Abstract
Four clonally related Escherichia coli strains were isolated successively from bile duct of a girl suffering from sclerosing cholangitis. One of them, selected after an imipenem-containing regimen, was resistant to carbapenems and to broad-spectrum cephalosporins due to a plasmid-mediated cephalosporinase, CMY-2, and the lack of outer membrane proteins OmpF and OmpC.
Resistance to carbapenems, while rare in Enterobacteriaceae, can be mediated by several mechanisms (4, 13). Production of β-lactamases capable of hydrolyzing carbapenems has been reported in Enterobacteriaceae: mostly in Enterobacter spp. and Serratia spp. (16). Carbapenem resistance may result also from production of large quantities of chromosomal and plasmid-mediated cephalosporinases combined with decreased drug permeability through the outer membrane. Involvement of chromosomal cephalosporinase is known for Enterobacter spp., Proteus rettgeri, and Citrobacter freundii, whereas plasmid-mediated cephalosporinases are involved in low-level resistance to carbapenems for rare isolates of Proteus mirabilis, Klebsiella pneumoniae, and Salmonella enterica serotype Wien (1, 3, 6, 8, 18, 21). In Escherichia coli, reports of low-level resistance to imipenem are exceptional, resulting from AmpC hyperproduction and loss of porins (14, 16, 17, 22). We report here the clinical and microbiological features associated with a carbapenem-resistant E. coli isolate that had been selected in vivo by an imipenem-containing regimen. A detailed molecular analysis of the antibiotic resistance mechanisms is provided.
Patient and strains.
E. coli isolates 1 to 4 were isolated from bile duct of a 10-year-old girl hospitalized several times at the Hôpital Bicêtre (Le Kremlin-Bicêtre, France), because she had sickle cell disease, autoimmune hepatitis, and sclerosing cholangitis. E. coli isolates 1 and 3 had been isolated in November 2001, whereas isolates 2 and 4 had been isolated in February 2003. These isolates were identified with the API20E gallery (BioMérieux, La-Balme-les-Grottes, France), and identification was confirmed by sequencing of 16S ribosomal DNA sequencing as described previously (2). E. coli isolates 2 and 3 were also obtained from blood cultures in February 2003 and September 2002, respectively. Several treatments with antibiotics were given from November 2001 to February 2003, including the β-lactams amoxicillin, cefotaxime, and imipenem. E. coli isolate 4 was recovered in the immediate follow-up of an imipenem-containing regimen (750 mg two times daily for 8 days). Then the patient was treated with ciprofloxacin for 2 weeks and underwent surgery of the bile duct.
Susceptibility testing.
Disk diffusion susceptibility testing with antibiotic-containing disks (Sanofi Diagnostics Pasteur, Marnes-La-Coquette, France) (http://www/sfm.asso.fr) was performed with and without cloxacillin (150 μg/ml), a β-lactam molecule that inhibits cephalosporinase activity (5). MICs were determined by an agar dilution technique as reported previously (19). E. coli isolate 1 was susceptible to β-lactams, whereas E. coli isolate 2 was resistant to cephalosporins and aztreonam (Table 1). The resistance to β-lactams of E. coli isolate 3 was similar to that of E. coli isolate 2 (although expressed at a much lower level), whereas E. coli isolate 4 was resistant to all β-lactams, including imipenem at a high level (with the exception of cefepime) (Table 1). Antimicrobial susceptibility testing on cloxacillin-containing plates indicated that the resistance levels of E. coli isolates 2, 3, and 4 to cephalosporins were decreased after cloxacillin addition (data not shown), suggesting the strong expression of a cephalosporinase.
TABLE 1.
MICs of β-lactams for E. coli clinical isolates 1 to 4, transconjugant E. coli DH10B(pLN), and reference strain E. coli DH10B
| β-Lactam(s) | MIC (μg/ml) for E. coli isolate or strain:
|
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | DH10B(pLN) | DH10B | |
| Amoxicillin | 4 | >512 | >512 | >512 | >512 | 2 |
| Ticarcillin | 2 | >512 | 256 | 512 | >512 | 2 |
| Ticarcillin + CLA | 1 | >512 | 256 | 512 | 256 | 1 |
| Piperacillin | 1 | >512 | 256 | 128 | >512 | 1 |
| Piperacillin + TZB | 1 | 128 | 8 | 128 | 256 | 1 |
| Cephalothin | 8 | >512 | >512 | >512 | >512 | 4 |
| Ceftazidime | 0.06 | >512 | 256 | >512 | 512 | 0.06 |
| Cefotaxime | 0.12 | 128 | 16 | >512 | 256 | 0.12 |
| Cefepime | <0.06 | 4 | 0.12 | 16 | 1 | <0.06 |
| Cefpirome | 0.06 | 8 | 1 | 128 | 4 | 0.06 |
| Moxalactam | 0.12 | 64 | 4 | >512 | 8 | 0.25 |
| Aztreonam | 0.06 | 512 | 16 | 512 | 512 | 0.12 |
| Imipenem | 0.06 | 0.25 | 0.25 | >32 | 0.25 | 0.12 |
| Meropenem | 0.06 | 0.12 | 0.03 | 8 | 0.12 | 0.06 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
Molecular investigation and biochemical analysis.
The pulsed-field gel electrophoresis technique with restriction enzyme XbaI was used for genotyping E. coli clinical isolates (20) and showed indistinguishable patterns between E. coli isolates 1 and 2 and between isolates 3 and 4, suggesting two clonal origins (data not shown). Conjugation experiments using a rifampin-resistant E. coli strain, DH10B, and selection on Mueller-Hinton agar plates containing 100 μg of rifampin per ml and 100 μg of ampicillin per ml (19) gave transconjugants, using E. coli isolates 2, 3, and 4 as donors. An identical 60-kb conjugative plasmid (pLN) was extracted from these transconjugants (19) that conferred resistance to penicillins and expanded-spectrum cephalosporins (Table 1). Standard PCR conditions were used to amplify several β-lactamase genes encoding plasmid-mediated cephalosporinases, including blaCMY-1/2, blaFOX-1, blaMOX-1 (18), and several clavulanic acid-susceptible narrow-spectrum and extended-spectrum β-lactamases (19). PCR amplification and sequencing identified the narrow-spectrum penicillinase gene blaTEM-1 and the plasmid-mediated cephalosporinase blaCMY-2 gene located on the 60-kb plasmid in E. coli isolates 2, 3, and 4 and their transconjugants, whereas no acquired β-lactamase gene was detected in E. coli isolate 1. Identification of the blaCMY-2 gene in E. coli isolates 2, 3, and 4 was consistent with their cephalosporin resistance profile.
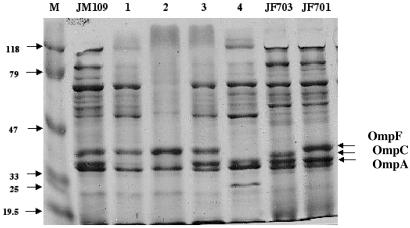
The outer membrane protein (OMP) profiles of the E. coli isolates were analyzed to explain imipenem resistance of E. coli isolate 4. OMP studies were performed with sodium dodecyl sulfate-containing polyacrylamide gel electrophoresis as described previously (15, 20) and E. coli control strains expressing either OmpC or OmpF (12). The OMP profiles of E. coli isolates 1 and 2 were identical, expressing two major porins, OmpF and OmpC, which comigrated, but missing porin OmpA (Fig. 1). Comparison of the OMP profiles of E. coli isolates 3 and 4 showed expression of OmpA in both strains, lack of OmpC in both strains, and lack of OmpF protein in E. coli isolate 4, whereas it was detected in E. coli isolate 3 (Fig. 1). Using whole-cell DNA of E. coli isolates 3 and 4 as a template and primers EcOmpFA (5′-CAGGTACTGCAAACGCTGC-3′) and EcOmpFB (5′-GTCAACATAGGTGGACAT G-3′) annealing at the ends of the OmpF gene of E. coli (15, 20), a 953-bp internal fragment of the OmpF gene was obtained (data not shown). Sequencing identified a wild-type OmpF gene for E. coli isolate 3, whereas that of E. coli isolate 4 had a 2-bp deletion located in the middle of the gene. This deletion introduced a premature stop codon in the protein leading to a truncated 187-amino-acid OmpF protein (normal size, 362 amino acids). Using primers EcOmpCA (5′-GTTAAAGTACTGTCCCTCCTG-3′) and EcOmpCB (5′-GAACTGGTAAACCAGACCCAG-3′), a 1,086-bp internal fragment of the OmpC gene of E. coli was amplified by using whole-cell DNA of E. coli isolates 1 and 2 as templates, whereas no amplification was obtained for E. coli isolates 3 and 4 (data not shown). Lack of an entire OmpF protein in E. coli isolate 4 in addition to lack of OmpC might explain additional resistance to imipenem.
FIG. 1.
OMP profiles of E. coli strains. OMPs were profiled by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Lanes 1 to 4 correspond to E. coli clinical isolates 1 to 4, respectively, and E. coli JM109 is a reference strain. E. coli isolates JF703 and JF701, expressing, respectively, OmpC or OmpF alone, are from reference 12. The molecular mass marker (M) and corresponding sizes (in kilodaltons) are indicated on the left. Horizontal arrows on the right indicate positions of the OMPs OmpC, OmpF, and OmpA.
Conclusions.
This report indicates that an imipenem-containing regimen may select for high-level imipenem resistance in E. coli. The conjugative property of the resistance plasmid explains its ability to transfer the β-lactam resistance marker to either clonally related (isolates 1 and 2) or non-clonally related (isolates 2 and 3) E. coli isolates. The plasmid encoded a cephalosporinase CMY-2 that is widespread in Enterobacteriaceae in human and animal isolates, especially in the United States (7, 10, 11, 18) and associated with TEM-type β-lactamase genes (7, 9, 10, 11, 20). Difference in susceptibility to several β-lactams (cefepime and moxalactam) between two unrelated CMY-2-positive E. coli isolates (isolates 2 and 3) may result from difference in another OMP, OmpA (Fig. 1).
This report indicates that plasmid-mediated cephalosporinases may constitute a reservoir of antibiotic resistance (18, 20), enhancing the probability to select for carbapenem resistance in E. coli. In those cases, use of carbapenems, including the novel commercially available ertapenem versus the cephalosporinase-resistant cefepime and cefpirome, is debatable. We showed as well that in vivo-acquired nucleotide substitutions in the OmpF gene contributed to resistance to carbapenems, as suggested by in vitro experiments (15). Selection of E. coli isolates with different antibiotic resistance patterns from bile duct parallels another study showing that molecular evolution of antibiotic resistance may have occurred in multiple liver cysts infected with E. coli (14).
From a general point of view, this case illustrates that high-level resistance to broad-spectrum β-lactams may occur in enterobacterial isolates responsible for chronic or recurrent infections from a two-step mechanism that arose successively: i.e., horizontal gene transfer and chromosomal mutations. Management of such infections may benefit from “on-line” surveillance of molecular mechanisms of resistance.
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, France, and by the European Community (6th PCRD, LSHM-CT-2003-503-335). L.P. is a researcher from the INSERM, France.
We thank G. A. Jacoby for the gift of E. coli isolates JF703 and JF701, used as controls.
REFERENCES
- 1.Armand-Lefèvre, L., V. Leflon-Guibout, J. Bredin, F. Barguellil, A. Amor, J.-M. Pagès, and M.-H. Nicolas-Chanoine. 2003. Imipenem resistance in Salmonella enterica serovar Wien related to porin loss and CMY-4 β-lactamase production. Antimicrob. Agents Chemother. 47:1165-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avidor, B., Y. Kletter, S. Abulafia, Y. Golan, M. Ephros, and M. Giladi. 1997. Molecular diagnosis of cat scratch disease: a two-step approach. J. Clin. Microbiol. 35:1924-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush, K. 2001. New β-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 32:1085-1089. [DOI] [PubMed] [Google Scholar]
- 5.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, V. T. B., G. Arlet, B. M. Ericsson, A. Tammelin, P. Courvalin, and T. Lambert. 2000. Emergence of imipenem resistance in Klebsiella pneumoniae owing to combination of plasmid-mediated CMY-4 and permeability alteration. J. Antimicrob. Chemother. 46:895-900. [DOI] [PubMed] [Google Scholar]
- 7.Carattoli, A., F. Tosini, W. P. Giles, M. E. Rupp, S. H. Hinrichs, F. J. Angulo, T. J. Barrett, and P. D. Fey. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow, J. W., and D. M. Shlaes. 1991. Imipenem resistance associated with the loss of a 40-kDa outer membrane protein in Enterobacter aerogenes. J. Antimicrob. Chemother. 28:499-504. [DOI] [PubMed] [Google Scholar]
- 9.Dunne, E. F., P. D. Fey, P. Kludt, R. Reporter, F. Mostashari, P. Shillam, J. Wicklund, C. Miller, B. Holland, K. Stamey, T. J. Barrett, J. K. Rasheed, F. C. Tenover, E. M. Ribot, and F. J. Angulo. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC β-lactamase. JAMA 24:3151-3156. [DOI] [PubMed] [Google Scholar]
- 10.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N. Engl. J. Med. 17:1242-1249. [DOI] [PubMed] [Google Scholar]
- 11.Hoyen, C. M., A. M. Hujer, K. M. Hujer, S. H. Marshall, L. Carias, P. Toltzis, L. B. Rice, and R. A. Bonomo. 2002. A clinical strain of Escherichia coli possessing CMY-2 plasmid-mediated Amp C β-lactamase: an emerging concern in pediatrics? Microb. Drug Resist. 8:329-333. [DOI] [PubMed] [Google Scholar]
- 12.Jacoby, G. A., and L. Sutton. 1985. β-Lactamases and β-lactam resistance in Escherichia coli. Antimicrob. Agents Chemother. 28:703-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlowsky, J. A., M. E. Jones, C. Thornsberry, I. R. Friedland, and D. F. Sahm. 2003. Trends in antimicrobial susceptibilities among Enterobacteriaceae isolated from hospitalized patients in the United States from 1998 to 2001. Antimicrob. Agents Chemother. 47:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low, A. S., F. M. MacKenzie, I. M. Gould, and I. R. Booth. 2001. Protected environments allow parallel evolution of a bacterial pathogen in a patient subjected to long-term antibiotic therapy. Mol. Microbiol. 42:619-630. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Martinez, L., M. C. Conejo, A. Pascual, S. Hernández-Allés, S. Ballesta, E. Ramirez De Arellano-Ramos, V. J. Benedi, and E. J. Perea. 2000. Activities of imipenem and cephalosporins against clonally related strains of Escherichia coli hyperproducing chromosomal β-lactamase and showing altered porin profiles. Antimicrob. Agents Chemother. 44:2534-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 17.Odeh, R., S. Kelkar, A. M. Hujer, R. A. Bonomo, P. C. Schrechenberger, and J. P. Quinn. 2002. Broad resistance due to plasmid-mediated AmpC β-lactamases in clinical isolates of Escherichia coli. Clin. Infect. Dis. 35:140-145. [DOI] [PubMed] [Google Scholar]
- 18.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., C. Héritier, I. Podglajen, W. Sougakoff, L. Gutmann, and P. Nordmann. 2003. Emergence in Klebsiella pneumoniae of a chromosome-encoded SHV β-lactamase that compromises the efficacy of imipenem. Antimicrob. Agents Chemother. 47:755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Smith Moland, E., J. A. Black, J. Ourada, M. D. Reisbig, N. D. Hanson, and K. S. Thomson. 2002. Occurrence of newer β-lactamases in Klebsiella pneumoniae isolates from 24 U.S. hospitals. Antimicrob. Agents Chemother. 46:3837-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stapleton, P. D., K. P. Shannon, and G. L. French. 1999. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 β-lactamase production and loss of an outer membrane protein. Antimicrob. Agents Chemother. 43:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]