Abstract
A reproducible and quantitative imaging biomarker is needed to standardize the evaluation of changes in bone scans of prostate cancer patients with skeletal metastasis. We performed a series of analytic validation studies to evaluate the performance of the automated bone scan index (BSI) as an imaging biomarker in patients with metastatic prostate cancer.
Methods
Three separate analytic studies were performed to evaluate the accuracy, precision, and reproducibility of the automated BSI. Simulation study: bone scan simulations with predefined tumor burdens were created to assess accuracy and precision. Fifty bone scans were simulated with a tumor burden ranging from low to high disease confluence (0.10–13.0 BSI). A second group of 50 scans was divided into 5 subgroups, each containing 10 simulated bone scans, corresponding to BSI values of 0.5, 1.0, 3.0, 5.0, and 10.0. Repeat bone scan study: to assess the reproducibility in a routine clinical setting, 2 repeat bone scans were obtained from metastatic prostate cancer patients after a single 600-MBq 99mTc-methylene diphosphonate injection. Follow-up bone scan study: 2 follow-up bone scans of metastatic prostate cancer patients were analyzed to determine the interobserver variability between the automated BSIs and the visual interpretations in assessing changes. The automated BSI was generated using the upgraded EXINI boneBSI software (version 2). The results were evaluated using linear regression, Pearson correlation, Cohen κ measurement, coefficient of variation, and SD.
Results
Linearity of the automated BSI interpretations in the range of 0.10–13.0 was confirmed, and Pearson correlation was observed at 0.995 (n = 50; 95% confidence interval, 0.99–0.99; P < 0.0001). The mean coefficient of variation was less than 20%. The mean BSI difference between the 2 repeat bone scans of 35 patients was 0.05 (SD = 0.15), with an upper confidence limit of 0.30. The interobserver agreement in the automated BSI interpretations was more consistent (κ = 0.96, P < 0.0001) than the qualitative visual assessment of the changes (κ = 0.70, P < 0.0001) was in the bone scans of 173 patients.
Conclusion
The automated BSI provides a consistent imaging biomarker capable of standardizing quantitative changes in the bone scans of patients with metastatic prostate cancer.
Keywords: bone scan index (BSI), imaging biomarker, bone scan, metastatic prostate cancer, analytical validation
Prostate cancer is a bone-tropic cancer, and nearly 85% of patients with fatal prostate cancer are reported to have bone metastases (1). In clinical practice, a bone scan is the most prevalent and cost-effective diagnostic imaging tool to detect the onset of skeletal metastasis in advanced prostate cancer patients (2). However, the clinical utility of on-treatment changes in the bone scans of advanced metastatic prostate patients remains limited, largely because of the lack of a consistent methodology for quantifying changes in bone scans.
The bone scan index (BSI), developed at the Memorial Sloan Kettering Cancer Center, is a fully quantitative analysis of skeletal metastasis in bone scans (3). The BSI represents the tumor burden in a bone scan as a percentage of the total skeletal mass and has shown clinical significance as a prognostic imaging biomarker (4,5). The labor-intensive process of manually calculating the BSI has prevented its widespread adoption in clinical practice.
The image-analysis program developed by EXINI Diagnostics for bone scan imaging automates the BSI calculation (6–8). The scan normalization and the iterative artificial neural network detect hotspots that are suspected metastatic lesions and generate BSI in a significantly shortened time span (<10 s). The automated BSI was shown to correlate with the manual BSI of newly diagnosed prostate cancer patients, and it was independently associated with overall survival (9).
However, the clinical qualification of the automated BSI as an imaging biomarker indicative of the treatment response depends on its analytic performance characteristics, which are yet to be validated. The analytic validation of the automated BSI against a known analytic standard and against the preanalytic variability of the routine bone scan procedure is essential to assess the automated BSI as a standardized quantitative platform for prospective clinical studies. We have performed analytic studies that incorporate computer simulations and clinical patients to evaluate the accuracy, precision, and reproducibility of the automated BSI. We hypothesized that with minimal manual supervision, the automated BSI could standardize the quantitative changes in bone scans of patients with metastatic prostate cancer.
MATERIALS AND METHODS
Study Design
Three analytic studies were performed to evaluate the performance characteristics of the automated BSI. The predefined objectives and endpoint analysis for each of the studies are summarized in Table 1. The overall aim of the studies was to test the hypothesis that the automated BSI can standardize quantitative changes in bone scans. Ethical permission and individual patient consent were obtained.
TABLE 1.
Summary of Analytic Studies to Evaluate Performance Characteristics of Automated BSI as a Consistent Imaging Biomarker to Standardize Quantitative Analysis of Bone Scans
| Analytic study | Objective | Design | Endpoint |
|---|---|---|---|
| Simulation study | Accuracy and precision | Simulation of bone scans with known phantom BSI as analytic standard | Measuring automated BSI against phantom BSI |
| Repeat bone scan study | Reproducibility | Metastatic patients with repeat bone scans | Measuring difference between 2 automated BSI interpretations |
| Follow-up bone scan study | Interobserver variability | Metastatic patients with 2 routine follow-up clinical bone scans | Measuring observer agreement in assessing automated BSI change |
Simulation Study
The objective of the simulation study was to assess the accuracy and precision of the automated BSI against the known tumor burdens of the simulated bone scans.
In the simulation study, 2 sets of 50 bone scans were simulated with known tumor burdens and a corresponding known phantom BSI. The localization of the simulated tumor was randomized. The first set of 50 simulated bone scans was created with focal tumor lesions ranging from low to high disease confluence (0.10–13.0 phantom BSI). Another set of 50 scans was divided into 5 subgroups, each containing 10 simulated bone scans, corresponding to phantom BSI values of 0.5, 1.0, 3.0, 5.0, and 10.0.
Bone Scan Simulation
The SIMIND Monte Carlo (10) program together with the XCAT phantom (11), representing a standard male, were used to simulate bone scans with predefined tumor burdens in the skeleton. Randomly distributed focal lesions were inserted in the XCAT phantom skeleton by a MATLAB script corresponding to a predefined BSI. The tumors were confined to the skeleton volume. The phantom BSI was calculated as described in the original study (3). A restriction was set so that no tumor was placed below the mid femur or below the mid humerus. A virtual scintillation camera with a 9-mm-thick crystal, 9.5% energy resolution at 140 keV, a 256 × 1,024 image matrix with a 2.4-mm pixel size, and a low-energy, high-resolution collimator was used for the simulations. To mimic real measurements, the simulations were performed with sufficient histories to avoid Monte Carlo noise. Poisson noise was added after the simulations, corresponding to measurements with a total of 1.5 million counts in the anterior image. Anterior and posterior whole-body images were simulated for every phantom. The relative activity concentration in the bone, kidneys, bone marrow, and tumors was set to 18, 9, 2.5, and 72, respectively, in relation to the remainder of the body.
Repeat Bone Scan Study
The objective of the repeat bone scan study was to determine the reproducibility threshold of the automated BSI in relation to the preanalytic variability of the routine clinical bone scan procedure.
In this study, a repeat whole-body bone scan was obtained from metastatic patients who were referred for a routine bone-scan procedure. A previous bone scan was used to determine the presence of skeletal metastasis. The first whole-body bone scan was obtained 3 h after a single intravenous injection of 600 MBq of 99mTc methylene diphosphonate. The repeat bone scan was obtained directly after the first.
Patient Bone Scan
Whole-body images with anterior and posterior views (scan speed, 10 cm/min; 256 × 1,024 matrix), were obtained using a γ-camera equipped with low-energy, high-resolution, parallel-hole collimators (Maxxus; GE Healthcare). Energy discrimination was provided by a 15% window centered on the 140-keV peak of 99mTc.
Follow-up Bone Scan Study
The objective of the follow-up bone scan study was to compare the interobserver agreement of the visual interpretations with that of the automated BSI interpretations in assessing changes between the 2 follow-up bone scans.
In the follow-up bone scan study, all prostate cancer patients with skeletal metastasis who underwent at least 2 whole-body examinations between January 2002 and December 2008 as part of their clinical routine follow-up were considered for inclusion. Patients with digitally stored images were included, and if a selected patient had more than 2 scans, the last 2 follow-up scans were used for the study.
The 2 follow-up bone scans for each patient were independently analyzed by 3 experienced nuclear medicine bone scan interpreters at 3 different interpretation sessions. At least 3 mo elapsed between each session. In the first session, each interpreter independently classified the patients according to the signs of progressive metastatic disease. Patients with a follow-up scan showing new lesions or lesions present in the first scan that were distinctly larger were classified as having signs of progression. In the second session, the 3 interpreters independently classified all the patients on the basis of the presence of 2 or more new lesions. In the third session, the 3 interpreters used EXINI boneBSI software (EXINI Diagnostics) to report changes in the automated BSI values between the 2 follow-up bone scans. The changes in the automated BSI were defined as an increase or decrease in the automated BSI equal to or greater than the reproducibility threshold obtained in the repeat bone scan study.
Automated BSI Analysis
The upgraded EXINI boneBSI software (version 2) was used to generate the automated BSI. The methodology of the automated platform has been described in detail in a previous study (9). In summary, the different anatomic regions of the skeleton are segmented, and the abnormal hotspots are detected and classified as metastatic lesions. The weight fraction of the skeleton for each metastatic hotspot is calculated, and the BSI is calculated as the sum of all such fractions.
Statistical Analysis
In the simulation study, automated BSI values were obtained for all the simulated bone scans and compared with the known standard, the phantom BSI, to assess analytic accuracy and precision. A linear regression model and Pearson correlation testing were used to evaluate the accuracy of the automated BSI values compared with the phantom BSIs for the first set of 50 simulated bone scans. To confirm the method linearity, we tested the assumptions regarding residuals normality (Shapiro–Wilk test) and the method’s homoscedasticity. Once the linear model was confirmed, the linear regression parameters were estimated, including a correlation coefficient (r), at a 95% confidence interval. In the second set of 50 simulated bone scans, the coefficient of variation and SD were used to determine the precision of the automated BSI at 5 different levels of the phantom BSI.
In the repeat bone scan study, the automated BSI was measured from both bone scans. The difference between the 2 automated BSI interpretations was calculated to compare the reproducibility threshold with the preanalytical variability of the routine clinical bone scan. The mean and SD of the difference between the automated BSI interpretations were used to calculate the upper confidence limit, or the 95th percentile value. This value represented the automated BSI reproducibility threshold for assessing consistent measurements of the changes in bone scans.
In the follow-up bone scan study, the interobserver agreement of the visual subjective interpretation versus the automated BSI was measured by Cohen κ agreement, which measures agreement beyond that expected by chance. κ agreement was evaluated pairwise among the 3 interpreters—A versus B, B versus C, and A versus C—for each of the interpretation sessions. The mean κ agreement of the 3 paired interpreters was then compared for each session.
No prior assumptions were made for the automated BSI performance to render power calculations. Statistical significance for each statistical test was set at 0.05. All statistical analyses were performed using SPSS, version 22 (IBM), for Windows (Microsoft).
RESULTS
Simulation Study
The automated BSI, the dependent variable, was obtained from both sets of simulated bone scans and measured against the known phantom BSI, which was considered the independent variable. In the first set of 50 simulated bone scans, the Shapiro–Wilk test confirmed that the residuals of the dependent variable were normally distributed (P = 0.850). In addition, the mean residual value of 0.00 with an SD of 0.25 confirmed homoscedasticity, showing constant variation across all values of the independent variable. Given that the residuals exhibited normality and homoscedasticity, the model was considered linear. The scatterplot with a linear fit line and the associated parameters for the linear regression in the range from 0.10 to 13.0 BSI are presented in Figure 1 and in Table 2, respectively. Pearson correlation was observed to be 0.995 (95% confidence interval, 0.99–0.99; P < 0.0001). Table 3 provides the coefficient of variation and SD of the automated BSI values at each of the predefined tumor burdens with varying localization for the second set of 50 simulated bone scans. The coefficient of variation at each of the 5 predefined phantom BSIs was less than 20%.
FIGURE 1.
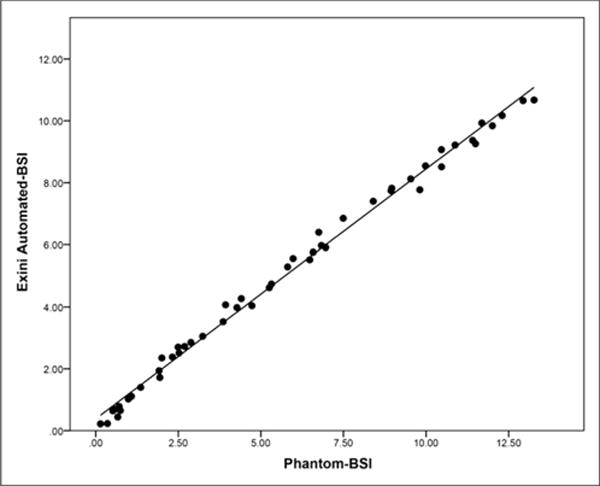
Scatterplot showing linearity of automated BSI vs. phantom BSI in first set of 50 simulated bone scans.
TABLE 2.
Parameters for Linear Regression Model in First Set of 50 Phantoms with Predefined BSI Range of 0.10–13.0
| Linearity measure | Value | 95% CI | P |
|---|---|---|---|
| r | 0.99 | 0.99–0.99 | <0.0001 |
| Slope | 0.80 | 0.78–0.83 | <0.0001 |
| Intercept | 0.38 | 0.25–0.51 | <0.0001 |
TABLE 3.
SD and Coefficient of Variation for Automated BSI in Second Set of 50 Phantoms at 5 Predefined Levels of Tumor Burden
| Parameter | Phantom BSI | ||||
|---|---|---|---|---|---|
| 0.5 (n = 10) | 1.0 (n = 10) | 3.0 (n = 10) | 5.0 (n = 10) | 10.0 (n = 10) | |
| SD | 0.10 | 0.13 | 0.26 | 0.17 | 0.29 |
| Coefficient of variation | 0.19 | 0.11 | 0.08 | 0.06 | 0.03 |
Repeat Bone Scan Study
Thirty-five patients provided consent and were enrolled in the repeat bone scan study. All bone scans were eligible for automated BSI analysis. The Bland–Altman plot of the differences between the automated BSI readings of the 35 repeat bone scans is illustrated in Figure 2. The mean BSI difference between the 2 repeat bone scans was 0.05 with an SD of 0.15. The reproducibility threshold of the automated BSI for consistent measurement of changes in bone scans, defined as the 95th percentile of the data, was observed at 0.30.
FIGURE 2.
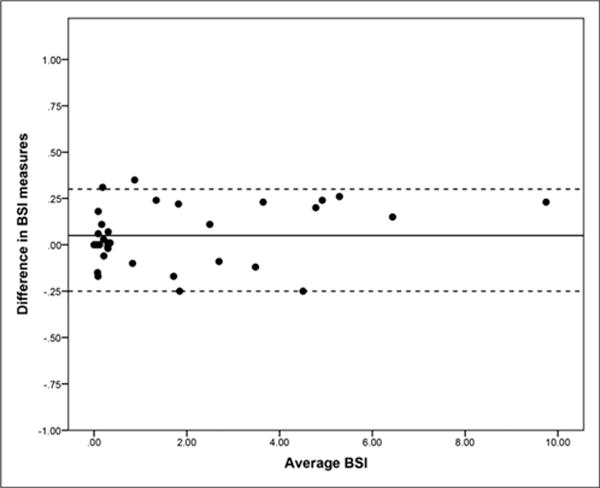
Bland–Altman plot to evaluate reproducibility of automated BSI interpretations from repeat bone scans of 35 metastatic patients. Mean BSI difference, 0.05 (solid horizontal line), with upper confidence limit of 0.30 and lower confidence limit of −0.25 (horizontal dotted lines).
Follow-up Bone Scan Study
The bone scans of 173 metastatic prostate cancer patients were eligible for the study. The 2 consecutive bone scans from all 173 patients were independently analyzed by 3 experienced nuclear medicine interpreters for all 3 interpretation sessions. The pairwise κ agreement at each session is demonstrated in Table 4. In sessions 1 and 2, the mean κ agreement was 0.70 and 0.66 (P < 0.0001), respectively. In session 3, the mean change in the automated BSI interpretations between the 2 follow-up bone scans of 173 patients was 1.84 (median, 0.42; interquartile range, −2.4 to 10.26). The change in the automated BSI was defined as the increase or decrease beyond the reproducibility threshold of 0.30 BSI. At 0.96 (P < 0.0001), the mean κ agreement of the 3 interpreters using automated BSIs was higher than it was for the first 2 sessions.
TABLE 4.
Pairwise Cohen κ Agreement Evaluating Interobserver Agreement Among 3 Independent Interpreters to Assess Changes in Bone Scans from 173 Patients with Metastatic Prostate Cancer
| Finding | Interpreter A vs. B | Interpreter A vs. C | Interpreter B vs. C |
|---|---|---|---|
| Increased burden | 0.56 | 0.90 | 0.65 |
| Two new lesions | 0.62 | 0.81 | 0.55 |
| Change in BSI | 0.96 | 0.97 | 0.96 |
DISCUSSION
Currently, there is no quantitative imaging biomarker in patients with metastatic prostate cancer, and the on-treatment changes in bone scan results are inadequately assessed in an interpreter-dependent visual analysis. The inherent variability of these kinds of assessments affects the clinical association of the intended biomarker with clinical endpoints. The Food and Drug Administration biomarker qualification review program explicitly states that the clinical validation of a biomarker is empirically significant only if the marker is measured consistently and reproducibly (12).
However, during the course of this study we found that, unlike those for blood-based biomarkers, the guidelines for analytically validating biomarkers in image diagnostics are not well defined. One of the most serious limitations in the effort to analytically validate an imaging biomarker has been the difficulty in procuring a true analytic standard: histologic confirmation of abnormal hotspots as metastatic lesions. Previous studies have circumvented the issue of the lack of an analytic standard by using a reference interpreter for the bone scan analysis (13). Using a manual assessment as a true analytic benchmark limits the analytic validation to the skills of the reference individual. To be independent of such limitations, the analytic standard has to represent the known burden of disease against which the true performance of the biomarker can be evaluated.
In the absence of a true analytic standard, we believe that the simulated bone scans from XCAT phantoms with predefined tumor burdens confined to the volume of the skeleton and the SIMIND Monte Carlo simulation of the γ-camera could be an alternative. The simulated bone scans with randomized tumor locations but known tumor burden can create real-patient scenarios to evaluate the performance of the automated BSI against a true analytic standard, which is the known ground truth.
The results of the simulation study demonstrated that, irrespective of varying tumor localization, the accuracy and precision of the automated BSI were maintained from low to high disease confluence of focal lesions, in the BSI range from 0.10 to 13.0. This result marks an improvement in the performance of EXINI’s current version of the automated BSI over that of the previous version. The first generation of the EXINI automated BSI platform was reported to underestimate the BSI values in patients with a high disease burden (9). The consistent linearity of the upgraded version of the automated BSI is vital to its intended clinical utility in patients with advanced metastatic prostate cancer. The changes in the automated BSI would be reproducible, with consistent accuracy and precision, and its association with clinical endpoints would be reliable.
In our simulated study, we also noted that the automated-BSI platform has a blind spot for lesions superimposed on the bladder. To avoid classification of tracer activity from the urinary bladder, the lesions simulated in the lower sacrum, coccyx, and pubic regions were sporadically not classified as metastatic lesions. An example is illustrated in Figure 3. The resulting automated BSI values for such simulations were slightly lower than the true phantom BSIs. Given the technical limitation of the platform, manual supervision should be considered when the bone scans of patients with hotspots in the lower sacrum, coccyx, or pubic region are being analyzed.
FIGURE 3.
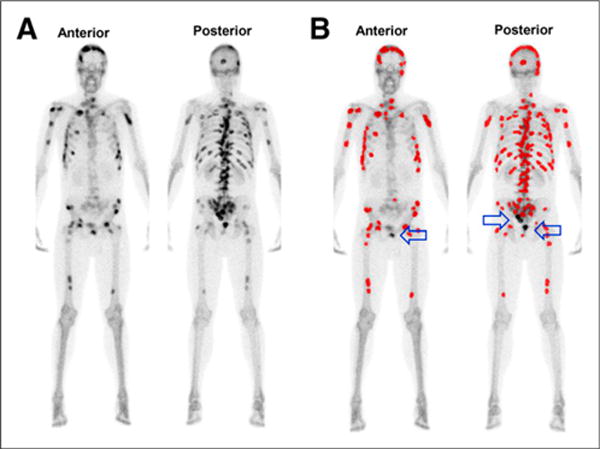
(A) Simulated bone scan with known tumor burden corresponding to phantom BSI of 10.0. (B) Lesions detected and classified as metastatic by automated platform for BSI calculation are highlighted in red; arrows indicate simulated lesions at blind spot of automated BSI platform.
Although providing a true analytic standard, simulated bone scans are limited in their representation of the variables associated with patient bone scans. The simulated bone scans represent a single individual, rendering the same activity concentration and varying only by tumor burden and tumor localization. Therefore, unlike the simulation study, the purpose of the repeat bone scan study in patients with skeletal metastasis was to evaluate the reproducibility of the automated BSI in the clinical environment, which is predisposed to the variables associated with the routine bone scan procedure. Variables such as image count corresponding to scanning time and interpatient-dependent attenuation factors can cause noise in BSI interpretations, and this noise can affect the reliability of the on-treatment BSI change as a quantitative biomarker. In previous studies, the on-treatment BSI change has been reported as the percentage of BSI change, BSI doubling time, and BSI difference (5,14,15), but none of the studies have accounted for the noise in BSIs that results from the inherent variability of the bone scan procedure.
Although limited in scale, our study attempts to empirically address the noise in the automated BSI associated with the preanalytic variability of the routine bone scan procedure. We propose that accounting for the reproducibility threshold of 0.30 will result in a consistent and reliable on-treatment BSI change that is clinically relevant. The preliminary data presented here warrant further validation of this threshold and its clinical implications.
In the follow-up bone scan study, the interpretation sessions that incorporated visual assessment of the changes in bone scans reported higher discrepancies among the 3 interpreters. The 3 interpreters in our study were from the same hospital, and their interpretation styles for bone scan assessment were probably more consistent than the styles of interpreters from different centers would be. Prior studies have reported similar discordance in visual assessment of bone scans. In a Swedish study, 37 interpreters from 18 hospitals showed considerable disagreement in bone scan assessment. The κ agreement among the interpreters ranged from 0.16 to 0.82, with a mean of 0.48 (8).
Compared with visual assessment of changes in bone scans, the interobserver agreement among the 3 nuclear medicine interpreters increased significantly using the automated BSI platform. The manual supervision of the automated BSI assessment in bone scans with abnormal hotspots located at the lower sacrum, coccyx, or pubic region were the cause of some ambiguities among the interpreters.
Furthermore, the automated BSI represented changes in bone scans as a continuous numeric variable. The gradient of this change could potentially have clinical utility in assessing both response and progression. Not only can a confirmed sequential increase in the automated BSI value augment the Prostate Cancer Working Group 2 recommendation of tracking 2 or more new lesions as an endpoint for radiographic progression, but the confirmed sequential decline in the treatment follow-up BSI value can also be evaluated as a biomarker indicative of efficacy response.
Despite the advantages of a fully quantitative imaging biomarker, the automated BSI assessment does not absolve the inherent limitations of bone scanning as a non–tumor-specific imaging modality. Uptake of 99mTc in a bone scan reflects the increase in osteoblastic activity in bone. As a result, the bone scan can show a temporary increase in activity in response to an effective treatment. Future clinical investigations are warranted to develop BSI guidelines, similar to those incorporated in Prostate Cancer Working Group 2 for bone scan progression, to determine the on-treatment BSI change that is clinically relevant.
CONCLUSION
We have demonstrated that with minimal manual supervision the automated BSI overcomes the limitations of qualitative visual assessment and provides an accurate, precise, and reproducible platform for standardizing quantitative changes in the bone scans of prostate cancer patients with skeletal metastasis. This study is the foundation for subsequent clinical investigations aimed at validating the clinical utility of changes in the automated BSI as a consistent, quantitative imaging biomarker indicative of treatment response.
Acknowledgments
This study was funded by grants from the Swedish Cancer Foundation, the Swedish Medical Research Council, the Gunnar Nilsson Cancer Foundation, Skåne University Hospital Cancer Foundation, and the ALF.
Footnotes
DISCLOSURE
No other conflict of interest relevant to this article was reported.
References
- 1.Jacobs SC. Spread of prostatic cancer to bone. Urology. 1983;21:337–344. doi: 10.1016/0090-4295(83)90147-4. [DOI] [PubMed] [Google Scholar]
- 2.Heidenreich A, Bastian PJ, Bellmunt J, et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent—update 2013. Eur Urol. 2014;65:124–137. doi: 10.1016/j.eururo.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 3.Imbriaco M, Larson SM, Yeung HW, et al. A new parameter for measuring metastatic bone involvement by prostate cancer: the bone scan index. Clin Cancer Res. 1998;4:1765–1772. [PubMed] [Google Scholar]
- 4.Sabbatini P, Larson SM, Kremer A, et al. Prognostic significance of extent of disease in bone in patients with androgen-independent prostate cancer. J Clin Oncol. 1999;17:948–957. doi: 10.1200/JCO.1999.17.3.948. [DOI] [PubMed] [Google Scholar]
- 5.Dennis ER, Jia X, Mezheristskiy IS, et al. Bone scan index: a quantitative treatment response biomarker for castration-resistant metastatic prostate cancer. J Clin Oncol. 2012;30:519–524. doi: 10.1200/JCO.2011.36.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadik M, Suurkula M, Hoglund P, Jarund A, Edenbrandt L. Improved classifications of planar whole-body bone scans using a computer-assisted diagnosis system: a multicenter, multiple-reader, multiple-case study. J Nucl Med. 2009;50:368–375. doi: 10.2967/jnumed.108.058883. [DOI] [PubMed] [Google Scholar]
- 7.Sadik M, Hamadeh I, Nordblom P, et al. Computer-assisted interpretation of planar whole-body bone scans. J Nucl Med. 2008;49:1958–1965. doi: 10.2967/jnumed.108.055061. [DOI] [PubMed] [Google Scholar]
- 8.Sadik M, Suurkula M, Hoglund P, Jarund A, Edenbrandt L. Quality of planar whole-body bone scan interpretations: a nationwide survey. Eur J Nucl Med Mol Imaging. 2008;35:1464–1472. doi: 10.1007/s00259-008-0721-5. [DOI] [PubMed] [Google Scholar]
- 9.Ulmert D, Kaboteh R, Fox JJ, et al. A novel automated platform for quantifying the extent of skeletal tumor involvement in prostate cancer patients using the bone scan index. Eur Urol. 2012;62:78–84. doi: 10.1016/j.eururo.2012.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ljungberg M, Strand SE. A Monte Carlo program for the simulation of scintillation camera characteristics. Comput Methods Programs Biomed. 1989;29:257–272. doi: 10.1016/0169-2607(89)90111-9. [DOI] [PubMed] [Google Scholar]
- 11.Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BM. 4D XCAT phantom for multimodality imaging research. Med Phys. 2010;37:4902–4915. doi: 10.1118/1.3480985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodsaid F, Frueh F. Biomarker qualification pilot process at the US Food and Drug Administration. AAPS J. 2007;9:E105–E108. doi: 10.1208/aapsj0901010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown MS, Chu GH, Kim HJ, et al. Computer-aided quantitative bone scan assessment of prostate cancer treatment response. Nucl Med Commun. 2012;33:384–394. doi: 10.1097/MNM.0b013e3283503ebf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong AJ, Kaboteh R, Carducci MA, et al. Assessment of the bone scan index in a randomized placebo-controlled trial of tasquinimod in men with metastatic castration-resistant prostate cancer (mCRPC) Urol Oncol. 2014;32:1308–1316. doi: 10.1016/j.urolonc.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaboteh R, Gjertsson P, Leek H, et al. Progression of bone metastases in patients with prostate cancer: automated detection of new lesions and calculation of bone-scan index. EJNMMI Res. 2013;3:64–70. doi: 10.1186/2191-219X-3-64. [DOI] [PMC free article] [PubMed] [Google Scholar]


