Abstract
Rationale
Recently, there has been significant interest in the therapeutic administration of miRNA mimics and inhibitors to treat cardiovascular disease. In particular, miR-27b has emerged as a regulatory hub in cholesterol and lipid metabolism and potential therapeutic target for treating atherosclerosis. Despite this, the impact of miR-27b on lipid levels in vivo remains to be determined. As such, here we set out to further characterize the role of miR-27b in regulating cholesterol metabolism in vitro and to determine the effect of miR-27b overexpression and inhibition on circulating and hepatic lipids in mice.
Methods and Results
Our results identify miR-27b as an important regulator of LDLR activity in human and mouse hepatic cells through direct targeting of LDLR and LDLRAP1. In addition, we report that modulation of miR-27b expression affects ABCA1 protein levels and cellular cholesterol efflux to ApoA1 in human hepatic Huh7 cells. Overexpression of pre-miR-27b in the livers of wild-type mice using AAV8 vectors increased pre-miR-27b levels 50–fold and reduced hepatic ABCA1 and LDLR expression by 50% and 20%, respectively, without changing circulating and hepatic cholesterol and triglycerides. To determine the effect of endogenous miR-27b on circulating lipids, wild-type mice were fed a Western diet for one month and injected with 5 mg/kg of LNA control or LNA anti-miR-27b oligonucleotides. Following two weeks of treatment, the expression of ABCA1 and LDLR were increased by 10–20% in the liver, demonstrating effective inhibition of miR-27b function. Intriguingly, no differences in circulating and hepatic lipids were observed between treatment groups.
Conclusions
The results presented here provide evidence that short-term modulation of miR-27b expression in wild-type mice regulates hepatic LDLR and ABCA1 expression but does not influence plasma and hepatic lipid levels.
Keywords: miRNAs, miR-27b, LDLR, ABCA1, lipid homeostasis, atherosclerosis
INTRODUCTION
Alterations in the control of cholesterol homeostasis can lead to pathological processes, including atherosclerosis, the most common cause of mortality in Western societies. Epidemiological studies have identified many environmental and genetic factors that contribute to atherogenesis. In particular, high levels of low-density lipoprotein (LDL) cholesterol and low levels of high-density lipoprotein (HDL) cholesterol are associated with increased cardiovascular disease (CVD) risk1,2. As a result, substantial therapeutic progress has resulted from the widespread use of statins3 and other lipid-lowering drugs aimed at lowering plasma LDL-cholesterol (LDL-C). Despite this, statins are not sufficient to prevent the progression of atherosclerosis in many individuals and there is considerable evidence that quantitatively important determinants of disease susceptibility remain to be identified4,5.
In humans, the majority of serum cholesterol is transported as cholesterol esters in LDL particles. To ensure that blood cholesterol levels are balanced, LDL is constantly internalized. The uptake of LDL and other ApoE/ApoB containing lipoproteins occurs through the LDL receptor (LDLR) and is a classic example of receptor-mediated endocytosis6,7. The circulating level of LDL is determined in large part by its rate of uptake through this pathway, as evidenced by mutations in LDLR or LDLR adaptor protein 1 (LDLRAP1), which lead to the massive accumulation of LDL in patients with familial hypercholesterolemia (FH)8,9. The expression of the LDLR is tightly controlled by feedback mechanisms that operate at both transcriptional and post-transcriptional levels. When intracellular levels of cholesterol are high, the ER-bound sterol regulatory element-binding proteins (SREBPs) coordinate the down-regulation of the LDLR, as well as 3-hydroxy-3methylglutaryl coenzyme A reductase (HMGCR), the rate-limiting enzyme of cholesterol biosynthesis10,11. Conversely, when sterol concentrations are low, SREBPs upregulate HMGCR and the LDLR, thereby enhancing LDL clearance from the plasma and ensuring that intracellular cholesterol levels are maintained10,11. Under these conditions, the liver X receptor (LXR)/ retinoid X receptor (RXR) heterodimers recruit corepressor complexes and actively repress genes involved in cholesterol efflux, such as the ATP-binding cassette (ABC) transporters, ABCA1 and ABCG1, thereby maintaining cellular cholesterol homeostasis12.
miRNAs are a class of small, non-coding RNAs that negatively regulate gene expression at the post-transcriptional level by promoting mRNA degradation and/or inhibiting mRNA translation13–15. Recently, numerous miRNAs have been implicated in the physiological processes underlying atherosclerosis and CVD, making them potential biomarkers and therapeutic targets for ameliorating cardiometabolic disorders. In particular, work from our lab and others have demonstrated a direct role for miR-33a/b in controlling cholesterol and fatty acid metabolism in concert with their SREBP host genes and have highlighted the therapeutic potential of modulating this miRNA family to increase ABCA1 expression, cellular cholesterol efflux, and circulating levels of HDL-C16,17.
In addition to miR-33, a number of other miRNAs have been associated with alterations in lipid homeostasis. Specifically, miR-27b has recently been identified as a regulatory hub in lipid metabolism, with many confirmed metabolic targets, including LPL, PPARγ, ANGPTL3, GPAM, ABCA1 and RXRα18–20. Additionally, studies have correlated miR-27b expression levels with clinical pathological factors and the prognosis of patients with atherosclerosis19. Furthermore, aberrant expression of miR-27b has been shown to be a predictor for unstable atherosclerotic plaques21,22. Despite these clinical observations, to date, no reports are available regarding the impact of miR-27b on lipid metabolism in vivo. Hence, here we set out to further analyze the effect of mIR-27b on cholesterol metabolism in vitro and determine the effect of miR-27b overexpression and inhibition on circulating lipid levels and hepatic gene expression in wild-type (WT) mice.
RESULTS
The liver enriched miR-27b is regulated by dietary lipids
miR-27b is a conserved member of the miR-23b~27b~24-1 miRNA cluster encoded within intron 14 of the C9orf3 gene on human chromosome 9 (Fig. S1A). In agreement with previous reports18, miR-27b expression is highly enriched in the liver and is regulated by dietary lipids. Specifically, qRT-PCR analysis revealed that mature miR-27b was increased in the livers of mice and non-human primates fed a high-fat diet (HFD) compared to those fed a chow diet (Fig. S1B–C). Taken together, these results corroborate recent reports demonstrating the regulation of the miR-27b transcript by dietary lipids and suggest that this regulation is evolutionarily conserved.
miR-27b directly targets the 3’UTR of human LDLR, LDLRAP1 and ABCA1
Recently, miR-27b has been identified as a key post-transcriptional regulator of cholesterol homeostasis19. Indeed, when we performed our own bioinformatics analyses, we found that miR-27b had predicted binding sites in the 3’UTRs of a myriad of lipid metabolism regulators (Table S1, Fig. 1A). To determine the specificity of miR-27b in regulating these pathways, we first overexpressed miR-27b in human hepatic (Huh7) cells and assessed the quantitative expression of lipid-related genes by qPCR technology. As expected, overexpression of miR-27b significantly repressed ABCA1 and GPAM mRNA levels (Fig. 1B), previously validated targets for miR-27b18,20. Intriguingly, we noticed that miR-27b also significantly decreased the expression of the LDLR and LDLRAP1, key players in the LDLR pathway that are essential for efficient clearance of circulating LDL-C. While ABCA1 has previously been confirmed as a direct target of miR-27b in human THP1 macrophages20, the role of miR-27b in directly regulating ABCA1 expression in the liver has not been investigated. Given that LDLR, LDLRAP1 and ABCA1 contribute to controlling the elevated LDL-C/HDL-C ratio, a prominent risk factor for developing CVD, we decided to investigate these targets further.
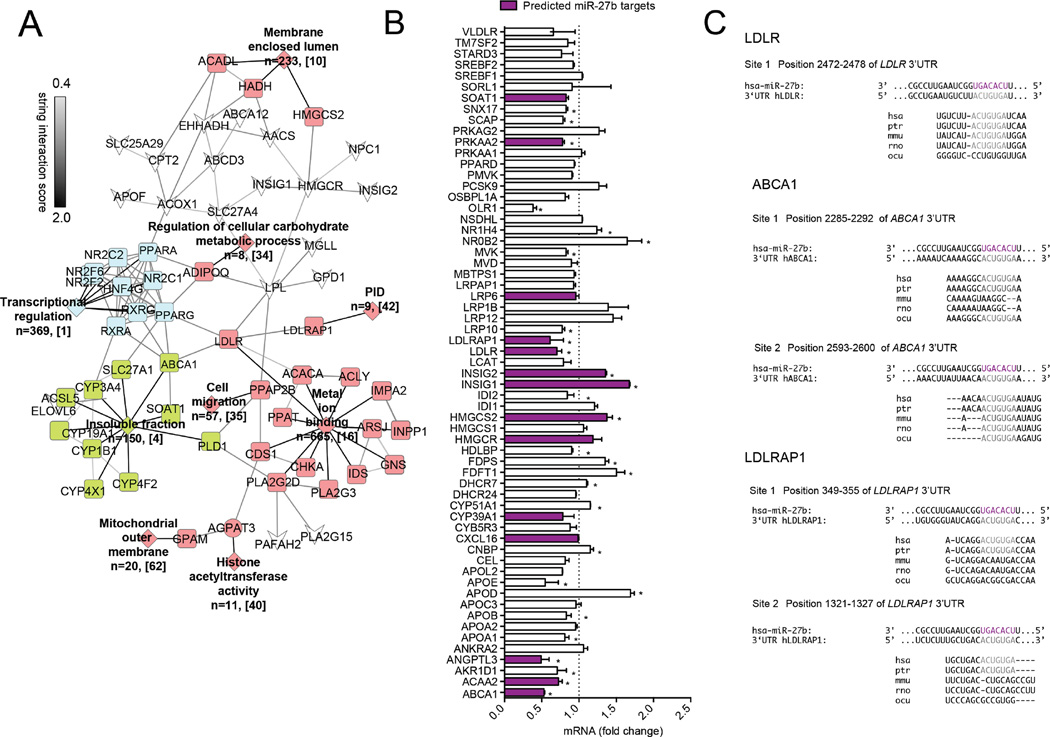
Figure 1. miR-27b is predicted to target a vast network of genes involved in lipid metabolism.
(A) Bioinformatic analysis of predicted target genes for miR-27b. Targets for miR-27b (predicted in Targetscan, miRWalk, and miRanda) were uploaded into DAVID for functional annotation cluster analysis. Functional clusters with an enrichment score of ≥ 1.0 are depicted in colored diamonds. n represents the number of genes within each cluster, while bracketed numbers represent each cluster number (Table S1). Grey lines between genes of different clusters indicate STRING interaction score. Genes not found within a functional annotation cluster are indicated by white inverted arrowheads. (B) Relative expression of 61 lipid metabolism genes in Huh7 cells overexpressing miR-27b. Values are expressed as fold-change compared to cells transfected with a control mimic (CM). Data are the mean ± SEM of 3 independent experiments. Purple bars represent predicted targets for miR-27b. *, P ≤ 0.05 compared to CM transfected cells (dashed line) by student’s t-test. (C) Location of predicted binding sites for miR-27b in the human 3’UTRs of LDLR, LDLRAP1, and ABCA1. The seed sequence of miR-27b is highlighted in purple, while complementary binding regions in the respective 3’UTR are highlighted in grey. Site conservation between species is shown below. Hsa, human; ptr, chimpanzee; mmu, mouse; rno, rat; ocu, rabbit.
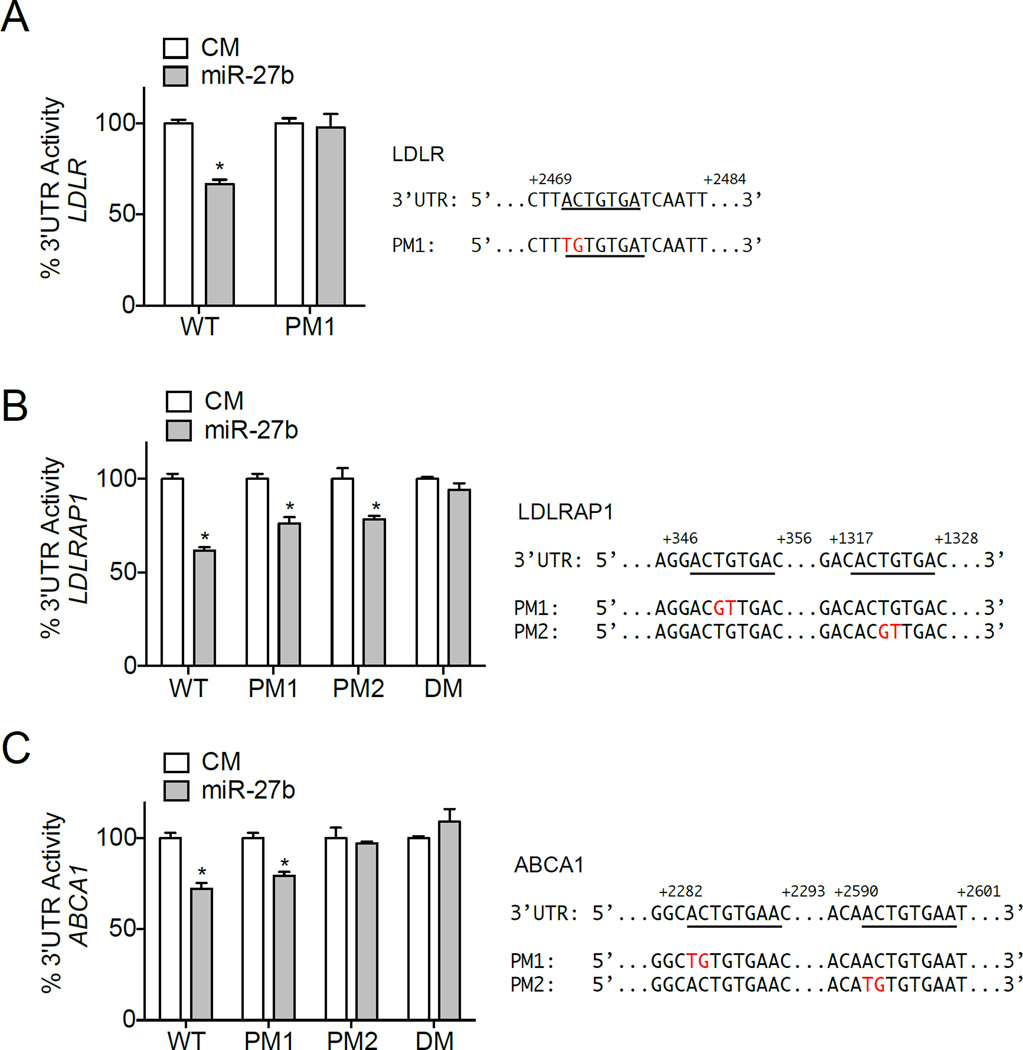
The human 3’UTR of LDLR has one conserved predicted miR-27b binding site, while the human 3’UTR of ABCA1 and LDLRAP1 have two predicted binding sites (Fig. 1B). Interestingly, site 2 in the 3’UTR of ABCA1 is highly conserved among species, while site 1 is only conserved in primates; the conservation of both predicted binding sites in the 3’UTR of LDLRAP1 is lost in rats and mice (Fig. 1B). To determine whether the 3’UTR of LDLR, LDLRAP1 and ABCA1 are direct targets of miR-27b, we performed luciferase reporter assays in COS7 cells. As shown in Fig. 2A–C, transfection of miR-27b significantly reduced LDLR, LDLRAP1 and ABCA1 3’UTR activity compared to control mimic (CM) transfected cells. Importantly, when we mutated the respective miR-27b binding sites in each 3’UTR, luciferase activity was significantly derepressed (Fig. 2A–C). Interestingly, mutation of both miR-27b binding sites (DM) was needed to fully rescue LDLRAP1 3’UTR luciferase activity, while mutation of only the conserved miR-27b binding site (PM2) was needed to rescue ABCA1 3’UTR activity (Fig. 2B, C). Taken together, these findings suggest that miR-27b directly interacts with the 3’UTR of human LDLR, LDLAP1, and ABCA1. As such, miR-27b might represent an ideal target to alter plasma levels of LDL and HDL cholesterol.
Figure 2. miR-27b specifically targets the 3’UTR of human LDLR, LDLRAP1, and ABCA1.
Luciferase reporter activity in COS7 cells transfected with a control mimic (CM), or miR-27b mimic (miR-27b) and the human 3’UTR of LDLR (A), LDLRAP1 (B), or ABCA1 (C) with or without the indicated point mutations (PM) in the miR-27b target sites. Double mutation (DM) indicates that two miR-27b binding sites were mutated in the same 3’UTR construct. Human LDLR, LDLRAP1, and ABCA1 3’UTR sequences are shown to the right. Underlined sequences indicate predicted miR-27b binding sites. Nucleotides highlighted in red indicate respective point mutations. WT, wild-type 3’UTR. Data are the mean ± SEM and representative of ≥ 3 experiments in triplicate. *, P ≤ 0.05 compared to CM-transfected cells by unpaired t-test.
miR-27b negatively regulates LDLR and LDLRAP1 expression and LDLR activity
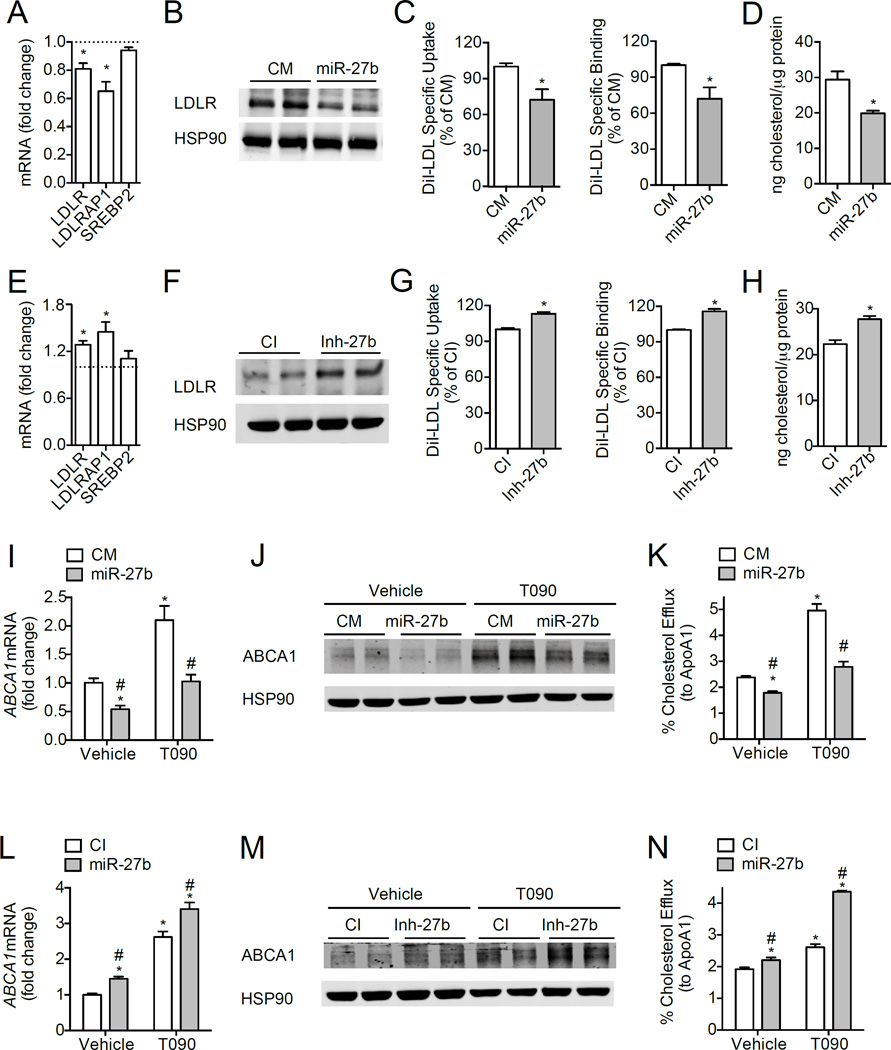
The hepatic LDLR plays a crucial role in controlling levels LDL-C in the blood, by facilitating the uptake of LDL-C in the liver1,2. To ascertain whether miR-27b overexpression or inhibition affects endogenous levels of LDLR and LDLRAP1, we assessed mRNA and protein expression of these target genes in human and mouse hepatic cells. As shown in Fig. 3A–B, transfection of human hepatic (Huh7) cells with a miR-27b mimic (miR-27b), but not a control mimic (CM), significantly decreased LDLR mRNA and protein levels, while the expression of SREBP2, a cholesterol-related gene not predicted to be targeted by miR-27b, was unaffected.. Conversely, inhibition of endogenous miR-27b significantly increased the expression of the LDLR (Fig. 3E), suggesting a physiological role for endogenous miR-27b in regulating levels of LDLR. Similar results were observed in mouse hepatic cells (Hepa) overexpressing or inhibiting miR-27b (Fig. S2A–B, S2D–E). In addition, miR-27b also regulated the expression of LDLRAP1 mRNA in Huh7 cells (Fig. 3A, E), suggesting that this miRNA might control LDLR activity in humans by direct targeting of the LDLR and by regulating its endocytosis.
Figure 3. Post-transcriptional regulation of LDLR, LDLRAP1 and ABCA1 by miR-27b in human hepatic cells.
(A–B) qRT-PCR (A) and Western blot (B) analysis of LDLR and LDLRAP1 in Huh7 cells transfected with 40 nM of control mimic (CM) or miR-27b mimic (miR-27b) for 48 h. Dashed line represents CM-transfected cells (A). HSP90 was used as a loading control (B). (C) Flow cytometry analysis of DiI-LDL specific uptake and binding in Huh7 cells transfected as in (A). (D) Intracellular cholesterol content in Huh7 cells transfected as in (A) and incubated with 30 µg/ml native LDL (nLDL) for 2 h at 37°C. (E–F) qRT-PCR (E) and Western blot analysis (F) of LDLR and LDLRAP1 in Huh7 cells transfected with 60 nM of control inhibitor (CI) or inhibitor of miR-27b (Inh-27b) for 48 h. Dashed line represents CI-transfected cells (E). HSP90 was used as a loading control (F). (G) Flow cytometry analysis of DiI-LDL specific uptake and binding in Huh7 cells transfected as in (E). (H) Intracellular cholesterol content in Huh7 cells transfected as in (E) and treated as in (D). (I–J) qRT-PCR (I) and Western blot (J) analysis of ABCA1 expression transfected with 40 nM CM or miR-27b and treated with 3 µM T090 for 12 h. (K) Cholesterol efflux to ApoA1 in Huh7 cells treated as in (I–J). (L–M) qRT-PCR (L) and Western blot (M) analysis of ABCA1 expression transfected with 60 nM CI or Inh-27b and treated with 3 µM T090 for 12 h. (N) Cholesterol efflux to ApoA1 in Huh7 cells treated as in (M–N). Data are the mean ± SEM and representative of ≥ 4 experiments in duplicate (A–B, E–F, I–J, and L–M) or ≥ 3 experiments in triplicate (C–D, G–H, K and N). *, P ≤ 0.05 compared to cells transfected with CM (A, C–D) or CI (E, G–H) by unpaired t-test. In panels (I, K–L, and N), statistical comparisons by one-way ANOVA with Bonferroni correction for multiple comparisons. *, P ≤ 0.05 compared to vehicle-treated cells transfected with CM or CI. #, P ≤ 0.05 compared to T090-treated cells transfected with CM or CI.
To determine the functional consequence of miR-27b-mediated regulation of LDLR and LDLRAP1, we next overexpressed or inhibited miR-27b and assessed DiI-LDL specific uptake and binding by flow cytometry. As shown in Fig. 3C and Fig. S2C, transfection of Huh7 and Hepa cells with miR-27b significantly decreased DiI-LDL specific uptake and binding. Intracellular cholesterol levels in miR-27b transfected cells were also significantly reduced after incubation with native LDL (nLDL) (Fig. 3D), consistent with miR-27b’s regulatory role in controlling LDLR-mediated endocytosis. Importantly, when we inhibited endogenous miR-27b using antisense inhibitors (Inh-27b) in Huh7 and Hepa cells, DiI-LDL specific binding, uptake and intracellular cholesterol levels were significantly increased in cells transfected with an inhibitor of miR-27b (Inh-27b) compared to cells transfected with a control inhibitor (CI) (Fig. 3G–H, Fig. S2F).
miR-27b regulates ABCA1 expression and cholesterol efflux in human hepatic cells
ABCA1 plays a major role in regulating cholesterol efflux from macrophages to ApoA1 and in the biogenesis of HDL in the liver, thereby controlling reverse cholesterol transport (RCT), a process that mediates the clearance of cholesterol from peripheral cells to the liver for excretion to the bile and feces23. To assess whether miR-27b post-transcriptionally regulates ABCA1, we transfected Huh7 cells with miR-27 mimics (miR-27b) or miR-27 antisense oligonucleotides (Inh-27b) and treated them with T0901317 (T090), an LXR agonist which induces ABCA1 expression. As expected, miR-27b overexpression strongly reduced the stimulation of ABCA1 mRNA and protein levels (Fig. 3I–J). Conversely, inhibition of endogenous miR-27b significantly increased ABCA1 expression in vehicle and T090–treated cells (Fig. 3L–M). To assess the functional consequence of inhibiting ABCA1 expression, we next determined whether miR-27b could regulate cholesterol efflux in human hepatic cells. Notably, overexpression and inhibition of miR-27b significantly decreased and increased, respectively, cholesterol efflux to ApoA1 (Fig. 3K,N). Thus, miR-27b appears to regulate hepatic cholesterol efflux and HDL biogenesis in the liver.
Modulation of miR-27b does not influence plasma and hepatic lipids in wild-type mice
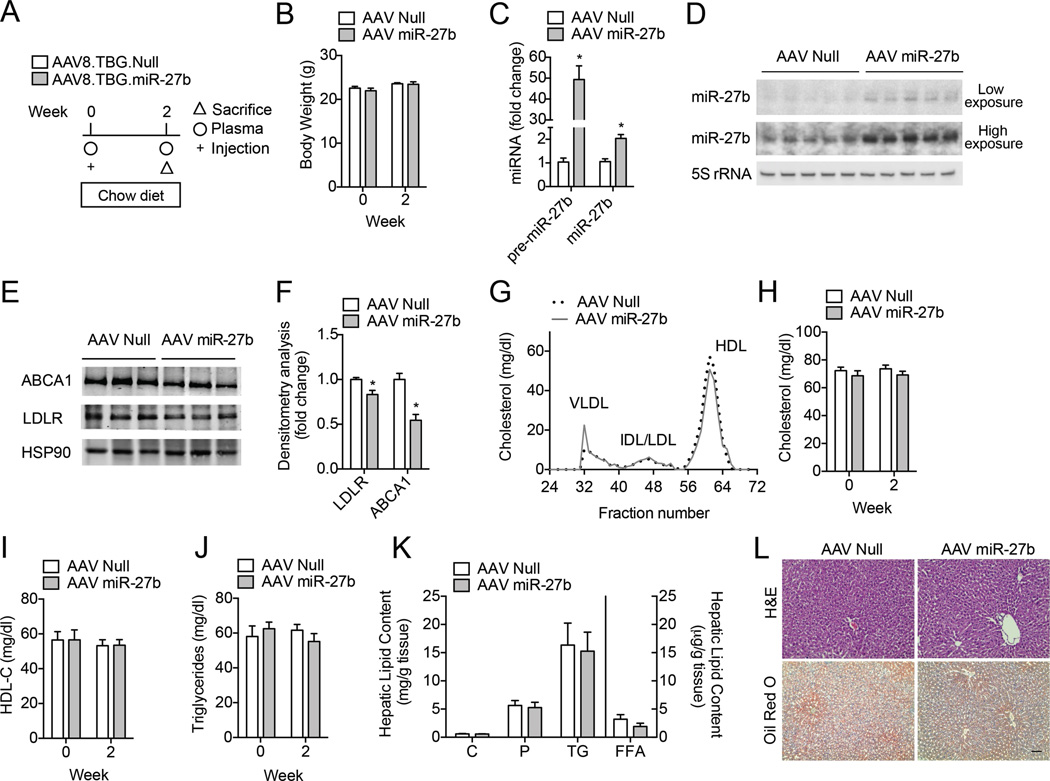
Because miR-27b alters both LDL uptake and cholesterol efflux in human hepatic cells, we next sought to determine the functional consequence of modulating miR-27b expression in vivo. As such, we first determined the functional contribution of increased miR-27b levels on plasma lipids in mice fed a chow diet. To specifically alter hepatic miRNA expression, we used an adeno-associated virus serotype 8 (AAV8) vector encoding the precursor form of miR-27b (pre-miR-27b, AAV miR-27b) or a control vector (AAV Null). AAV8 vectors have previously been evaluated for liver directed gene transfer in murine models and show no signs of liver toxicity24,25. For the chow diet studies, AAV Null or AAV miR-27b vectors (5 × 1012 GC/kg) were delivered to 8-week old male C57BL/6 (wild-type, WT) mice (n = 10 per group) via retro-orbital injection; plasma lipids were measured after two weeks (Fig. 4A). To determine the efficacy of miR-27b overexpression, we first measured the expression of hepatic miR-27b and its target genes after two weeks of treatment. As expected, pre-miR-27b and mature miR-27b expression levels were significantly higher in mice treated with AAV miR-27b compared to mice treated with AAV Null, while no changes in body weight between the two treatment groups were observed (Fig. 4B–D). Consistent with our in vitro results, both LDLR and ABCA1 protein levels were significantly reduced in the livers of mice treated with AAV miR-27b compared to those treated with AAV Null (Fig. 4E–F). Given that decreased hepatic expression of ABCA1 and LDLR would be predicted to reduce HDL biogenesis and increase circulating LDL-C, respectively, we next analyzed lipoproteins by fast performance liquid chromatography (FPLC). Surprisingly, hepatic overexpression of miR-27b had no effect on plasma cholesterol in the VLDL, IDL/LDL or HDL fractions after two weeks of treatment (Fig. 4G). Consistent with this, circulating total and HDL-C, as well as triglycerides, were similar in mice treated with AAV Null or AAV miR-27b (Fig. 4H–J). Furthermore, hepatic levels of cholesterol (C), phospholipids (P), triglycerides (TGs), and free fatty acids (FFAs), were unchanged in mice over-expressing miR-27b, suggesting that miR-27b does not influence lipid levels in the liver or circulation in WT mice (Fig. 4K–L).
Figure 4. Overexpression of miR-27b reduces hepatic LDLR and ABCA1 protein expression in wild-type mice.
(A) Experimental outline of eight-week old wild-type (WT) mice treated with 5 × 1012 GC/kg AAV Null or AAV miR-27b (n = 10 per group) and fed a chow diet. (B) Body weight of AAV Null and AAV miR-27b treated mice at baseline (Week 0) and two weeks after treatment. (C–D) qRT-PCR (C) and representative Northern blot (D) analysis of pre-miR-27b and miR-27b expression in the livers of mice treated with AAV Null and AAV miR-27b. 5S rRNA was used as a loading control. *, P ≤ 0.05 compared to AAV Null by unpaired t-test. (E–F) Representative Western blot analysis of hepatic ABCA1 and LDLR in the livers of mice after two weeks of treatment. HSP90 was used as a loading control. Quantification of blots compared to HSP90 is shown in (F). Numbers represent fold-change compared to AAV Null treated mice. *, P ≤ 0.05 compared to AAV Null by unpaired t-test. (G) Cholesterol content of FPLC-fractionated lipoproteins (n = 5 per group) of mice treated as in (A). (H–J) Total cholesterol, HDL-C, and triglycerides in the plasma of mice (n = 10 per group) treated with AAV Null or AAV miR-27b. (K) Total cholesterol (C), phospholipids (P), triglycerides (TG) and free fatty acids (FFA) in the livers of mice (n = 5 per group) treated as in (A). (L) Representative liver sections of mice treated with AAV Null or AAV miR-27b and stained with H&E or Oil red O. Scale bar = 70 µm. All data are the mean ± SEM.
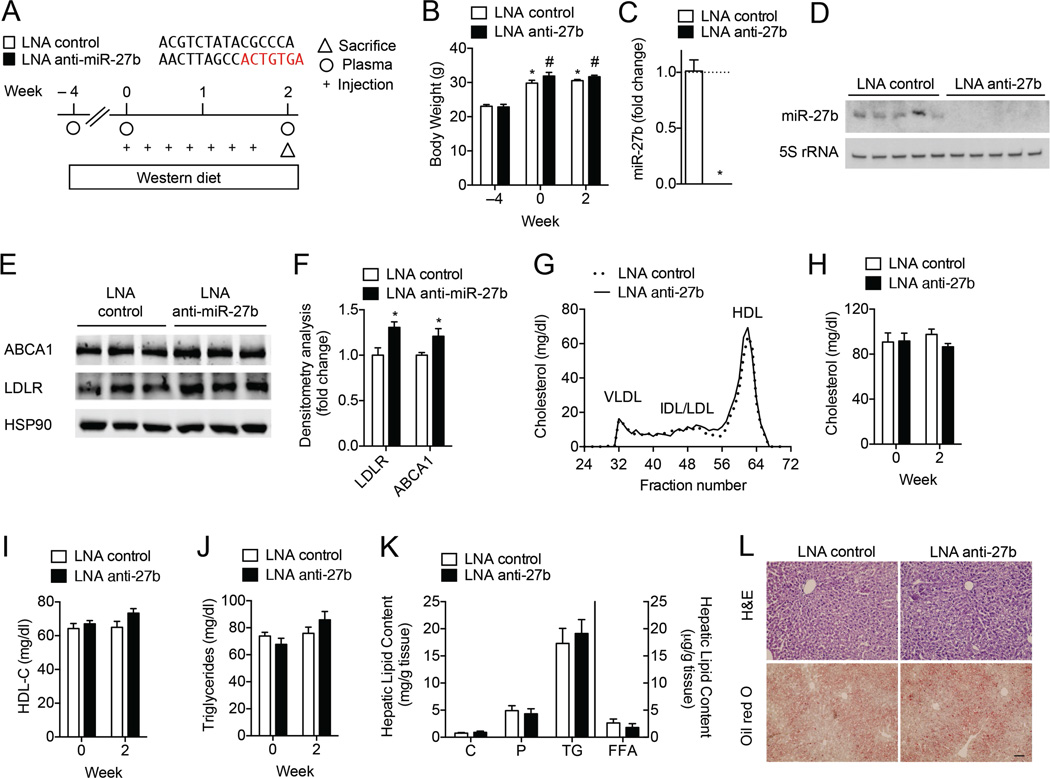
To gain insight into the physiological role of miR-27b in vivo, we next silenced miR-27b using locked nucleic acid (LNA) antisense inhibitors. Because the rate of hepatic LDL clearance is 40-fold greater in WT mice than in humans26, we challenged mice with a Western diet (WD) one month prior to treatment to increase circulating levels of LDL-C (Fig. 5A). After two weeks of treatment with 5 mg/kg LNA control antisense inhibitors (LNA control) or LNA antisense inhibitors against miR-27b (LNA anti-miR-27b), hepatic miR-27b levels were abolished (Fig. 5C–D). Additionally, no changes in body weight were observed between the different treatment groups (Fig. 5B). In accordance with decreased levels of miR-27b, hepatic LDLR and ABCA1 protein expression were modestly, but significantly increased, in LNA anti-miR-27b treated mice compared to controls (Fig. 5E–F). Surprisingly, when we analyzed fasting plasma lipid levels by FPLC, we observed no statistically significant differences in the cholesterol content of fractionated lipoproteins. Accordingly, total plasma cholesterol, HDL-C, and triglycerides were not significantly different in mice treated with LNA control or LNA anti-miR-27b (Fig. 5G–J). Moreover, the hepatic lipid content was similar in both treatment groups (Fig. 5K–L). Collectively, data in Figs. 4 and 5 suggest that inhibition of miR-27b may not represent an ideal therapeutic strategy to reduce levels of LDL-C and increase levels of HDL-C, a beneficial outcome for combating cardiometabolic disorders.
Figure 5. Inhibition of miR-27b levels does not influence plasma cholesterol levels in wild-type mice.
(A) Experimental outline of eight-week old wild-type (WT) mice fed a Western diet (Week −4) and treated with 5 mg/kg LNA control or LNA anti-miR-27b (LNA anti-27b) every two days for two weeks (n = 8 per group). (B) Body weight of LNA control and LNA anti-miR-27b treated mice at baseline (Week −4), at the start of treatment (Week 0) and two weeks after treatment. Statistical analysis by two-way ANOVA with Bonferroni correction for multiple comparisons. *, P ≤ 0.05 compared to LNA control treated mice at Week −4. #, P ≤ 0.05 compared to LNA anti-miR-27b treated mice at Week −4. (C–D) qRT-PCR (C) and representative Northern blot (D) analysis of miR-27b expression in the livers of mice treated with LNA control and LNA anti-miR-27b. 5S rRNA was used as a loading control. *, P ≤ 0.05 compared to LNA control by unpaired t-test. (E–F) Representative Western blot analysis of hepatic ABCA1 and LDLR in the livers of mice after two weeks of treatment. HSP90 was used as a loading control. Quantification of blots compared to HSP90 is shown in (F). Numbers represent fold-change compared to LNA control treated mice. *, P ≤ 0.05 compared to LNA control by unpaired t-test. (G) Cholesterol content of FPLC-fractionated lipoproteins (n = 8 per group) of mice treated as in (A). (H–J) Total cholesterol, HDL-C, and triglycerides in the plasma of mice (n = 8 per group) treated with AAV Null or AAV miR-27b. (K) Total cholesterol (C), phospholipids (P), triglycerides (TG) and free fatty acids (FFA) in the livers of mice (n = 8 per group) treated as in (A). (L) Representative liver sections of mice treated with LNA control or LNA anti-miR-27b and statined with H&E or Oil red O. Scale bar = 70 µm. All data are the mean ± SEM.
DISCUSSION
Over the years, in silico and in vitro analyses have implicated miR-27b in numerous cellular processes that regulate atherosclerosis, including lipid metabolism, insulin resistance and type-2 diabetes. In particular, many genes associated with cholesterol homeostasis have been identified as predicted targets for miR-27b. Despite this, few have been functionally validated in relevant metabolic cell types and in vivo experimentation is lacking. In the present study, we investigate, for the first time, the role of miR-27b in controlling target gene expression and lipid metabolism in vivo. Specifically, we demonstrate that acute overexpression and inhibition of hepatic miR-27b does not significantly affect circulating and hepatic lipids in mice, suggesting that modulation of miR-27b levels may not be an effective therapeutic strategy for treating dyslipidemias.
miR-27b is a member of the miR-27 family, of which there are two isoforms, miR-27a and miR-27b. While miR-27a is an intergenic miRNA located on human chromosome 19, miR-27b is located within intron 14 of the C9orf3 gene and is a member of the miR-23b~27b~24-1 cluster. Recently, miR-27a/b have been shown to directly and indirectly target many lipid-metabolism transcription factors, including RXRα, PPARγ, ABCA1, GPAM, LPL, CD36 and ACAT120,27–30. In addition to these, here, we identify and describe two additional targets for miR-27b, namely LDLR and LDLRAP1. By inhibiting the expression of LDLR and LDLRAP1, miR-27b reduces DiI-LDL uptake and binding, and concomitantly decreases intracellular cholesterol concentrations in human hepatic cells. During the preparation of this manuscript, Alvarez et. al. demonstrated that miR-27a also controls cholesterol uptake by direct regulation of LDLR and LDLRAP1 in HepG2 cells, thereby implicating both miR-27 family members in the control of circulating LDL-C levels (Alvarez et al. Atherosclerosis. 2015, In Press). However, given that hepatic miR-27b expression levels are higher than miR-27a18, it is likely that this miRNA contributes more to regulating LDLR endocytosis in vivo. Indeed, when we overexpressed and inhibited miR-27b in mice, we found a significant decrease and increase, respectively, in hepatic LDLR expression. Despite this, no differences were observed in circulating levels of LDL-C and total cholesterol. Given that WT mice carry most of their cholesterol in HDL26, it is possible that appreciable differences in plasma LDL-C were masked in our mouse model. Alternatively, miR-27b may contribute to the regulation of other processes that control circulating levels of LDL-C, such as VLDL secretion. It was previously shown that miR-27b directly regulates GPAM, an enzyme that catalyzes the first committed step in de novo triglyceride synthesis31; as such, miR-27b-mediated increases in LDL-C (through inhibition of LDLR) may be balanced by decreases in triglyceride availability for VLDL production. No significant differences in circulating or hepatic triglycerides, however, were observed when miR-27b levels were altered. Nevertheless, future studies using humanized mouse models of familial hypercholesterolemia and non-human primates are needed to definitely characterize the role of miR-27b in regulating plasma LDL-C in vivo.
In addition to LDLR and LDLRAP1, we also validate miR-27b as a negative regulator of ABCA1 expression and cellular cholesterol efflux to ApoA1 in Huh7 cells. In agreement with this, miR-27a/b were recently shown to directly regulate ABCA1 expression and cholesterol efflux in several macrophage and hepatic cell lines20,30, suggesting that miR-27 controls RCT and may be useful as a therapeutic strategy for altering levels of circulating HDL-C. As such, we sought to determine whether acute modulation of miR-27b levels regulated hepatic ABCA1 expression and plasma HDL-C in vivo. Unexpectedly, overexpression of hepatic miR-27b did not alter levels of HDL-C, despite causing a ~50% reduction in ABCA1 protein levels. Similarly, no significant differences in HDL-C were observed when we inhibited endogenous levels of miR-27b in mice fed a WD. miR-27b has previously been shown to decrease endothelial lipase activity by targeting ANGPTL318; therefore, it is plausible that changes in circulating HDL-C are offset by this pathway. Interestingly, Rotllan et al and Marquart et al found that similar increases in hepatic ABCA1 expression following therapeutic silencing of miR-33 in WD-fed Ldlr−/− mice did not alter plasma HDL-C levels either, however the effects on atheroma development were controversial32,33. Nonetheless, miR-27b may still be a viable therapeutic option to increase the capacity of HDL as an acceptor for peripheral cholesterol efflux and thus reduce the burden of atherosclerosis. As such, before disregarding anti-miR-27b treatment as a viable option to treat CVD, future experiments analyzing the effects of miR-27b on HDL functionality are warranted.
Many groups have emphasized the regulatory potential of miR-27b in controlling lipid metabolism. However, they also reflect how difficult it is to determine the biological function of miR-27b, as not all predicted and/or confirmed metabolic targets of miRNAs contribute to a phenotype. Ascertaining the biological function of miRNAs in regulating a physiological process, therefore, is complex and relies on systematic, unbiased experiments in living cells or organisms. Our study unequivocally stresses the importance of manipulating miRNA expression in vivo in order to determine the mechanisms by which a miRNA fine-tunes regulatory pathways and controls whole-body metabolism. Studies using anti-sense oligonucleotides and tissue-specific knockout mice are essential for defining the role of miR-27b as a master regulator of lipid metabolism in the future.
METHODS
Materials
Chemicals were obtained from Sigma-Aldrich unless otherwise noted. The synthetic LXR ligand T0901217 (T090) was purchased from Cayman Chemical. Human ApoA1 was obtained from Meridian Life Sciences. Lipoprotein-deficient serum (LPDS) was prepared from FBS delipidated with 4% fumed silica. 1,1’-Dioctadecyl-3,3,3,3’-tetramethylindocarbocyanineperclorate (DiI) was purchased from Molecular Probes (Invitrogen). A mouse monoclonal antibody against ABCA1 (#ab18180) was purchased from Abcam. A rabbit polyclonal antibody against LDLR (#1007665) was obtained from Cayman Chemical and a mouse monoclonal antibody against HSP90 (#610418) was purchased from BD Bioscience. Secondary fluorescently labeled antibodies were from Molecular Probes (Invitrogen). miRNA mimics and inhibitors were obtained from Dharmacon. For in vivo experiments, miRCURY locked nucleic acid (LNA)™ miRNA inhibitors against mmu-miR-27b or scrambled control were purchased from Exiqon.
Cell culture
Mouse (Hepa) hepatic cells and monkey kidney fibroblast (COS7) cells were obtained from American Type Tissue Collection. The human hepatocellular carcinoma cell line, Huh7, was a kind gift from Dr. Edward Fisher (NYU School of Medicine). Huh7, Hepa and COS7 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) and 2% penicillin-streptomycin in 10 cm2 dishes at 37°C and 5% CO2. For DiI-LDL uptake and binding experiments, Huh7 cells were incubated in DMEM containing 10% LPDS supplemented with 30 µg/ml DiI-LDL cholesterol. For analysis of LDLR protein expression, Huh7 and Hepa cells were plated in DMEM containing 10% LPDS and treated as described below.
miRNA mimic/inhibitor transfections
For mimic and inhibitor transfections, Huh7 and Hepa cells were transfected with 40 nM miR-27b mimics (miR-27b) or with 60 nM miR-27b inhibitors (Inh-27b) (Dharmacon) utilizing RNAimax (Invitrogen). All experimental control samples were treated with an equal concentration of a non-targeting control mimic sequence (CM, Dharmacon) or inhibitor negative control sequence (CI, Dharmacon) for use as controls for non-sequence-specific effects in miRNA experiments. Verification of miR-27b over-expression and inhibition was determined using qRT-PCR, as described below.
RNA isolation and quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. For mRNA quantification, cDNA was synthesized using iScript RT Supermix (Bio-Rad), following the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) analysis was performed in triplicate using iQ SYBR green Supermix (BioRad) on an iCycler Real-Time Detection System (Biorad). The mRNA level was normalized to GAPDH or 18S as a house keeping gene. For miRNA quantification, total RNA was reverse transcribed using the miScript II RT Kit (Qiagen). Primers specific for pre-miR-27b and miR-27b (Qiagen) were used and values normalized to SNORD68 (Qiagen) as a housekeeping gene.
For mouse tissues, total liver RNA from C57BL/6 mice fed a chow or Western diet was isolated using the Bullet Blender Homogenizer (Next Advance) in TRIzol. 1 µg of total RNA was reverse transcribed and gene/miRNA expression assessed as above.
PCR Array Gene Expression Profiling
Total RNA was extracted from Huh7 cells over-expressing either a control mimic (CM) or miR-27b mimic, as described above. Reverse transcription was performed on 1 µg of total RNA, as described above. qRT-PCR analysis of 84 lipid metabolism related genes was performed using the Human Lipoprotein and Cholesterol PCR Array (SABiosciences) as per the manufacture’s instructions. The complete list of genes analyzed is available online at http://www.sabiosciences.com. Data analysis was performed using the manufacturer’s integrated web-based software package for the PCR Array System using ΔΔCt based fold-change calculations. Data is the mean of three independent experiments and is represented by fold-change compared to control mimic (CM) ± SEM.
Western blot analysis
Cells were lysed in ice-cold buffer containing 50 mM Tris-HCl, pH 7.5, 125 mM NaCl, 1% NP-40, 5.3 mM NaF, 1.5 mM NaP, 1 mM orthovanadate and 1 mg/ml of protease inhibitor cocktail (Roche) and 0.25 mg/ml AEBSF (Roche). Cell lysates were rotated at 4 °C for 1 h before the insoluble material was removed by centrifugation at 12,000 × g for 10 min. After normalizing for equal protein concentration, cell lysates were resuspended in SDS sample buffer before separation by SDS-PAGE. Following overnight transfer of the proteins onto nitrocellulose membranes, the membranes were probed with the following antibodies: ABCA1 (1:1000), LDLR (1:500), and HSP90 (1:1000). Protein bands were visualized using the Odyssey Infrared Imaging System (LI-COR Biotechnology). Densitometry analysis of the gels was carried out using ImageJ software from the NIH (http://rsbweb.nih.gov/ij/).
Northern blot analysis
miRNA expression was assessed by Northern blot analysis as previously described34. Briefly, total RNA (5 ug) was separated on a 15% acrylamide TBE 8M urea gel and blotted onto a Hybond N+ nylon filter (Amersham Biosciences). DNA oligonucleotides complementary to mature miR-27b (5’-GCAGAACTTAGCCACTGTGAA-3’) were end-labeled with [a-32P] ATP and T4 polynucleotide kinase (New England Biolabs) to generate high-specific activity probes. Hybridization was carried out according to the ExpressHyb (Clontech) protocol. Following overnight membrane hybridization with specific radiolabeled probes, membranes were washed once for 30 min at 42 °C in 4× SSC/0.5% SDS and subjected to autoradiography. Blots were reprobed for 5s rRNA (5′-CAGGCCCGACCCTGCTTAGCTTCCGAGAGATCAGACGAGAT-3′) to control for equal loading.
LDL receptor activity assays
Human LDL was isolated and labeled with the fluorescent probe DiI as previously reported35. Huh7 cells were transfected in 6-well plates with miRNA mimics and inhibitors in DMEM containing 10% LPDS for 48 h. Then, cells were washed once in 1× PBS and incubated in fresh media containing DiI-LDL (30 µg cholesterol/ml). Non-specific uptake was determined in extra wells containing a 50-fold excess of unlabeled native LDL (nLDL). Cells were incubated for 2 h at 37 °C to allow for DiI-LDL uptake unless otherwise noted. In other instances, cells were incubated for 90 min at 4 °C to assess DiI-LDL binding. At the end of the incubation period, cells were washed, resuspended in 1 ml of PBS and analyzed by flow cytometry (FACScalibur, Becton Dickinson), as previously described36. The results are expressed in terms of specific median intensity of fluorescence (M.I.F.) after subtracting autofluorescence of cells incubated in the absence of DiI-LDL.
3’UTR luciferase reporter assays
cDNA fragments corresponding to the entire 3’UTR of human LDLR, ABCA1 and LDLRAP1 were amplified by RT-PCR from total RNA extracted from HepG2 cells with XhoI and NotI linkers. The PCR product was directionally cloned downstream of the Renilla luciferase open reading frame of the psiCHECK2™ vector (Promega) that also contains a constitutively expressed firefly luciferase gene, which is used to normalize transfections. Point mutations in the seed region of the predicted miR-27b binding sites within the 3’UTR of LDLR, ABCA1 and LDLRAP1 were generated using the Multisite-Quickchange Kit (Stratagene), according to the manufacturer’s protocol. All constructs were confirmed by sequencing. COS7 cells were plated into 12-well plates and co-transfected with 1 µg of the indicated 3’UTR luciferase reporter vectors and 40 nM of a miR-27b mimic or control mimic (CM) (Dharmacon) utilizing Lipofectamine 2000 (Invitrogen), as previously described17. Luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Renilla luciferase activity was normalized to the corresponding firefly luciferase activity and plotted as a percentage of the control (cells co-transfected with the corresponding concentration of control mimic). Experiments were performed in triplicate wells of a 12-well plate and repeated at least three times.
Cholesterol efflux assays
Cholesterol efflux assays were performed as previously described37. Briefly Huh7 cells were seeded at a density of 2 × 105 cells per well and transfected with either a control mimic (CM), miR-27b mimic, control inhibitor (CI), or miR-27b inhibitor (Inh-27b). Following 48h of transfection, cells were loaded with 0.5 µCi/ml 3H-cholesterol for 24 h. 12 h after loading, cells were incubated with 3 µM T090 to increase the expression of ABCA1. Then, cells were washed twice with PBS and incubated in DMEM supplemented with 2 mg/ml fatty-acid free BSA (FAFA-media) in the presence of an ACAT inhibitor (2 µmol/L) for 4 h prior to the addition of 50 µg/ml human ApoA1 in FAFA-media with or without the indicated treatments. Supernatants were collected after 6h and expressed as a percentage of total cell 3H-cholesterol content (total effluxed 3H-cholesterol+cell-associated 3H-cholesterol).
Cellular cholesterol measurements
Huh7 cells were seeded at a density of 5 × 105 cells/well and transfected with either a control mimic (CM) or miR-27b mimic or a control inhibitor (CI) or miR-27b inhibitor (Inh-27b). Following 48 h transfection, cells were incubated with 30 µg/ml nLDL for 2 h. Intracellular cholesterol content was measured using the Amplex Red Cholesterol Assay Kit (Molecular Probes, Invitrogen), according to the manufacturer’s instructions.
AAV8 pre-miR-27b vector
mmu-pre-miR-27b (accession MI000142) was subcloned into an AAV8 vector and its expression was regulated under the control of a liver-specific thyroxine-binding globulin (TBG) promoter. The AAV8 particles (AAV8.TBG.PI.mir27b.rBG) were generated at the University of Pennsylvania’s Penn Vector Core. An empty AAV8 (AAV8.TBG.PI.Null.bGH), also provided by the Penn Vector Core, was used as a control in all experiments.
Mouse Studies
Eight-week-old male C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA) and kept under constant temperature and humidity in a 12 h controlled dark/light cycle. For high-fat diet (HFD) studies, eight-week old male mice (n = 5 per group) were placed on a chow diet or HFD containing 0.3% cholesterol and 21% (wt/wt) fat (Dyets, Inc) for three weeks. Liver samples were collected as previously described17 and stored at −80°C until total RNA was harvested for miRNA expression analysis.
For miR-27b overexpression studies, C57BL/6 mice were randomized into 2 groups: non-targeting AAV8 (AAV-Null, n=10) and pre-miR-27b AAV8 (AAV- 27b, n = 10 per group). Mice fed a chow diet were treated once with 5 × 1012 GC/kg AAV Null or 5 × 1012 GC/kg in PBS by retro-orbital injection. Blood samples were collected at baseline (week 0) and 2 weeks after treatment for lipid analysis and lipoprotein profile measurements (see below). Then mice were sacrificed, and hepatic gene expression and liver histology were analyzed (see above). All animal experiments were approved by the Institutional Animal Care Use Committee of New York University Medical Center.
For miR-27b inhibition experiments, eight-week old male C57BL/6 mice fed a chow diet were randomized into 2 groups: LNA control (n = 8) and LNA anti-miR-27b (n = 8) and fed a Western diet for one month [(WD) 9.5% casein, 0.3% DL-Methionine, 15% cornstarch, 40% sucrose, 5% cellulose, 21% anhydrous milk fat, 3.5% mineral mix, 1% vitamin mix, 0.4% calcium carbonate and 0.3% cholesterol]. Mice received i.p. injections of 5 mg/kg LNA control (5’-ACGTGCTATACGCCCA-3’) or LNA anti-miR-27b (5’-AACTTAGCCACTGTGA-3’) oligonucleotides every two days for a total of two weeks. Twenty-four hours after the final injection, mice were sacrificed and hepatic gene expression analyzed (see above). All animal experiments were approved by the Institutional Animal Care Use Committee of New York University Medical Center. Following sacrifice, liver miRNA and gene expression were analyzed as above.
Plasma Lipid Analysis and Lipoprotein Profile Measurements
Mice were fasted for 12–14 h before blood samples were collected by retro-orbital venous plexus puncture. Plasma was separated by centrifugation and stored at −80 °C. Total plasma cholesterol and HDL-cholesterol were enzymatically measured using the Amplex Red Cholesterol Assay Kit (Molecular Probes, Invitrogen), according to the manufacture’s instructions. Total triglycerides were measured with the Wako Diagnostics Triglycerides Reagent. The lipid distribution in plasma lipoprotein fractions was assessed by fast-performance liquid chromatography (FPLC) gel filtration with 2 Superose 6 HR 10/30 columns (Pharmacia) as previously described17. Cholesterol in each fraction was enzymatically measured using the Amplex Red Cholesterol Assay Kit (Molecular Probes, Invitrogen).
Hepatic lipid analysis
Total lipid extracts from livers (50 mg) from AAV Null, AAV miR-27b, LNA control or LNA anti-miR-27b mice were obtained using a modified Folch method. The different lipid classes (triglycerides, phosphatidyl choline, free fatty acids, and total cholesterol) were quantified from chloroform extracts using enzymatic kits, as previously described38.
Non-human Primate Studies
Male rhesus monkeys (Macaca mulatta), 7–13 years old, were fed a standard (TestDiet #5038; Purina Mills) or high fat/high sucrose diet (42% kcal in fat, Custom formula #07802; Harlan, Teklad, Madison, WI) for two years (n = 5 per group) and maintained as previously described39. Animal procedures were approved by the Animal Care and Use Committee of the NIA Intramural Research Program. Following sacrifice, liver RNA was isolated using the Bullet Blender Homogenizer (Next Advance) in TRIzol. For mRNA quantification, 1 µg of total RNA was reverse transcribed using iScript RT Supermix (BioRad) and iQ SYBR green Supermix (BioRad). Quantification of miR-27b was assessed as described above.
Statistics
Animal sample size for each study was chosen based on literature documentation of similar well-characterized experiments17,40,39,41. The number of animals used in each study is listed in the figure legends and main text. No inclusion/exclusion criteria were used and studies were not blinded to investigators, unless otherwise noted. All data are expressed as mean ±SEM. Statistical differences were measured using an unpaired two-sided Student’s t test or one-way or two-way ANOVA with Bonferroni correction for multiple comparisons. A non-parametric test (Mann-Whitney) was used when data did not pass the normality test. A value of P ≤ 0.05 was considered statistically significant. Data analysis was performed using GraphPad Prism Software Version 5.0a (GraphPad, San Diego, CA).
Supplementary Material
HIGHLIGHTS.
-
►
miR-27b directly targets LDLR and LDLRAP1
-
►
miR-27b regulates LDLR expression and activity in human and mouse hepatic cells
-
►
miR-27b controls ABCA1 expression and cellular cholesterol efflux in Huh7 cells
-
►
Modulation of miR-27b levels in mice affects hepatic LDLR and ABCA1 expression
-
►
miR-27b does not regulate circulating and hepatic lipids in mice
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (R01HL107953, R01HL107953-04S1, and R01HL106063 to CF-H; R01HL105945 to YS; 1F31AG043318 to LG; and R01HL107794 to AB), the American Heart Association (15SDG23000025 to CMR and GRNT20460189 to AB), the Howard Hughes Medical Institute International Student Research Fellowship (to EA), and the Foundation Leducq Transatlantic Network of Excellence in Cardiovascular Research (to CFH).
COMPETING FINANCIAL INTERESTS
CFH and LG have a patent (PCT/US2014/042196) on the use of miR-27b inhibitors to treat dyslipidemias and cardiovascular disease.
Abbreviations
- ABCA1
ATP binding cassette transporter A1
- ABCG1
ATP binding cassette transporter G1
- CVD
cardiovascular disease
- HDL
high-density lipoprotein
- HMGCR
3-hydroxy-3methylglutaryl coenzyme A reductase
- LDL
low-density lipoprotein
- LDL-C
LDL-cholesterol
- LDLR
LDL receptor
- LDLRAP1
LDLR adapter protein 1
- LNA
locked nucleic acid
- LXR
liver X receptor
- miRNA
microRNA
- RXR
retinoid X receptor
- SREBP
sterol regulatory element-binding protein
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 3.Gould AL, Rossouw JE, Santanello NC, Heyse JF, Furberg CD. Cholesterol reduction yields clinical benefit: impact of statin trials. Circulation. 1998;97:946–952. doi: 10.1161/01.cir.97.10.946. [DOI] [PubMed] [Google Scholar]
- 4.Sjouke B, Kusters DM, Kastelein JJ, Hovingh GK. Familial hypercholesterolemia: present and future management. Current cardiology reports. 2011;13:527–536. doi: 10.1007/s11886-011-0219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hennekens CH. Increasing burden of cardiovascular disease: current knowledge and future directions for research on risk factors. Circulation. 1998;97:1095–1102. doi: 10.1161/01.cir.97.11.1095. [DOI] [PubMed] [Google Scholar]
- 6.Brown MS, Goldstein JL. Receptor-mediated control of cholesterol metabolism. Science. 1976;191:150–154. doi: 10.1126/science.174194. [DOI] [PubMed] [Google Scholar]
- 7.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 8.Brown MS, Goldstein JL. Familial hypercholesterolemia: defective binding of lipoproteins to cultured fibroblasts associated with impaired regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:788–792. doi: 10.1073/pnas.71.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 10.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 12.Beaven SW, Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med. 2006;57:313–329. doi: 10.1146/annurev.med.57.121304.131428. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 16.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vickers KC, et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen WJ, Yin K, Zhao GJ, Fu YC, Tang CK. The magic and mystery of microRNA-27 in atherosclerosis. Atherosclerosis. 2012;222:314–323. doi: 10.1016/j.atherosclerosis.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Zhang M, et al. MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis. 2014;234:54–64. doi: 10.1016/j.atherosclerosis.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Li T, et al. Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin Chim Acta. 2011;412:66–70. doi: 10.1016/j.cca.2010.09.029. [DOI] [PubMed] [Google Scholar]
- 22.Staszel T, et al. Role of microRNAs in endothelial cell pathophysiology. Pol Arch Med Wewn. 2011;121:361–366. [PubMed] [Google Scholar]
- 23.Oram JF, Vaughan AM. ABCA1-mediated transport of cellular cholesterol and phospholipids to HDL apolipoproteins. Curr Opin Lipidol. 2000;11:253–260. doi: 10.1097/00041433-200006000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, et al. Systematic evaluation of AAV vectors for liver directed gene transfer in murine models. Mol Ther. 2010;18:118–125. doi: 10.1038/mt.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kassim SH, et al. Adeno-associated virus serotype 8 gene therapy leads to significant lowering of plasma cholesterol levels in humanized mouse models of homozygous and heterozygous familial hypercholesterolemia. Hum Gene Ther. 2013;24:19–26. doi: 10.1089/hum.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietschy JM, Turley SD, Spady DK. Role of liver in the maintenance of cholesterol and low density lipoprotein homeostasis in different animal species, including humans. J Lipid Res. 1993;34:1637–1659. [PubMed] [Google Scholar]
- 27.Kim SY, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun. 2010;392:323–328. doi: 10.1016/j.bbrc.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Karbiener M, et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun. 2009;390:247–251. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 29.Kida K, et al. PPARalpha is regulated by miR-21 and miR-27b in human liver. Pharm Res. 2011;28:2467–2476. doi: 10.1007/s11095-011-0473-y. [DOI] [PubMed] [Google Scholar]
- 30.Shirasaki T, et al. MicroRNA-27a Regulates Lipid Metabolism and Inhibits Hepatitis C Virus Replication in Human Hepatoma Cells. J Virol. 2013;87:5270–5286. doi: 10.1128/JVI.03022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Baro MR, Lewin TM, Coleman RA. Regulation of Triglyceride Metabolism. II. Function of mitochondrial GPAT1 in the regulation of triacylglycerol biosynthesis and insulin action. Am J Physiol Gastrointest Liver Physiol. 2007;292 doi: 10.1152/ajpgi.00553.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C. Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr−/− mice--brief report. Arterioscler Thromb Vasc Biol. 2013;33:1973–1977. doi: 10.1161/ATVBAHA.113.301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marquart TJ, Wu J, Lusis AJ, Baldan A. Anti-miR-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2013;33:455–458. doi: 10.1161/ATVBAHA.112.300639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 35.Calvo D, Gomez-Coronado D, Suarez Y, Lasuncion MA, Vega MA. Human CD36 is a high affinity receptor for the native lipoproteins HDL, LDL, and VLDL. J Lipid Res. 1998;39:777–788. [PubMed] [Google Scholar]
- 36.Suarez Y, et al. Synergistic upregulation of low-density lipoprotein receptor activity by tamoxifen and lovastatin. Cardiovascular research. 2004;64:346–355. doi: 10.1016/j.cardiores.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 37.Ramirez CM, et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen RM, Marquart TJ, Jesse JJ, Baldan A. Control of very low-density lipoprotein secretion by N-ethylmaleimide-sensitive factor and miR-33. Circ Res. 2014;115 doi: 10.1161/CIRCRESAHA.115.303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattison JA, et al. Resveratrol prevents high fat/sucrose diet-induced central arterial wall inflammation and stiffening in nonhuman primates. Cell Metab. 2014;20:183–190. doi: 10.1016/j.cmet.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 41.Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci U S A. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.