Bcl6 in B cells is to regulate cell cycle, DNA damage response and to suppress a host of signal pathways via cooperating with SMRT, NCOR and BCOR (Basso et al., 2010; Crotty et al., 2010; Hatzi et al., 2013; Huang et al., 2013; Klein and Dalla-Favera, 2008). Bcl6 also has important functions in macrophage, in which it restricts Toll like receptors (TLR)-induced inflammatory reactions to re-establish quiescence by cistrome-based antagonism of NF-κB (Barish et al., 2010; Barish et al., 2012). Chromatin immunoprecipitation coupled with deep sequencing (ChIP-Seq) analysis of Bcl6 and NF-κB bindings in macrophage shows extensive colocalization between Bcl6 and NF-κB, suggesting a direct intersection between Bcl6 and NF-κB in a dynamic balance from activation to quiescence (Barish et al., 2010).
Considering the molecular mechanisms underlying Bcl6 function in Tfh cells have been elusive, we assessed genome-wide analysis of Bcl6 binding sites in primary Tfh cells, and integrated this data set with the occupancy of multiple TFs, global map of epigenetic modifications, and transcriptome analysis of Bcl6 responsive genes. We have identified Bcl6 as a repressor against IL-7R-STAT5 signaling cascade during Tfh development.
Results
Bcl6 exhibits unique binding motifs in Tfh cells
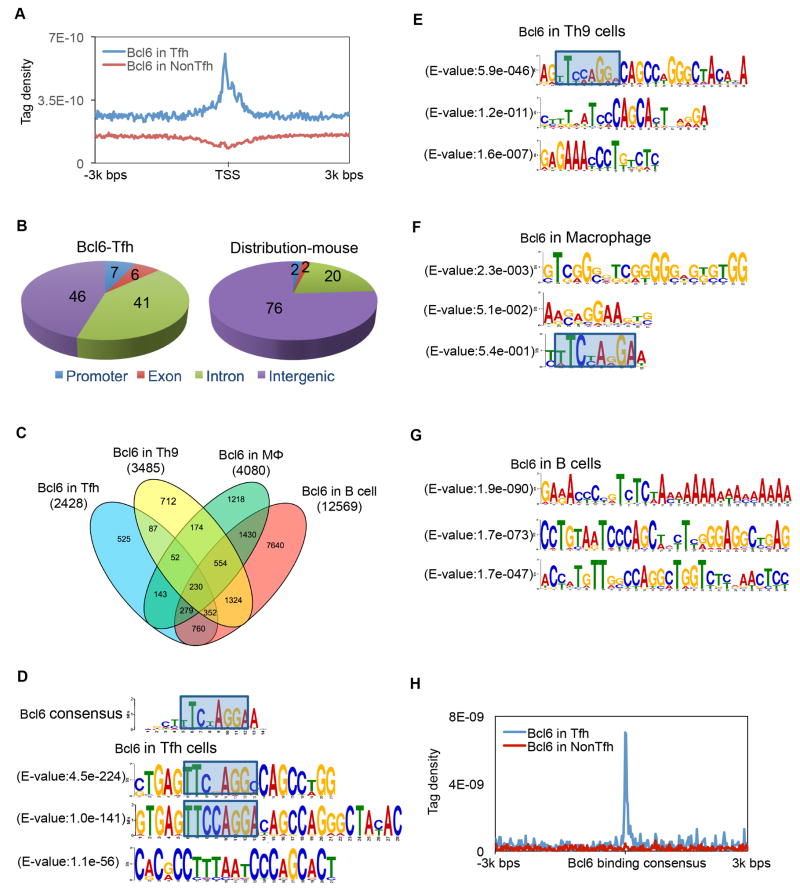
To understand how Bcl6 mediates Tfh cell differentiation, we performed ChIP-seq analysis for Bcl6 using in vivo-generated Tfh cells from Bcl6-RFP reporter mice as previously described (Liu et al., 2012), with non-Tfh cells as a control. A total of 5191 Bcl6 binding peaks were identified in Tfh cells (Table S1). Most Bcl6 binding sites were localized to intron (41%) and intergenic regions (46%), while ~7% of them were located to promoter regions (between 3kb upstream and downstream of the transcription start site) (Figure 1A–B). Previous studies have demonstrated that Bcl6 exerts different roles in Th9, macrophage and B cells (Barish et al., 2010; Barish et al., 2012; Hatzi et al., 2013; Huang et al., 2013; Liao et al., 2014). When we compared Bcl6 binding sites among T, B cells and macrophages, we found that Bcl6 preferentially bound to promoter regions in Th9 (promoter regions 18%, exons 6%, introns 38% and intergenic 38%) and B cells (promoter regions 16%, exons 9%, introns 40% and intergenic 35%), strikingly different from macrophage (promoter 3%, exons 3%, introns 42% and intergenic 50%) and Tfh cells (promoter regions 7%, exons 6%, introns 41% and intergenic 46%) (Figure 1B and Figure S1A). Analysis of Bcl6-bound genes revealed that only 230 genes were shared in all four types of cells (Figure 1C and Table S2), suggesting that Bcl6 regulates gene transcription in a cell-type-specific manner. Not surprisingly, when examining Bcl6 binding motifs, after exclusion of random repeat motifs, we found that top Bcl6 binding motifs in both Tfh and Th9 cells contains classical Bcl6 consensus element 5′-TTCNAGG(A/C)-3′, which is different from those in macrophages and B cells (Figure 1D–G). Further alignment of Bcl6 binding consensus with Bcl6-occupied sequences confirmed Bcl6 binding specificity in Tfh cells (Figure 1H). Notably, none of the top three binding motifs in B cells contains the Bcl6 core element. These data suggest divergent functions of Bcl6 among T cells, macrophages and B cells.
Figure 1. Preferential Bcl6 binding motifs in Tfh, Th9, Macrophage and B cells.
A. Bcl6 binding in Tfh vs. Non-Tfh cells. TSS, transcription start site. The tag density of Bcl6 binding around TSS of all RefSeq (mm9) genes, uniquely mapped tags were summarized in 20 bp windows. All window tag counts were normalized by the total number of bases in the windows and the total read number of the given sample.
B. Distribution of Bcl6 binding peaks in Tfh cells, and distribution of the genetic features across the whole mouse genome (mm9).
C. Venn diagram of genes bound by Bcl6 in Tfh, Th9, B cells, and macrophages. ChIP-seq data for Bcl6 binding in Th9 were derived from GenBank under accession GSE41317-GSM1234367 (Liao et al., 2014), macrophage were derived from GenBank under accession GSE16723 (Barish et al., 2010), and Bcl6 binding in B cells was from GSE43350 (Huang et al., 2013).
D. Classical Bcl6 binding consensus, and Bcl6 binding motifs in Tfh cell.
E. Bcl6 binding motifs in Th9 cell (original sequence reads were reanalyzed and mapped to mouse genome mm9).
F. Bcl6 binding motifs in macrophages.
G. Bcl6 binding motifs in B cells.
H. Alignment of Bcl6 binding consensus on overall ChIP-seq signal in Tfh and non-Tfh cells.
Bcl6 binding correlates with decreased 5hmC
T cell fate specification is regulated by epigenetic modifications of both histones and DNA (Lu et al., 2011; Tsagaratou et al., 2014; Wei et al., 2009). To test whether the recruitment of Bcl6 correlates with histone modification changes, we used our previous ChIP-seq data sets for trimethylated histone H3 lysine 4 (H3K4me3) and trimethylated histone H3 lysine 27 (H3K27me3) and compared them with Bcl6-bound areas (Liu et al., 2014). As shown in Figure S1B and C, in both Tfh and naïve CD4+ T cells, the intensity of H3K4me3 and H3K27me3 in Bcl6-occupied regions remained very low and unchanged, suggesting Bcl6 binding is not associated with altered modifications of H3K4me3 or H3K27me3.
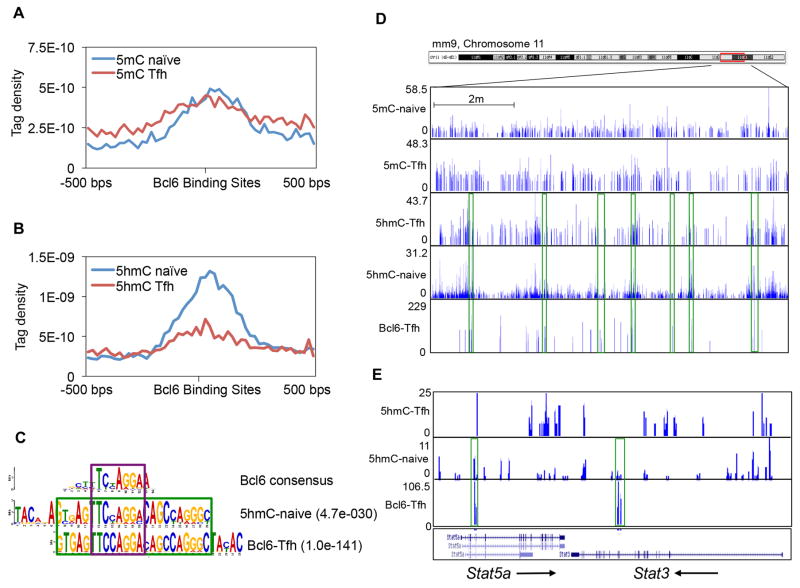
To further examine the association between Bcl6 and DNA methylation, we performed methylated and hydroxymethylated DNA immunoprecipitation (MeDIP/hMeDIP)-seq assays to map genome-wide modification profiles of both 5mC and 5hmC in naïve CD4+ T and Tfh cells (Table S1). The resulting 5mC profile defined 48133 peaks in naïve T cells and 33063 peaks in Tfh cells (Table S1). Importantly, no difference was found in 5mC enrichment between Tfh and naïve T cells in the 5191 Bcl6-bound areas (Figure 2A). In contrast, a great reduction of 5hmC was found in Tfh cells in Bcl6 binding sites as compared to that in naïve T cells (Figure 2B), as well as Th1, Th2, Th17 and inducible regulatory T (iTreg) cells (Ichiyama et al., 2015) (Figure S1D). Analysis of genome-wide 5hmC reduction at Bcl6 binding sites in Tfh cells defined 696 regions (distributed among 645 gene loci) (Table S3). Among these, 98 genes were transcriptionally controlled by Bcl6, and 15 were Bcl6-targeted Tfh-specific genes (data not shown). Random examination of sequencing tracks along mouse chromosome 11 illustrated obvious co-localization of Bcl6 with 5hmC peaks at the genome-wide level (Figure 2C and D). For example, at the Stat5a and Stat3 gene loci, Bcl6 peaks only co-localized with 5hmC peaks in naïve T cells due to the absence of 5hmC enrichment in Tfh cells (Figure 2E). Furthermore, Comparison of 5hmC enrichment at Stat5a, Runx3, and Mgll gene loci among T cell subsets confirmed the above statement (Figure S2A).
Figure 2. Bcl6 recruitment to target genes in Tfh cells reduced the level of 5-hydroxymethylcytosine (5hmC).
A. At Bcl6-bound regions, 5-methylcytosine (5mC) enrichments were compared between Naïve T and Tfh cells.
B. At Bcl6-bound regions, 5-hydroxymethylcytosine (5hmC) enrichments were compared between Naïve T and Tfh cells.
C. Classical Bcl6 binding consensus, 5hmC-enrichment motif in Naïve T cells, and Bcl6 binding motif in Tfh cells.
D. ChIP-Seq binding tracks for 5mC, 5hmC, and Bcl6 at selected chromosome 11 regions in Naïve and Tfh cells. Green frames represent the co-localization of peaks.
F. ChIP-Seq binding peaks for 5hmC and Bcl6 at selected Stat5a and Stat3 gene loci in Naïve and Tfh cells. Green frames represent the co-localization of peaks.
Our aforementioned analysis suggested that the reduction of 5hmC in Tfh cell is dependent on Bcl6. To test this hypothesis, we isolated CD4+ CD44hi T cells from Bcl6fl/fl/CD4-Cre and control mice immunized with KLH/CFA, and performed hMeDIP-PCR assays. As exemplified at gene loci including Stat5a, Runx3, and Mgll, the levels of 5hmC in both WT and Bcl6-deficient non-Tfh cells were enhanced (Figure S2B). To further elucidate the mechanism underlying Bcl6-dependent 5hmC reduction, we first measured expression levels of methylcytosine dioxygenase Tet genes (Ten-eleven-translocation proteins) including Tet1, Tet2 and Tet3 in naïve, Tfh and non-Tfh cells, and observed substantial reduction of Tet1 expression in Tfh cells in comparison with Tet2 and Tet3 (Figure S2C). We then focused on functional analysis of Tet1 by conducting ChIP assay in Bcl6-sufficient Tfh and non-Tfh cells as well as Bcl6-deficient non-Tfh cells. As shown in Figure S2D, Tet1 recruitment was increased at all three Stat5a, Runx3, and Mgll loci in WT non-Tfh cells, as compared with Tfh cells, and was even more significant in KO non-Tfh cells. Collectively, these data suggested a critical role of Bcl6 in inhibiting Tet1 recruitment and 5hmC modification.
STAT5 and Bcl6 share binding sequences
Transcription factors including BATF, IRF4, c-Maf, and STAT3 have been shown important in Tfh cells (Liu et al., 2013). We thus compared the published data on the binding of BATF, IRF4, c-Maf, and STAT3 (Ciofani et al., 2012) to Bcl6 data. Motif analysis of Bcl6 binding revealed that Bcl6 did not share consensus binding motifs with BATF, IRF4, c-Maf, or STAT3 (Figure 1D and data not shown), although a small number of Bcl6 binding sites in Tfh cells could be the direct binding sites of BATF, IRF4, C-Maf or STAT3 in Th17 cells (Table S4). These observations suggest Bcl6 may function together with BATF, IRF4, C-Maf, or STAT3 in shaping Tfh cell polarization.
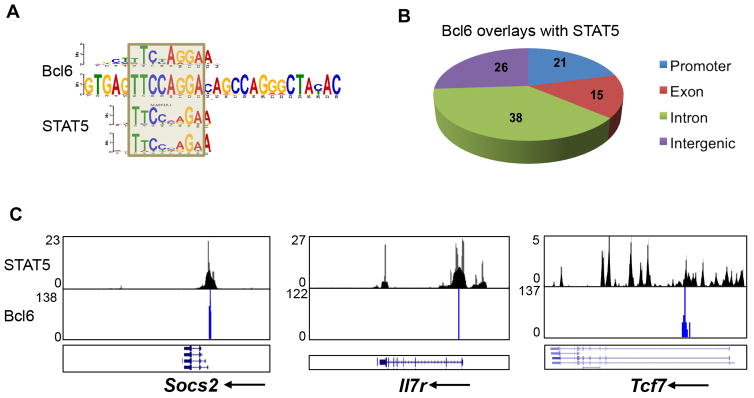
Further motif analysis showed that the core element in Bcl6 binding consensus in Tfh cells was almost the same as STAT5 binding motif, except for one nucleotide difference (Figure 3A). Because STAT5 inhibits Tfh cell generation (Johnston et al., 2012; Nurieva et al., 2012), and Tfh cell was shown to lack STAT5 signaling (Johnston et al., 2012), our observation implicated potential competition between Bcl6 and STAT5 in regulating Tfh cell differentiation. We then examined and compared Bcl6 binding sites in Tfh cells and STAT5 binding peaks in Th17 cells at the genome-wide level (Yang et al., 2011), and found that they potentially share a great amount of binding sites, among which the majority (Proximal Promoter 21% + Exon 15% + Intron 38%) was located in gene bodies (Figure 3B) and a total of 424 genes could be co-regulated by Bcl6 and STAT5 (Table S5). As exemplified in Figure S3A–D and Figure 3C, although co-occupancy of STAT5 and Bcl6 was barely found in Th-related signature gene loci, strong co-localization was seen in Socs2, Il7r and Tcf7. To confirm our ChIP-seq data, we conducted ChIP-PCR with anti-Bcl6, anti-p-STAT5α, and IgG control antibody. In comparison with control-IgG enrichment, p-STAT5 was observed bound to Il7r, Socs2 and Tcf7 regions in IL-2- or IL-7-treated non-Tfh cells, while Bcl6 bound to the same location in Tfh cells (Figure S4). Thus, these findings implicate reciprocal roles of Bcl6 and STAT5 in Tfh versus non-Tfh cell polarization, respectively.
Figure 3. Preferential Bcl6 binding to STAT5 motifs in T cells.
MEME-ChIP (http://meme.nbcr.net/meme/tools/meme-chip) (default parameters) were applied to analyze the consensus sequences within Bcl6 binding sites in Tfh cells and STAT5 binding sites in Th17 cells (Yang et al., 2011). The binding sites of both Bcl6 and Stat5 were first defined using the SICER V1.1 program. A binding site is defined as one or several consecutive windows of 200 bp of the genome where a significant enrichment of sequencing reads against the background appears. If any part of one TF binding site overlaps with the other one, it is defined as the overlay of the two binding sites.
A. Comparison of Bcl6 binding motifs in Tfh cells with STAT5 binding motifs in T cells. ChIP-Seq data for STAT5 binding in T cells was derived from GSE26553-GSM652878 (Yang et al., 2011).
B. Distribution of Bcl6/STAT5 co-occupancies in proximal promoter, exon, intron, and intergenic regions
C. Bcl6- and STAT5-colocalized peaks located at gene loci including Socs2, Il7r, and Tcf7
Transcriptional changes mediated by Bcl6 in Tfh cells
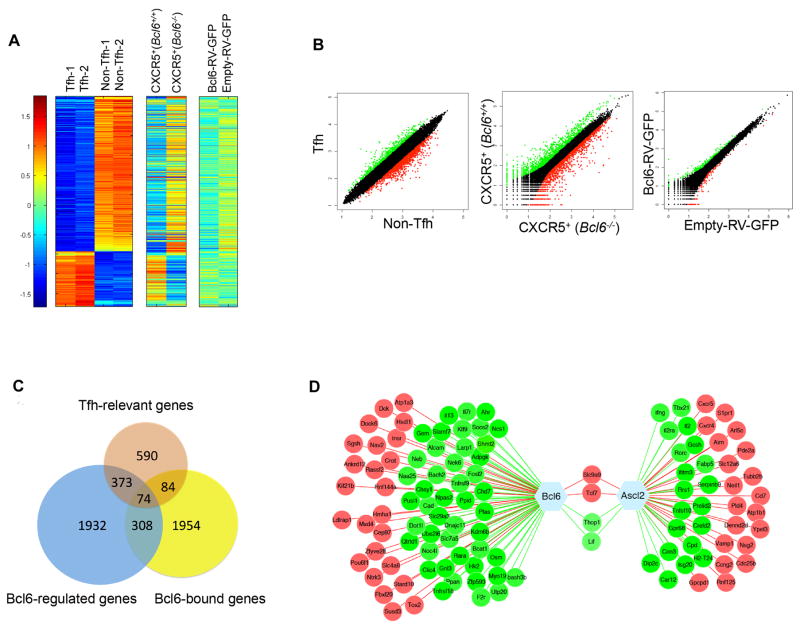
From our published microarray data obtained using Bcl6-RFP reporter mice (Liu et al., 2012), we have previously identified ~1100 Tfh-relevant genes, among which there were more downregulated genes than upregulated ones (left panel, Figure 4A and B). To further define genes that are transcriptionally regulated by Bcl6, we transferred WT or Bcl6-deficient OT-II cells into congenic recipient mice. Four days after ovalbumin (OVA)/CFA immunization, we harvested CXCR5+ donor cells-as we had reported, Bcl6 deficiency did not ameliorate CXCR5 expression at this early stage- and then performed RNA-Seq assays (data not shown). RNA-Seq analysis revealed that Bcl6 played a pivotal role in regulating Tfh-related transcriptome and the absence of Bcl6 altered Tfh-gene profiling dramatically (middle panel, Figure 4A and B). Additionally, we also conducted RNA-seq analysis on T cells infected in vitro with Bcl6- or vector-overexpressing retrovirus. Bcl6 was sufficient globally in regulating gene expression landscape, although the fold changes for each gene were relatively modest (right panel, Figure 4A and B). These data clearly indicate a critical transcriptional regulation by Bcl6.
Figure 4. Bcl6-dependent transcriptional regulation of Tfh-relevant genes.
A. Heat map of ~1100 Tfh-relevant genes regulated in CXCR5hi Bcl6-RFPhi Tfh and CXCR5lo Bcl6-RFPlo Non-Tfh cells (student T test on Tfh vs non-Tfh microarray data analysis and identified ~1100 genes with at least 2 fold difference (Heat map of ~1100 TFH-relevant genes (p<0.05, student T test; fold change >2), Microarray accession GSE40068 (Liu et al., 2012), left column), donor-derived CXCR5+ Bcl6+/+ OT-II and CXCR5+ Bcl6−/− OT-II cells (RNA-Seq, middle column), and Bcl6-RV-GFP- or control vector-transduced T cells cultured under Th0 condition for 4 days (RNA-Seq, right column). The color-coding applies to gene expression level (log2) with 0 as median.
B. Scatterplot of the average signal of Tfh versus Non-Tfh cells gene expression microarray data, Day-3-donor-derived CXCR5+ Bcl6+/+ versus CXCR5+ Bcl6−/− OT-II cells gene expression RNA-Seq data, and Bcl6-RV-GFP+ versus Empty-RV-GFP+ T cell gene expression RNA-Seq data.
C. Venn diagram of Tfh-relevant genes, Bcl6-regulated genes and Bcl6-bound genes.
D. Bcl6- and Ascl2 (accession GSE52840 (Liu et al., 2014))-targeted Tfh-specific genes. Green represents downregulation of gene expression; red represents upregulation of gene expression.
After combining three datasets including ChIP-seq, RNA-sequencing of WT and Bcl6 mutant OT-II cells in vivo, and Tfh-relevant genes from microarray, we identified a total number of 74 genes that were directly regulated by Bcl6 in Tfh cells (Figure 4C and Table S6). Among these Bcl6 target Tfh -specific genes, 26 were upregulated, and 48 were downregulated by Bcl6 (Figure 4D and Table S5). Functional analysis and comparison of these genes with Ascl2-targets (44 genes) further confirmed our previous conclusion that Bcl6 and Ascl2 play distinct roles during Tfh differentiation (Liu et al., 2014). Bcl6 and Ascl2 co-regulate only 4 genes including Slc9a9, Tcf7, Thop1, and Lif, among which transcriptional factor TCF-1 (Tcf7) was demonstrated as a critical regulator for Tfh cell generation in two recent studies (Figure 4D)(Choi et al., 2015; Kubo, 2015; Xu et al., 2015). Bcl6 target genes in Tfh cells were more likely involved in cell death and survival, cell cycle and growth, etc., while Ascl2 target genes were associated with cell differentiation, migration and effector function (Figure S5A–D). Out of the 74 Tfh-specific Bcl6 target genes, the top 15 most upregulated and downregulated ones are listed in Figure S6A. Interestingly, Il13 gene was most repressed by Bcl6, consistent with the excessive Th2-associated inflammation in Bcl6-mutant mice (Ye et al., 1997). Notably, Th17-related Ahr and Fosl2 were among the top suppressed genes (Ciofani et al., 2012; Veldhoen et al., 2008), indicating an indirect role of Bcl6 in shaping the Th17 program. Interestingly, a reduction of 5hmC level is also found at the center of Bcl6 binding sites located to Tfh-specific Bcl6 target genes in Tfh cells as compared to that in naïve T cells (Figure S6B). In the list of Bcl6-upregulated genes, Mxd4, encoded by a basic helix-loop-helix-leucine zipper family member was able to inhibit IL-2 expression in activated T cells (Figure S6C).
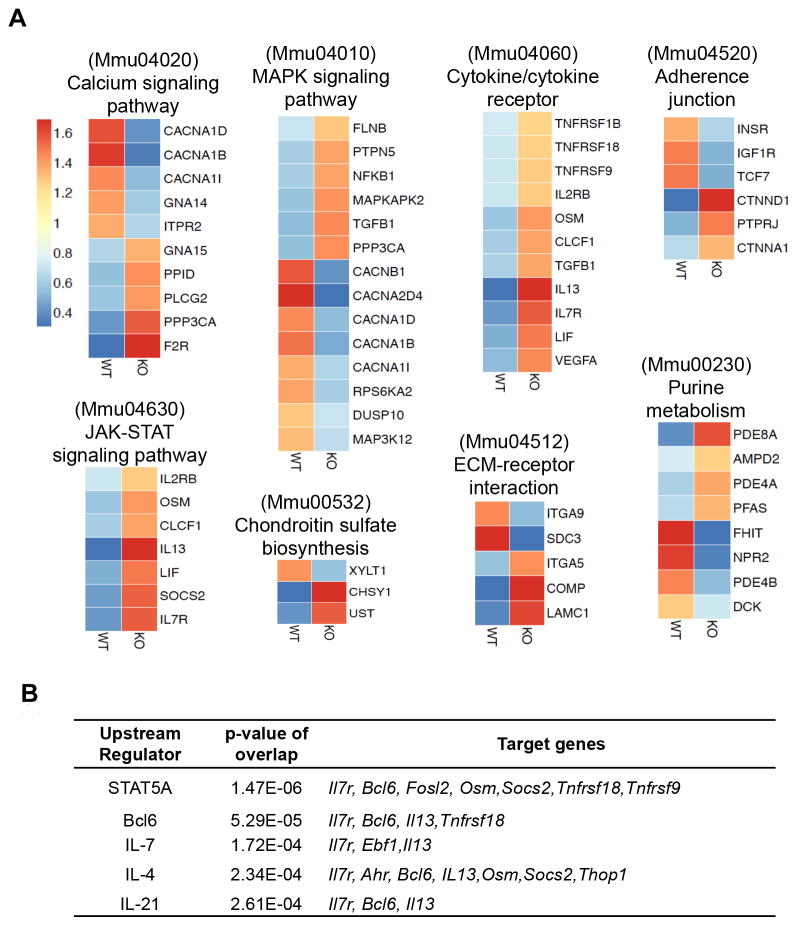
Bcl6-mediated regulatory modules in Tfh cells
If Tfh-relevant genes were not taken into consideration, combination of Bcl6-regulated genes and Bcl6-bound genes generated a list of 382 putative Bcl6 target genes (Figure 4C and Table S7), indicating that beyond regulating 74 Tfh-specific genes, Bcl6 exerts a broader biological function in Tfh cell physiology. Pathway analysis of 382 genes using DAVID (The Database for Annotation, Visualization and Integrated Discovery v6.7) (Huang da et al., 2009) revealed that Bcl6 target genes were strongly associated with cellular activation and maintenance, signal transduction, cell adherence and metabolism, and were specifically enriched for genes involved in calcium and MAPK signaling pathway, JAK-STAT signal transduction, cytokine/cytokine receptors, and purine metabolism (Figure 5A). Importantly the top two upstream regulators of Bcl6-targeted genes, predicted using Ingenuity Systems, were STAT5A and Bcl6, followed by other regulators including IL-7, IL-4, and IL-21 (Figure 5B), suggesting again Bcl6 and STAT5 may cooperate the regulation of Tfh cell polarization and function. Taken together, validations and classification of Bcl6 target genes have established a unique Bcl6-centered network that orchestrated Tfh cell differentiation.
Figure 5. Bcl6-mediated regulatory modules for Tfh cell development.
A. Kyoto Encyclopedia Gene and Genome (KEGG) pathway analysis of Bcl6 directly regulated genes using DAVID. A heatmap of normalized gene expression levels was shown. Blue represents downregulation of gene expression and red represents upregulation of gene expression, with 1 as the median.
B. Upstream regulators prediction performed using the ingenuity analysis tool (IPA) on Bcl6-target genes with the value p<0.05.
Bcl6 suppresses IL-7R expression during Tfh cell generation
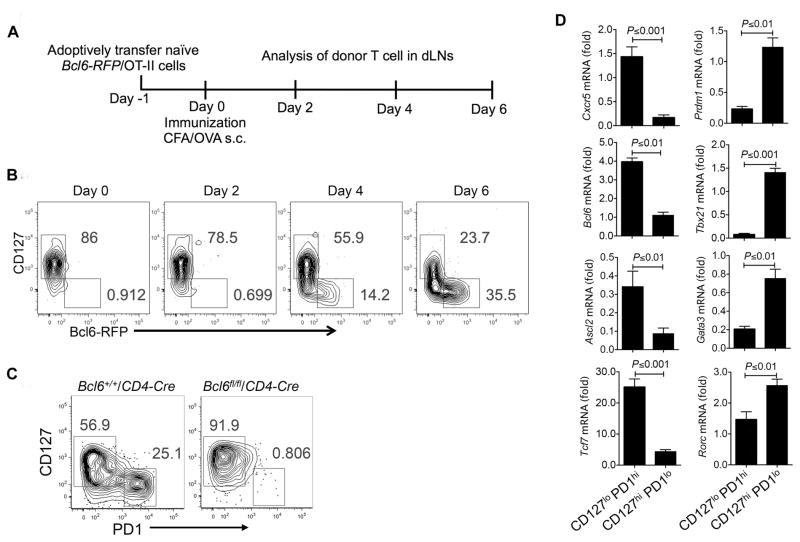
In the above analysis, IL-7R was predicted as direct downstream targets of five signaling pathways during Tfh cell development, including Bcl6- and STAT5-related pathways (Figure 5B). RNA-seq of Bcl6-mutant OT-II cells together with Bcl6 ChIP-seq data defined IL-7R as one of the most repressed Tfh-relevant genes by Bcl6 (Figure S6A, D and E). Moreover, ectopic expression of Bcl6 in activated T cells in vitro strongly inhibited IL-7R expression (Figure S6D). We thus reasoned that Bcl6 suppression of IL-7R might be important in Tfh cell differentiation. We then examined IL-7R protein and Bcl6 expression during Tfh ontogeny in vivo by transferring naïve Bcl6-RFP/OT-II cell into congenic recipient mice, followed with OVA/CFA immunization. As shown in Figure 6A and B, donor-derived OT-II cells gradually gained expression of Bcl6-RFP, while CD127 (IL-7R) expression was reduced. Of the note, the expression levels of CD127 in Bcl6-RFP+ Tfh cells were extremely low. To confirm the above observation, we measured expression of CD127 in donor-derived OT-II cells from congenic mice immunized with OVA/CFA s.c., and found that CD127 (IL-7R) expression was inversely correlated with all three classical Tfh cell markers PD-1, CXCR5 and Bcl6 (Figure S6F and G). To understand whether Bcl6 is necessary for repressing IL-7R gene expression, we generated a new strain of Bcl6 conditional knockout mice and bred them with CD4-Cre mice (Figure S7A). These mice were immunized with keyhole limpet hemocyanin (KLH)/CFA for seven days, with Bcl6+/+/CD4-Cre littermate as controls. Strikingly, the disruption of the Bcl6 gene in T cells resulted in all of CD4+CD44+ activated T cells becoming CD127hi cells (Figure 6C). Therefore, these results suggested that IL-7R could be a novel marker for non-Tfh cells and that Bcl6-dependent IL-7R repression might be critical for Tfh cell differentiation. To further test this idea, we sorted two populations of CD4+CD44hi CD127hiPD1lo and CD4+CD44hi CD127loPD1hi cells from KLH/CFA-immunized WT mice and quantified mRNA expression of Tfh-related genes (Figure 6C and D). Cxcr5, Bcl6, Ascl2, and Tcf7 were all highly expressed in CD127loPD1hi Tfh cells, while Prdm1, Tbx21, Gata3 and Rorc were preferentially expressed in CD127hiPD1lo non-Tfh cells (Figure 6D).
Figure 6. Reciprocal expression of IL-7R and Bcl6 during differentiation of activated CD4+ T cells into distinct T cell subsets.
A–B Naïve Bcl6-RFP/OT-II CD4+ T cells were transferred into naïve congenic CD45.1 mice, which were subsequently immunized with OVA/CFA subcutaneously. Data are a representative of two independent experiments.
A. Experimental design for measuring Bcl6-RFP and IL-7R expression in donor-derived Bcl6-RFP/OT-II cells in vivo.
B. Flow cytometry analysis of CD127 and Bcl6-RFP expression in donor-derived Bcl6-RFP/OT-II cell in a time-dependent manner.
C. Flow cytometry measurement of CD127 and PD1 expression on activated CD44+ CD4+ T cell in draining LNs from Bcl6+/+/CD4-Cre and Bcl6fl/fl/CD4-Cre mice immunized with KLH/CFA subcutaneously for eight days. Data are a representative of two independent experiments.
D. Two populations of CD127hi PD1lo and CD127lo PD1hi were sorted from CD44+ CD4+ T cell pools in dLNs of Bcl6+/+/CD4-Cre mice immunized with KLH emulsified in CFA subcutaneously. Quantitative RT-PCR measurement of Cxcr5, Bcl6, Ascl2, Tcf7, Prdm1, Tbx21, Gata3, and Rorc mRNA expression in sorted two populations of cells.
Data are a representative of two independent experiments. Bar graph displayed as mean ± SD. n = 3 per group.
IL-7R/STAT5 axis negatively regulates Tfh cells
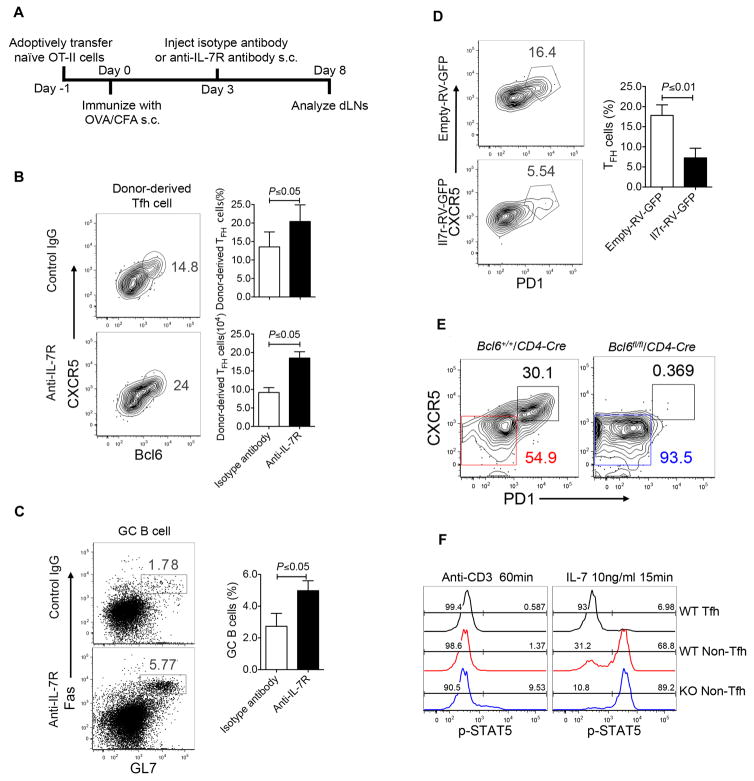
Our aforementioned results showed that IL-7R expression is inversely correlated with Tfh commitment, we hypothesized that IL-7 signaling might inhibit Tfh cell generation. To directly test that, we performed gain- and loss-of-function experiments. As shown in Figure 7A, when naïve OT-II cell were transferred into congenic mice that were subsequently immunized with OVA/CFA together with anti-IL-7R or an isotype control antibody, IL-7R blockade did not impair OT-II cell expansion (Figure S7B), but did enhance the development of Tfh cell (Figure 7B). Accordingly, germinal center (GC) reaction was also elevated in anti-IL-7R-treated mice (Figure 7C).
Figure 7. Bcl6 protected Tfh cells from negative regulation of IL-7R/STAT5 axis.
A. Experimental design for testing IL-7R function in vivo.
B–C. Naïve OT-II cells were adoptively transferred into naïve congenic mice, followed with subcutaneous OVA/CFA immunization. At day 3 after immunization, anti-IL-7R or isotype control antibodies (200μg/mouse) were injected subcutaneously into mice, respectively. Data are a representative of two independent experiments. n = 3 per group.
B. At day8 post immunization, flow cytometry analysis of donor-derived cells in dLNs with CXCR5 and Bcl6 staining. Bar graph displayed as mean ± SD. n = 4 per group.
C. GC B cells (GL-7hi Fashi) in recipient mice. Bar graph displayed as mean ± SD. n = 4 per group.
D. Il7r-RV-GFP- or control vector- transduced OT-II cells were adoptively transferred into naïve congenic mice, respectively, followed with subcutaneous OVA/CFA immunization for seven days. Flow cytometry analysis of donor-derived cells from dLNs with CXCR5 and PD1 staining. Data are a representative of two independent experiments. Bar graph displayed as mean ± SD. n = 4 per group.
E–F. Bcl6+/+/CD4-Cre and Bcl6fl/fl/CD4-Cre mice were subcutaneously immunized with KLH/CFA. Data are a representative of two independent experiments. n = 4 per group.
E. At day 8, flow cytometry analysis of Tfh and Non-Tfh cells from dLNs with staining CXCR5 and PD1.
F. Three populations of cells including CD4+ CD44hi CXCR5hi PD1hi Tfh cells from Bcl6+/+/CD4-Cre mice, CD4+ CD44hi CXCR5lo PD1lo Non-Tfh cells from both Bcl6+/+/CD4-Cre and Bcl6fl/fl/CD4-Cre mice were sorted, re-stimulated with plate-coated anti-CD3 or IL-7, and subject to intracellular measurement of STAT5 phosphorylation by flow cytometry. Data are a representative of two independent experiments.
Next, we transferred Il7r- or vector-transduced OT-II cells into congenic mice followed with subcutaneous OVA/CFA immunization for eight days. Although the expansion of IL-7R-overexpressing OT-II was slightly increased in numbers (Figure S7C), ectopic expression of IL-7R strongly inhibited Tfh cell differentiation (Figure 7D). Together, these data demonstrated that IL-7R plays a negative role in Tfh cell differentiation.
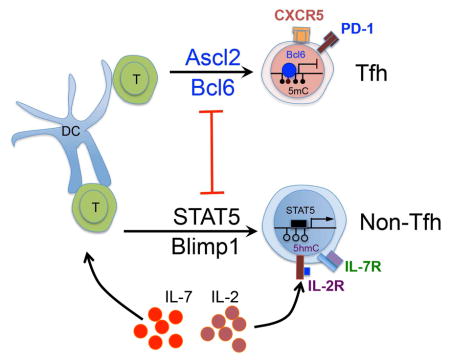
As documented above, IL-7R negatively regulated Tfh differentiation, and Bcl6 functions to suppress IL-7R expression during Tfh cell differentiation, we asked whether Bcl6 also antagonizes IL-7R-downstream-STAT5 activation in the process of Tfh differentiation. Bcl6+/+/CD4-Cre and Bcl6fl/fl/CD4-Cre mice were immunized with KLH protein emulsified with complete Freund’s adjuvant. Seven days after immunization, Tfh and non-Tfh cells were examined. As expected, CXCR5hiPD1hi Tfh cells were completely abolished in Bcl6fl/fl/CD4-Cre mice (Figure 7E). When three populations of cells including CXCR5hi PD1hi Tfh and CXCR5lo PD1lo non-Tfh cells from Bcl6+/+/CD4-Cre mice, and CXCR5lo PD1lo non-Tfh cells from Bcl6fl/fl/CD4-Cre mice were sorted and stimulated by either anti-CD3 or IL-7, assessment of STAT5 activation was conducted. Although STAT5 in Tfh cells remained non-phosphorylated, Bcl6 deficiency in non-Tfh cells led to strongly enhanced STAT5 phosphorylation (Figure 7F). The elevated STAT5 activation in Bcl6-deficient T cells was in part associated with the increase of Il7r and Stat5a mRNA expression (Figure S7D and E). These results collectively demonstrated that Bcl6 functions to suppress IL-7R/STAT5 signaling axis in promoting Tfh cell differentiation.
Discussion
Despite the discovery of Bcl6 as the core transcriptional factor for Tfh cells (Cannons et al., 2013; Crotty, 2011), little was known on how Bcl6 directs the development and function of Tfh cells. Here, we built a Bcl6-dependent transcriptional network by combining the gene expression profiling and ChIP-seq data sets, and further validating the role of IL-7R signaling during Tfh cell response. Our data demonstrate that Bcl6 promoted Tfh cell generation, at least in part through antagonizing the IL-7R/STAT5 axis. In comparison with published studies on Bcl6 function in multiple cell types (Barish et al., 2010; Barish et al., 2012; Basso et al., 2010; Ci et al., 2009; Huang et al., 2013; Liao et al., 2014), our findings highlighting the functional specificity of Bcl6 in mouse Tfh cell were supported by preferential DNA-binding motifs and distinct sets of target genes.
Bcl6 functions critically in various types of cells. Bcl6-deficient mice develop immune inflammation in multiple organs characterized with deregulated innate immunity, infiltration of eosinophil and Th2-mediated hyper immune responses, but has a defective GC reaction as a result of failed formation of both Tfh and GC B cells(Johnston et al., 2009; Nurieva et al., 2009; Phan and Dalla-Favera, 2004; Ye et al., 1997; Yu et al., 2009). In a recent study, knock-in point mutation in Bcl6 BTB domain dissociates the binding of BTB domain with its co-repressor NCOR, SMRT and BCOR, manifested as the failure of GC B cell formation, whereas Tfh cells and macrophage remain intrinsically unaffected by this mutation (Huang et al., 2013). What distinguished this selectivity remained unclear. Our present study analyzing global binding profiles of Bcl6 in Tfh, Th9, B cells and macrophages provided critical insights into functional diversity of Bcl6. In both mouse Tfh and Th9 cells, Bcl6 bound preferentially to consensus sequences containing classical Bcl6 binding motifs, which was in strikingly different from that in B cells, although to a less extent in macrophages. For instance, Bcl6 is able to bind to Prdm1 (Blimp1) gene locus in B cell and human T cell to inhibit its expression (Crotty et al., 2010; Hatzi et al., 2015), while no Bcl6 binding was found at the Prdm1 locus in mouse Tfh (Supplementary Figure 5A) or Th9 cells (Data not shown). Among Bcl6 target genes in Tfh cells, many of them were involved in fundamental biological activities, as such calcium-signaling pathway (Cacna1d, Cacna1b, Cacna1i, Itpr2, Ppp3ca), cell adherence junction (Insr, Igf1r, Ctnnd1, Ptprj, Ctnna1), and purine metabolism (Pdeba, Ampd2, Pde4a, Pfas, Fhit). Of the note, despite recent finding that transcriptional factor TCF-1 (Tcf-7) regulates Bcl6 expression in Tfh cell (Hatzi et al., 2015; Kubo, 2015; Xu et al., 2015), our ChIP-seq data demonstrated that TCF-1 was a common downstream target of both Bcl6 and Ascl2 (Figure 4D), indicating a complex interaction of transcription factors in the course of Tfh cell response. Also, Bcl6 was able to bind to Il13 locus in T cells, and to repress its expression at a magnitude of hundreds folds. This finding for the first time provided direct explanation for the elevation of Th2-mediated immunity in Bcl6-null mice (Ye et al., 1997). Bcl6 restrains inflammatory responses in macrophage through directly counteracting TLR4-induced transcriptome, NF-κB-cistromes, and histone acetylation (Barish et al., 2010). In Tfh cells, however, Bcl6 binding to gene loci was associated with decreased Tet1 recruitment specifically, and Bcl6 binding regions appears to have repressive epigenetic modification with low levels of 5hmC.
It has been described that Bcl6 acted as “master regulator” for Tfh cell commitment through repression of other T cell lineages including Th1, Th2, and Th17 (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). Bcl6 deficiency results in massive enhancement of inflammatory cytokine production in T cells (Figure 4A and B). However, a surprisingly few of Bcl6 binding sites were observed in Th signature gene loci, which raised the question of how Bcl6 repressed Th-relevent genes. Our observation that substantial amount of STAT5a peaks were enriched on majorities of these genes leaves us a hint that Bcl6 might restrain Th cell via controlling STAT5 activation (Supplementary Figure 5). Indeed, our genome-wide expression profiling and ChIP-seq analysis showed that Bcl6 suppresses both IL-7R (Supplementary Figure 9 and 13A) and STAT5 (Supplementary Figure 3A, 3B, and Supplementary Figure 13B) expression; Bcl6 also played a role blocking IL-2-triggered STAT5 activation through elevation of Mxd4 (Supplementary Figure 9A and B). Accordingly, Bcl6-mediated IL-2/IL-7 signaling inhibition in Tfh cells led to the silencing of STAT5 phosphorylation. Moreover, our results showed that Bcl6 and STAT5 bound reciprocally to overlapping sites in many common genes in Tfh and non-Tfh cells, respectively (Figure 3C and Supplementary Figure 6). Collectively, opposition between Bcl6 and STAT5 offered mechanistic basis for bifurcating Tfh vs. non-Tfh cell development.
IL-7R is expressed on naïve and memory T cell for their homeostatic survival (Kaech et al., 2003; Kaech et al., 2002; Li et al., 2003; Schluns et al., 2000). TCR engagement in activated T cell induces IL-7R downregulation via IL-2-mediated signaling pathway (Xue et al., 2002). Nonetheless, because Tfh cells are lack of IL-2 receptors including IL-2Ra and IL-2Rb (Johnston et al., 2012; Liu et al., 2014), repression of IL-7R by Bcl6 turns to be a dominant pathway, which prevents Tfh differentiation from STAT5-mediated regulation. After full commitment, Tfh memory precursor cells are proposed to gain IL-7R expression by reducing Bcl6, and prepare for memory generation (Kitano et al., 2011). Given that Bcl6 is necessary for maintenance of long-lived memory T cell (Ichii et al., 2002), further research is thus needed to clarify this discrepancy.
A recent study has identified genome-wide Bcl6 binding profiles in human tonsil Tfh cells (Hatzi et al., 2015). Analysis of their human data reached several consistent conclusions with our mouse Bcl6 binding data. For example, Bcl6 did not directly regulate expression of signature gene such as Rorc, Il-4 et al; IL-7R expression was substantially suppressed by Bcl6; STAT binding sites were enriched among Bcl6 binding sites. However, a significant difference in Bcl6 binding profiles existed between mouse and human Tfh cells. Bcl6 was shown to directly bind 8523 genomic loci and interact physically with AP-1 in human tonsil Tfh cells (Hatzi et al., 2015), while only 5191 Bcl6 binding sites were identified in CXCR5hi Bcl6-RFPhi Tfh cells sorted from mouse peripheral lymphocyte pools. In addition, we did not find an enrichment of AP-1 binding sites among mouse Bcl6 binding sites. It is well known that Bcl6 functions through cooperating with different partners in different cell types. It is understandable that the Bcl6 binding profiles in human tonsil CD45RO+ CD4+ CXCR5hi PD1hi T cells could be different from mouse peripheral CXCR5hi Bcl6+ cells. In addition, the discrepancies of Bcl6 binding profiles might also be caused by the fact that transcriptional regulation of mouse Tfh cell differs from that of human Tfh cell. For example, human, but not mouse, naïve CD4+ T cell can be polarized into Tfh cells by either TGF-β/IL-12 or TGF-β/IL-23 (SMAD2/STAT4/STAT6) (Schmitt et al., 2013; Schmitt et al., 2014).
In summary, by combining and comparing transcriptome and ChIP-seq data from T cell, B cell and macrophage, we have identified Tfh-specific Bcl6-mediated regulatory modules, among which some key components were tested by gain- or loss-of function assays. Therefore, our study has great implications for vaccine development, and clinical treatment of antibody-mediated autoimmune disorders.
Methods
Mice
C57BL/6, CD4-Cre, OT-II, Rosa26-FLP1, and B6SJLmice were from Jackson Laboratories. Bcl6-RFP and Bcl6−/− mice were maintained as previously described (Liu et al., 2012; Nurieva et al., 2009). The Bcl6fl/fl mice were created by homologous recombination. Bcl6fl/fl mice were bred with CD4-Cre mice to generate Bcl6fl/fl/CD4-Cre and control mice Bcl6+/+/CD4-Cre. Mice were maintained in specific pathogen-free animal facility and used in accordance with governmental and institutional guidelines for animal welfare. Age- and sex-matched donor or recipient mice were selected and included in different groups, typically 3–5 mice per group.
Immunization and Flow cytometry
Mice and their wild-type controls (6–8 week old; 3–5 mice per group) were immunized with KLH or OVA (0.5 mg/ml) emulsified in CFA (0.5 mg/ml) subcutaneously (s.c) (100 μl each mouse). Thymic, splenic and lymph node cells were stained with fluorescence-labeled antibodies, analyzed with a Fortesa and Flowjo. Intracellular staining was carried out with kits from ebioscience.
Retroviral transduction
Naïve CD4+CD25−CD44lowCD62Lhigh T cells from Bcl6−/−/OT-II, OT-II or C57BL6 mice were FACS-sorted and activated with plate-bound anti-CD3e (clone, 2C11) and anti-CD28 (clone, 37.51) under neutral conditions. 24 hours after activation, cells were infected by retroviruses Il7r-RV-GFP, Mxd4-RV-GFP, Bcl6-RV-GFP, or control empty vector (Empty-RV-GFP). 2 days after infection, GFP cells were FACS sorted for adoptive transfer, or followed by re-stimulation with pre-coated anti-CD3e. Cell transfer numbers were 1–4×105 for GFP+ T cells.
RNA-Seq
Total cellular RNA was extracted from sorted donor-derived CXCR5+ Bcl6−/−/OT-II, CXCR5+ Bcl6+/+/OT-II, Bcl6-RV-GFP+ or Empty-RV-GFP+ infected cells, with TRIzol reagent (Invitrogen). Sequence reads were obtained using the SOLiD v4 sequencing platform in order to generate a gene expression profile. To identify differential expression, the RNA-Seq reads were successfully mapped to mouse genome without quality problems. Read counts were normalized, fold changes were calculated for all possible comparisons, and a two-fold cutoff was used to select genes with expression changes. A padding procedure was used to calculate fold changes in avoidance of blowout due to near zero counts.
ChIP-Seq, MeDIP and hMeDIP
ChIP-Seq was performed as described previously (Wei et al., 2009). Briefly, in vivo-generated CXCR5hi Bcl6-RFPhi Tfh and CXCR5lo Bcl6-RFPlo Non-Tfh cells were sorted from Bcl6-RFP mice immunized with KLH/CFA, stimulated with plate-coated anti-CD3e, and fixed by 1% paraformaldehyde, and followed with digestion with Mnase cocktail (Activemotif). Chromatin from 2×106 cells was used for each ChIP experiment. Antibody against Bcl6 pAb (SC858 (N-3), Santa Cruz) was used. The methylated DNA immunoprecipitation (MeDIP) and hydroxymethylated DNA immunoprecipitation (hMeDIP) assay was performed as previously described (Ichiyama et al., 2015; Vucic et al., 2009). In brief, the genomic DNA from naïve CD4+ T cells and Tfh cells was purified and sonicated. DNA fragments (4 ug) were denatured and incubated with antibodies against 5mC (Eurogentec), 5hmC (Active Motif) or control IgG at 4°C overnight. Antibody-DNA complexes were captured by protein A agarose/Salmon sperm DNA. The immunoprecipitated DNA was purified and subjected to sequencing library preparation. All sequencing libraries were prepared using the TruSeq ChIP Sample Prep Kit (Illumina) according to the manufacture’s protocol. The DNA libraries were then sequenced with Illumina HiSeq 2500 at the Zhongshan Ophthalmic Center core facility.
Sequenced reads of 50 bp were obtained using the CASAVA 1.8.2 package (Illumina). All reads were mapped to the mouse genome mm9 and those uniquely mapped reads were subjected to further peak identification process. SICER_V1.1 (Zang et al., 2009) was used to identify significant peaks (FDR of 1%) with Input DNA (ChIP-Seq) or IgG IP (MeDIP-Seq) as the controls. Output of the peak files were converted to browser-extensible data (BED) files and viewed with UCSC genome browser. The MEME-ChIP (Machanick and Bailey, 2011) was used to perform the consensus binding motif analysis for Bcl6. To calculate the tag density of Bcl6 binding or epigenetic modifications around TSS or the centers of Bcl6 binding sites, uniquely mapped tags were summarized in 20 bp windows, and all window tag counts were normalized by the total number of bases in the windows and the total read number of the given sample.
Real-time RT-PCR analysis
Total RNA was extracted with Trizol reagent (Invitrogen). Oligo (dT), and MMLV reverse transcriptase (Invitrogen) were used to generate cDNA. Gene expressions were examined by iQ™ SYBR real-time kit (Bio-Rad Laboratories). The data were normalized to a reference gene beta-actin. The primer pairs for real time RT-PCR analysis of Bcl6, Ascl2, CXCR5, Blimp1, Gata3, T-bet, RORγ, Tet1, Tet2, and Tet3 were previously described (Ichiyama et al., 2015; Liu et al., 2012). Primer pair for detection of mouse Tcf7 is: forward, 5′-GGGAAAAACATCAAGAATCCACCA-3′; reverse, 5′-CTGGGCTAGCAAGCAGTTCT-3′.
Supplementary Material
Highlights.
Bcl6 exhibits specific binding in mouse Tfh cells.
Bcl6 binding correlates with decreased 5hmC.
Bcl6 and STAT5 target the same binding sites.
Bcl6 suppresses the IL-7R/STAT5 axis during Tfh cell generation.
Acknowledgments
We thank MD Anderson Genetic Engineering Facility for their assistance in the generation of Bcl6 conditional knockout mice, Weiqi Dang from Institute of Pathology and Southwest Cancer Center, Southwest Hospital for their help in mouse maintenance, and the Dong lab members for their help. The work was supported in part by starting package from Southwest Hospital (to X.L), the National Basic Research Program of China (2013CB967001 and 2015CB964601 to L.W.), Chinese Ministry of Science and Technology “973” program grant (No. 2014CB542501 to W. J.) and research grants from National Institutes of Health (NIH; Al106654 to C.D.). C.D. is a Bayer Chair Professor at Tsinghua University.
Footnotes
Accession Numbers
The ChIP-seq and RNA-se data were deposited in GEO, with accession number of GSE72188.
Author contribution
X.L. and C.D. conceptualized the study and designed the experiments. X.L. and L.W. performed the experiments, and wrote the manuscript; L.Z., H.L., X.Y., X.W., and T.C. analyzed the ChIP-Seq and RNA-Seq data. Q.Z., S.T., K.C.N. and K.I. helped with sample preparation for ChIP-Seq; W.J., Q.T., X.Z. and X.B. helped to conceive this study.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri MS, Becerra D, Kim J, Tseng TW, Tai LJ, Leblanc M, Diehl C, Cerchietti L, et al. The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab. 2012;15:554–562. doi: 10.1016/j.cmet.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Saito M, Sumazin P, Margolin AA, Wang K, Lim WK, Kitagawa Y, Schneider C, Alvarez MJ, Califano A, Dalla-Favera R. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115:975–984. doi: 10.1182/blood-2009-06-227017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Forster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. The Journal of experimental medicine. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons JL, Lu KT, Schwartzberg PL. T follicular helper cell diversity and plasticity. Trends Immunol. 2013;34:200–207. doi: 10.1016/j.it.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zincfinger protein, is a sequence-specific transcriptional repressor. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, Crotty S. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nature immunology. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ci W, Polo JM, Cerchietti L, Shaknovich R, Wang L, Yang SN, Ye K, Farinha P, Horsman DE, Gascoyne RD, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113:5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nature immunology. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- Dong C, Temann UA, Flavell RA. Cutting edge: critical role of inducible costimulator in germinal center reactions. Journal of immunology. 2001;166:3659–3662. doi: 10.4049/jimmunol.166.6.3659. [DOI] [PubMed] [Google Scholar]
- Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, Zumbo P, Kirouac K, Bhaskara S, Polo JM, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013;4:578–588. doi: 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K, Nance JP, Kroenke MA, Bothwell M, Haddad EK, Melnick A, Crotty S. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. The Journal of experimental medicine. 2015;212:539–553. doi: 10.1084/jem.20141380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Hatzi K, Melnick A. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nature immunology. 2013;14:380–388. doi: 10.1038/ni.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ichii H, Sakamoto A, Hatano M, Okada S, Toyama H, Taki S, Arima M, Kuroda Y, Tokuhisa T. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nature immunology. 2002;3:558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, et al. The methylcytosine dioxygenase tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015;42:613–626. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. The Journal of experimental medicine. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nature immunology. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. The Journal of experimental medicine. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, Okada T. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–972. doi: 10.1016/j.immuni.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Kubo M. TCF-1 and LEF-1 help launch the T(FH) program. Nature immunology. 2015;16:900–901. doi: 10.1038/ni.3254. [DOI] [PubMed] [Google Scholar]
- Li J, Huston G, Swain SL. IL-7 promotes the transition of CD4 effectors to persistent memory cells. The Journal of experimental medicine. 2003;198:1807–1815. doi: 10.1084/jem.20030725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Spolski R, Li P, Du N, West EE, Ren M, Mitra S, Leonard WJ. Opposing actions of IL-2 and IL-21 on Th9 differentiation correlate with their differential regulation of BCL6 expression. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3508–3513. doi: 10.1073/pnas.1301138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Nurieva RI, Dong C. Transcriptional regulation of follicular Thelper (Tfh) cells. Immunol Rev. 2013;252:139–145. doi: 10.1111/imr.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. The Journal of experimental medicine. 2012;209:1841–1852. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KT, Kanno Y, Cannons JL, Handon R, Bible P, Elkahloun AG, Anderson SM, Wei L, Sun H, O’Shea JJ, Schwartzberg PL. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 2011;35:622–632. doi: 10.1016/j.immuni.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis P, Opstelten D. Functional anatomy of germinal centers. Am J Anat. 1984;170:421–435. doi: 10.1002/aja.1001700315. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, Huang H, Wen R, Wang J, Li HS, et al. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J Biol Chem. 2012;287:11234–11239. doi: 10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- Schluns KS, Kieper WC, Jameson SC, Lefrancois L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nature immunology. 2000;1:426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- Schmitt N, Bustamante J, Bourdery L, Bentebibel SE, Boisson-Dupuis S, Hamlin F, Tran MV, Blankenship D, Pascual V, Savino DA, et al. IL-12 receptor beta1 deficiency alters in vivo T follicular helper cell response in humans. Blood. 2013;121:3375–3385. doi: 10.1182/blood-2012-08-448902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N, Liu Y, Bentebibel SE, Munagala I, Bourdery L, Venuprasad K, Banchereau J, Ueno H. The cytokine TGF-beta co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nature immunology. 2014;15:856–865. doi: 10.1038/ni.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- Tsagaratou A, Aijo T, Lio CW, Yue X, Huang Y, Jacobsen SE, Lahdesmaki H, Rao A. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E3306–3315. doi: 10.1073/pnas.1412327111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Vinuesa CG, Cyster JG. How T cells earn the follicular rite of passage. Immunity. 2011;35:671–680. doi: 10.1016/j.immuni.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Vucic EA, Wilson IM, Campbell JM, Lam WL. Methylation analysis by DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;556:141–153. doi: 10.1007/978-1-60327-192-9_10. [DOI] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X, He R, Hao Y, Wang H, Zhao T, et al. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nature immunology. 2015;16:991–999. doi: 10.1038/ni.3229. [DOI] [PubMed] [Google Scholar]
- Xue HH, Kovanen PE, Pise-Masison CA, Berg M, Radovich MF, Brady JN, Leonard WJ. IL-2 negatively regulates IL-7 receptor alpha chain expression in activated T lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13759–13764. doi: 10.1073/pnas.212214999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nature immunology. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Yu D, Vinuesa CG. The elusive identity of T follicular helper cells. Trends Immunol. 2010;31:377–383. doi: 10.1016/j.it.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.