Abstract
Background & aims
Several animal studies have emphasized the role of gut microbiota in non-alcoholic fatty liver disease (NAFLD). However, data about gut dysbiosis in human NAFLD remains scarce in the literature, especially studies including the whole spectrum of NAFLD lesions. We aimed to evaluate the association between gut dysbiosis and severe NAFLD lesions, i.e. non-alcoholic steatohepatitis (NASH) and fibrosis, in a well-characterized population of adult NAFLD.
Methods
57 patients with biopsy-proven NAFLD were enrolled. The taxonomic composition of gut microbiota was determined using 16S ribosomal RNA gene sequencing of stool samples.
Results
30 patients had F0/1 fibrosis stage at liver biopsy (10 with NASH), and 27 patients had significant F≥2 fibrosis (25 with NASH). Bacteroides abundance was significantly increased in NASH and F≥2 patients, whereas Prevotella abundance was decreased. Ruminococcus abundance was significantly higher in F≥2 patients. By multivariate analysis, Bacteroides abundance was independently associated with NASH and Ruminococcus with F≥2 fibrosis. Stratification according to the abundance of these 2 bacteria generated 3 patient subgroups with increasing severity of NAFLD lesions. Based on imputed metagenomic profiles, KEGG pathways significantly related to NASH and fibrosis F≥2 were mostly related to carbohydrate, lipid, and amino acid metabolism.
Conclusion
NAFLD severity associates with gut dysbiosis and a shift in metabolic function of the gut microbiota. We identified Bacteroides as independently associated with NASH and Ruminococcus with significant fibrosis. Thus, gut microbiota analysis adds information to classical predictors of NAFLD severity and suggests novel metabolic targets for pre/probiotics therapies.
Keywords: Non-alcoholic steatohepatitis, liver fibrosis, gut microbiome, Bacteroides, Ruminococcus
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD), the liver manifestation of the metabolic syndrome, is characterized by a wide spectrum of liver phenotypes ranging from simple steatosis to non-alcoholic steatohepatitis (NASH), the aggressive form of the disease leading to liver fibrosis and finally cirrhosis with its life threatening complications. The evolution of NAFLD to cirrhosis is not mandatory: around 20-30% of NAFLD patients develop NASH with only some of them further evolving to fibrosis and then cirrhosis (1, 2). NAFLD is a complex disease driven by the interaction between host genetic background and environmental factors. Genetic polymorphisms explain a small part of the inter-individual variability in hepatic phenotypes observed in NAFLD patients. As an example, the well-known I148M variant of the PNPLA3 gene is associated only with a 3.5-fold greater risk of NASH, and a 3.2-fold higher risk of developing liver fibrosis (3). Several other factors that influence the course of the disease have been identified such as epigenetics (4), hormonal status (5), or nutrition (6). However, despite progress in knowledge about NAFLD pathogenesis, the fact that some NAFLD patients develop NASH/fibrosis, while most of them do not, remains incompletely understood.
Recently, the gut microbiota has gained great attention in metabolic diseases since gut dysbiosis has been demonstrated in obesity (7, 8), the metabolic syndrome (9, 10), diabetes (11, 12), and cardiovascular diseases (13). Recent animal studies have placed the gut microbiota as a potentially important player in the pathogenesis of NAFLD (14, 15). However, data linking gut dysbiosis with the severity of NAFLD remains poorly documented in humans. Only a few series with generally small sample sizes, heterogeneous populations (adult versus children), and different methods for gut microbiota evaluation (qPCR versus pyrosequencing) are available in the literature (16-18). In addition, because liver biopsy was not available in all patients to phenotype liver lesions, under-diagnosis of NASH was possible, especially in obese patients (18). Finally, these studies focused on NASH and hence, very few patients with liver fibrosis were included, limiting assessment of the association between gut dysbiosis and fibrosis in NAFLD.
The aim of the present study was to evaluate if the severity of NAFLD lesions, i.e. NASH and fibrosis, is associated with gut dysbiosis, in a well-characterized and well-balanced population of biopsy-proven NAFLD patients.
PATIENTS AND METHODS
Patients
Patients with biopsy-proven NAFLD were consecutively included from October 2012 to September 2013 at Angers University Hospital (France). NAFLD was defined as liver steatosis on liver biopsy after exclusion of concomitant steatosis-inducing drugs, excessive alcohol consumption (>210 g/week in men or >140 g/week in women), chronic hepatitis B or C infection, and histological evidence of other concomitant chronic liver disease. Patients were excluded if they had cirrhosis complications (ascites, variceal bleeding, systemic infection, or hepatocellular carcinoma), history of chronic inflammatory bowel disease or bariatric surgery, or if they had been treated with antibiotics within the 2 months before inclusion. The study protocol conformed to the ethical guidelines of the current Declaration of Helsinki and was approved by the local ethics committee. All patients gave informed written consent before participating to the study.
Liver histology
Pathological examination of liver biopsy was performed by an expert of the NASH-CRN network (CG) who was blinded for patient data. Steatosis, lobular inflammation, ballooning, and liver fibrosis were semi-quantitatively evaluated according to the NASH CRN scoring system (19): F0 = no fibrosis, F1 = perisinusoidal or portal/periportal fibrosis, F2 = perisinusoidal and portal/periportal fibrosis, F3 = bridging fibrosis, and F4 = cirrhosis. As recently recommended (20), NASH was defined as the presence of each of the 3 following conditions: steatosis grade ≥1, lobular inflammation grade ≥1, and ballooning grade ≥1. ‘Significant fibrosis’ was defined as fibrosis stage F≥2.
Stool sample collection, microbial DNA extraction, amplicon library construction and sequencing
Stool samples were collected the day of the liver biopsy and immediately frozen at −80°C. Genomic DNA was isolated from stool samples using the PowerSoil DNA isolation kit (Mobio Laboratories) following the manufacturer's protocol.
DNA sequencing
Amplicon libraries were constructed with Illumina sequencing-compatible and barcode-indexed bacterial/archael PCR primers 515F and 806R, which target the V4 region of 16S rRNA gene (21). All PCR reactions were performed with Kappa HiFi using the manufacturer's protocol (Kappa Biosystems) and approximately 50 ng of extracted DNA per reaction. Reactions were held at 94°C for 3 min followed by 35 cycles of 94°C × 45 sec, 50°C × 60 sec, and 72°C × 90 sec. A final 10 min 72°C extension completed the reactions. All amplicons were purified by gel extraction (E-Gel; Invitrogen). The purified amplicons were then pooled in equimolar concentrations and the final concentration of the library was determined by Qubit (Invitrogen). Amplicon libraries were mixed with 30% PhiX control DNA. Sequencing was performed on a MiSeq instrument (Illumina) using a 250 × 2 V2 kit.
Clustering MiSeq reads into operational taxonomic units
Paired end reads were first merged and de-multiplexed into patient samples using Qiime version 1.9 (22). Subsequent processing of amplicon sequences was performed with UPARSE version 7.0 (23), and included read error correction, de-replication, chimera filtering, and finally de novo clustering into operational taxonomic units (OTUs) at a 97% identity cut-off. Taxonomic affiliation of each OTUs was performed with QIIME against the Greengenes database version 13.8.
Inferred metagenomics prediction of stool samples
A predicted functional composition of the gut microbiome was inferred for each stool samples using PICRUSt. Based on the fact that phylogeny and function are closely linked, this method accurately predicts the abundance of gene families from the 16S rRNA information (24). A previous study has shown that the PICRUSt imputed and shotgun sequenced metagenomes have very good correlation with an average Spearman coefficient around 0.8 (24). Briefly, metagenome inference was performed with 16S rRNA gene sequences clustered at a 97% identity threshold using closed reference of the Greengenes version 13.5 database. The resulting OTU table was then normalized by 16S rRNA gene copy number and predicted gene family abundance was inferred for each sample. Significant functional differences between patient classes were assessed with LEfSE (25) using a p value ≤ 0.05 and a LDA score >2.
Statistics
Quantitative variables were expressed as median with 1st and 3rd quartiles into brackets. Raw observation counts in taxa summary plots were normalized by calculating relative abundance. Qualitative variables were compared using the Fisher's exact test and quantitative variable using the Mann Whitney test. A p value less than 0.05 was considered statistically significant. Statistical analyses were performed using SPSS version 18.0 software (IBM, Armonk, NY, USA).
RESULTS
Patients
57 NAFLD patients were included in the study. Their characteristics are detailed in Table 1. Median age was 60 years and 34 patients (60%) were male. Forty six patients (81%) had a metabolic syndrome and 23 (40%) were diabetics under treatment. Thirty patients (53%) had F0/1 fibrosis stage on liver biopsy, of whom 10 had NASH. The remaining 27 patients (47%) had significant fibrosis (F≥2). Two patients with F≥2 fibrosis stage (respectively F3 and F4) had no NASH because no ballooning was demonstrated on liver biopsy. Fibrosis stage repartition from F0 to F4 was, respectively; 16, 14, 15, 6, and 6 patients.
Table 1.
patient characteristics at inclusion
| All | Fibrosis F0/1 |
Fibrosis F≥2 | p | ||
|---|---|---|---|---|---|
| No NASH | NASH | ||||
| Patients (n) | 57 | 20 | 10 | 27 | - |
| Age (years) | 60 (51-66) | 55 (47-64) | 61 (52-70) | 62 (56-67) | 0.283 |
| Sex (M/F) | 34/23 | 13/7 | 6/4 | 15/12 | 0.808 |
| BMI (kg/m2) | 31 (28-34) | 30 (26-34) | 31 (28-32) | 32 (29-35) | 0.268 |
| Diabetes (no/yes)a | 30/27 | 18/2 | 5/5 | 7/20 | <0.001 |
| Elevated blood pressure (no/yes)b | 14/43 | 10/10 | 1/9 | 3/24 | 0.005 |
| Elevated triglycerides (no/yes)c | 28/29 | 15/5 | 4/6 | 9/18 | 0.015 |
| Reduced HDL-cholesterol (no/yes)d | 14/43 | 6/14 | 1/9 | 7/20 | 0.475 |
| Metabolic syndrome (no/yes) | 11/46 | 8/12 | 0/10 | 3/24 | 0.011 |
| AST (IU/l) | 40 (32-59) | 36 (30-43) | 32 (20-44) | 56 (37-71) | 0.002 |
| ALT (IU/l) | 64 (39-89) | 62 (29-83) | 44 (32-86) | 69 (55-102) | 0.129 |
| GammaGT (IU/l) | 80 (40-124) | 71 (39-127) | 53 (29-117) | 93 (60-159) | 0.238 |
| Us CRP (mg/l) | 1.8 (1.0-4.3) | 1.4 (0.6-2.5) | 2.9 (1.1-4.2) | 1.8 (1.3-5.8) | 0.115 |
Quantitative results are expressed as median with 1st and 3rd quartiles into brackets
Either anti-diabetic treatment or fasting glycemia ≥7.0 mmol/l
According to the International Diabetes Federation definition:
Antihypertensive treatment or elevated blood pressure with systolic ≥130 mm Hg and/or diastolic ≥85 mm Hg
Lipid-lowering treatment or triglycerides ≥1.7 mmol/l
Lipid-lowering treatment or HDL-cholesterol <1.1 mmol/l in men or <1.3 mmol/l in women
Gut microbiota
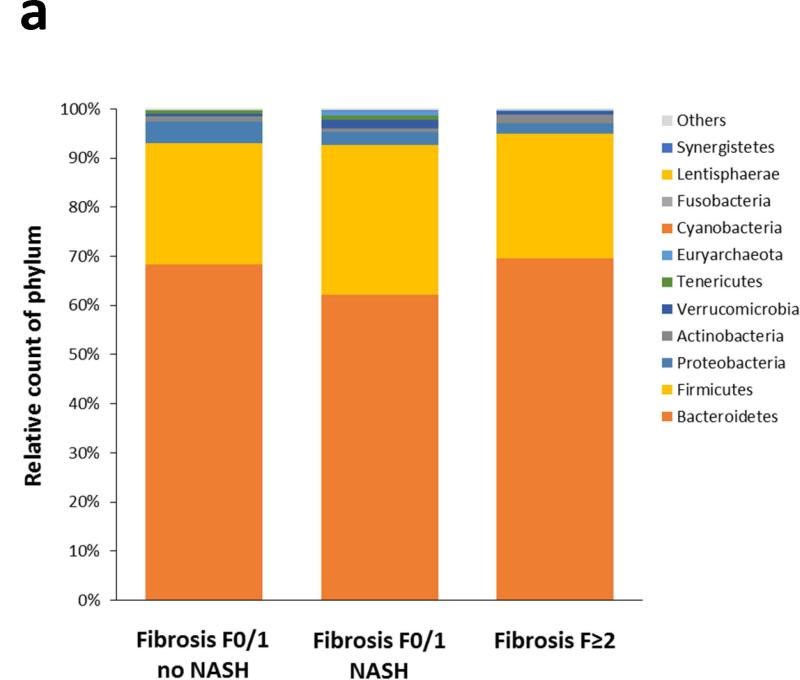
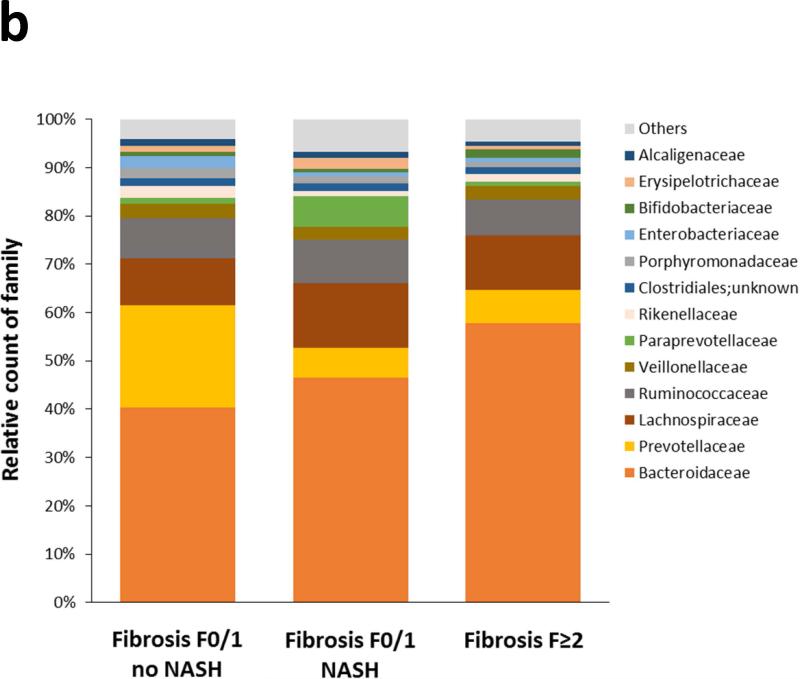
After paired end read merging and error correction of 16S rRNA sequencing, a total of 10,896,711 high quality sequences were obtained from the 57 stool samples with a mean of 191,170 ± 12,585 sequences per sample (range: 21,978 – 554,352). Based on ≥97% sequence identity, amplicons were clustered into 2,371 OTUs of whom 2,269 were finally assigned using the Greengenes database and 102 unassigned. The OTU richness and phylogenic diversity from the gut microbiota were not associated with NAFLD severity (see Figures s1 and s2 in Supplementary Material). Twelve bacteria phyla, 65 families and 133 genera were identified in the gut microbiomes in this study. At the phylum level, the taxonomic composition of the gut microbiomes showed no difference according to increasing NAFLD severity (Figure 1a). Significant differences started to appear at the family level (Figure 1b): Bacteroidaceae increased with the severity of liver lesions, whereas Prevotellaceae and Erysipelotrichaceae decreased.
Figure 1. Taxonomic composition of the gut microbiota as a function of NAFLD severity.
No significant difference was observed at the phylum level (Figure 1a). Significant differences appeared from the family level (Figure 1b).
Gut dysbiosis and NASH
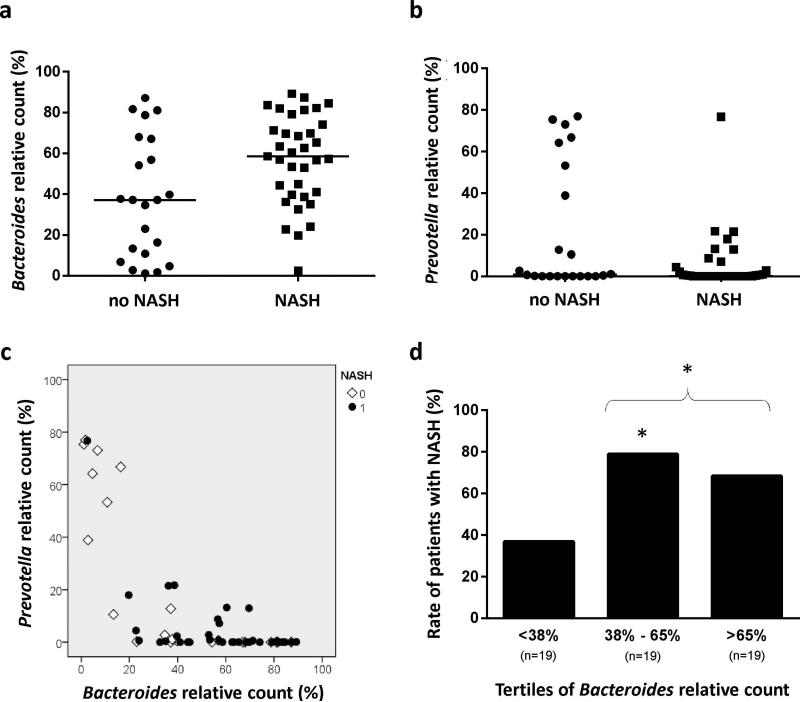
Two genera, Bacteroides and Prevotella, significantly differed between patients with NASH and those without (Table 2). Compared to those without NASH, patients with NASH had higher abundance of Bacteroides and lower abundance of Prevotella (Figures 2a and 2b). As previously described (26), these 2 genera act as competitors with an inverse relationship between their respective abundance (Figure 2c). Multivariate analysis adjusted on metabolic factors (BMI, diabetes, elevated blood pressure, elevated triglycerides, reduced HDL-cholesterol, metabolic syndrome) showed that Bacteroides abundance was independently associated with NASH (Table s1 in Supplementary Material). The study sample of 57 patients was divided according to the tertiles of Bacteroides count (Figure 2d): patients in the tertiles 2 and 3 had a 2-fold increase in NASH compared to those in the first tertile (74% vs 37%, p=0.010).
Table 2. Mean abundance of gut microbiome taxa in patients with or without NASH.
Phyla, families, and genera with >1% occurrence in the whole population are presented.
| Bacteria | No NASH (n=22) | NASH (n=35) | pa |
|---|---|---|---|
| Actinobacteria | 0.9 | 1.6 | 0.818 |
| Bifidobacteriaceae | 0.9 | 1.6 | 0.511 |
| Bifidobacterium | 0.9 | 1.6 | 0.511 |
| Bacteroidetes | 67.3 | 68.1 | 0.768 |
| Bacteroidaceae | 38.3 | 56.9 | 0.013 |
| Bacteroides | 38.3 | 56.9 | 0.013 |
| Porphyromonadaceae | 2.0 | 1.2 | 0.577 |
| Parabacteroides | 2.0 | 1.2 | 0.577 |
| Prevotellaceae | 21.7 | 5.5 | 0.053 |
| Prevotella | 21.7 | 5.5 | 0.053 |
| Rikenellaceae | 2.2 | 1.5 | 0.987 |
| Paraprevotellaceae | 1.0 | 2.4 | 0.882 |
| Firmicutes | 26.2 | 26.1 | 0.987 |
| Clostridiales; unknownb | 2.0 | 1.3 | 0.376 |
| Lachnospiraceae | 10.7 | 11.3 | 0.491 |
| Blautia | 1.6 | 1.9 | 0.149 |
| Unknown b | 5.4 | 5.4 | 0.451 |
| Ruminococcaceae | 8.6 | 7.8 | 0.544 |
| Ruminococcus | 0.8 | 1.4 | 0.235 |
| Unknown b | 7.0 | 5.7 | 0.376 |
| Veillonellaceae | 2.8 | 2.9 | 0.123 |
| Megasphaera | 1.5 | 1.5 | 0.650 |
| Erysipelotrichaceae | 1.1 | 1.2 | 0.272 |
| Proteobacteria | 4.0 | 2.4 | 0.491 |
| Alcaligenaceae | 1.3 | 0.9 | 1.000 |
| Sutterella | 1.3 | 0.9 | 1.000 |
| Enterobacteriaceae | 2.2 | 1.0 | 0.325 |
| Unknown b | 1.6 | 0.8 | 0.491 |
by Mann-Whitney test
16S rRNA sequence distinct from any known genera in this family/genus
Figure 2. Relationship between NASH at liver biopsy and Bacteroides or Prevotella abundance in the gut.
Figures 2a/2b: NASH patients had higher abundance of gut Bacteroides (p=0.013) and lower abundance of Prevotella (p=0.053). Figure 2c: As these bacteria act as competitors, Bacteroides and Prevotella abundance had an inverse relationship. Figure 2d: Rate of NASH patients as a function of the tertiles of Bacteroides relative count. The rate of NASH was significantly lower in patients with low abundance of Bacteroides (1st tertile). * p≤0.02 vs 1st tertile.
Gut dysbiosis and significant F≥2 fibrosis
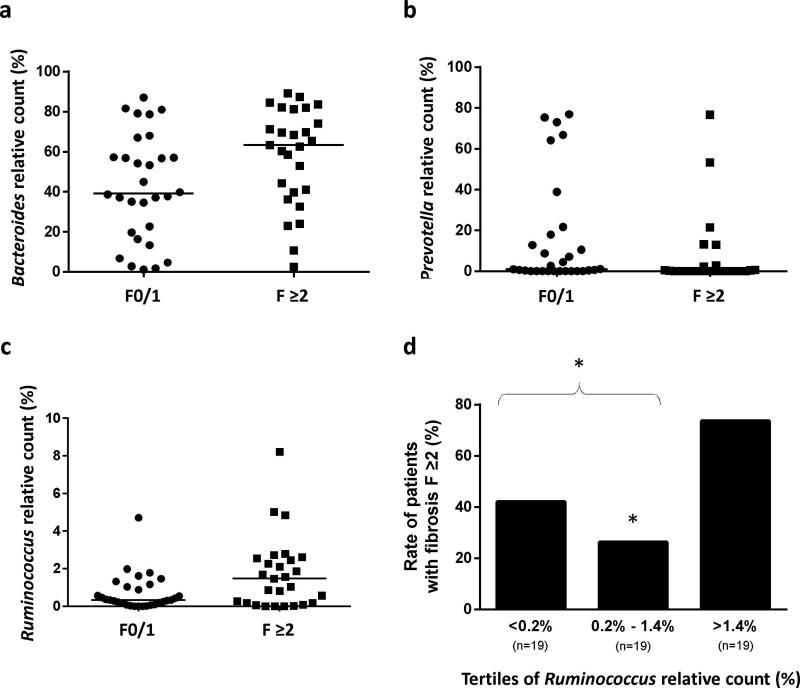
The 3 genera Bacteroides, Prevotella, and Ruminococcus significantly differed between patients with F0/1 fibrosis and those with significant F≥2 fibrosis (Table 3). Compared to F0/1 patients, those with F≥2 fibrosis had higher abundances of Bacteroides and Ruminococcus, and lower abundance of Prevotella (Figure 3a-c). Multivariate analysis adjusted on metabolic factors showed that Ruminococcus abundance was independently associated with fibrosis F≥2 (Table s2 in Supplementary Material). The study sample was divided according to the tertiles of Ruminococcus count (Figure 3d): patients in the third tertile had a 2-fold increase in fibrosis F≥2 compared to those in the tertiles 1 and 2 (74% vs 34%, p=0.010).
Table 3. Mean abundance of gut microbiome taxa in patients with no/mild fibrosis (F0/1 stage) or significant F≥2 fibrosis.
Phyla, families, and genera with >1% occurrence in the whole population are presented.
| Bacteria | F0/1 (n=30) | F≥2 (n=27) | pa |
|---|---|---|---|
| Actinobacteria | 0.9 | 1.8 | 0.987 |
| Bifidobacteriaceae | 0.9 | 1.8 | 0.949 |
| Bifidobacterium | 0.9 | 1.8 | 0.949 |
| Bacteroidetes | 66.2 | 69.6 | 0.388 |
| Bacteroidaceae | 42.4 | 57.8 | 0.018 |
| Bacteroides | 42.4 | 57.8 | 0.018 |
| Porphyromonadaceae | 1.9 | 1.0 | 0.231 |
| Parabacteroides | 1.9 | 1.0 | 0.231 |
| Prevotellaceae | 16.2 | 6.8 | 0.017 |
| Prevotella | 16.2 | 6.8 | 0.017 |
| Rikenellaceae | 2.0 | 1.6 | 0.949 |
| Paraprevotellaceae | 2.8 | 0.8 | 0.386 |
| Firmicutes | 26.7 | 25.4 | 0.798 |
| Clostridiales; unknownb | 1.7 | 1.4 | 0.270 |
| Lachnospiraceae | 10.9 | 11.3 | 0.774 |
| Blautia | 1.9 | 1.6 | 0.975 |
| Unknown b | 4.9 | 5.9 | 0.397 |
| Ruminococcaceae | 8.6 | 7.5 | 0.576 |
| Ruminococcus | 0.7 | 1.7 | 0.037 |
| Unknown b | 7.2 | 5.1 | 0.250 |
| Veillonellaceae | 2.9 | 2.8 | 0.620 |
| Megasphaera | 1.2 | 1.9 | 0.891 |
| Erysipelotrichaceae | 1.6 | 0.7 | 0.010 |
| Proteobacteria | 3.8 | 2.1 | 0.129 |
| Alcaligenaceae | 1.4 | 0.8 | 0.482 |
| Sutterella | 1.4 | 0.8 | 0.482 |
| Enterobacteriaceae | 1.9 | 1.0 | 0.099 |
| Unknown b | 1.5 | 0.7 | 0.128 |
by Mann-Whitney test
16S rRNA sequence distinct from any known genera in this family/genus
Figure 3. Relationship between significant F≥2 fibrosis at liver biopsy and Bacteroides, Prevotella, or Ruminococcus abundance in the gut.
Figures 3a-c: Patients with fibrosis F≥2 had higher abundance of gut Bacteroides (p=0.018) and Ruminococcus (p=0.037), and lower abundance of Prevotella (p=0.017). Figure 3d: Rate of F≥2 patients as a function of the tertiles of Ruminococcus relative count. The rate of F≥2 fibrosis was significantly higher in patients with a high abundance of Ruminococcus (3rd tertile). * p≤0.01 vs 3rd tertile.
Bacteroides, Ruminococcus, and increasing severity of NAFLD lesions
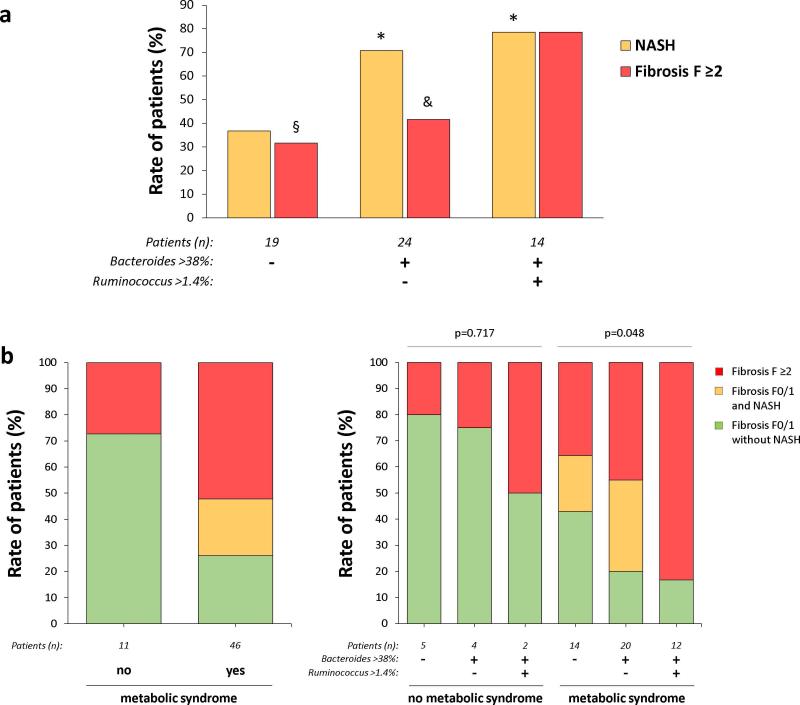
As Bacteroides were independently associated with NASH and Ruminococcus with fibrosis F≥2, we evaluated the severity of NAFLD lesions according to 3 subgroups defined by the level of these 2 bacteria: Bacteroides <38% (tertile 1), Bacteroides ≥38% (tertiles 1-2) and Ruminococcus ≤1.4% (tertiles 1-2), Bacteroides ≥38% and Ruminococcus >1.4% (tertile 3). The rate of NASH in these 3 subgroups was, respectively: 37%, 71%, and 79% (p=0.024, Figure 4a). The rate of fibrosis F≥2 was, respectively: 32%, 42%, and 79% (p=0.021, Figure 4a). Thus, using the abundance of Bacteroides and Ruminococcus, it was possible to define 3 distinct subgroups with increasing NAFLD severity: low NASH/low fibrosis, high NASH/low fibrosis, and high NASH/high fibrosis (Figure 4a).
Figure 4. Severity of NAFLD lesions according to Bacteroides and Ruminococcus abundance.
Figure 4a: Rate of patients with NASH or fibrosis F≥2 according to Bacteroides and Ruminoccocus abundance (* p<0.04 vs 1st subgroup, § p=0,013 vs 3rd subgroup, & p=0,043 vs 3rd subgroup). Figure 4b: Severity of NAFLD lesions according to the metabolic syndrome status alone or stratified according to the abundance of Bacteroides and Ruminococcus.
As previously described (27), the presence of the metabolic syndrome was strongly associated with more severe NAFLD lesions (Figure 4b). We evaluated whether the levels of Bacteroides and Ruminococcus help to stratify the 2 metabolic syndrome categories (no/yes) in several subgroups with increasing severity of NAFLD. Three of the 11 patients without metabolic syndrome had fibrosis F≥2. Stratification of these 11 patients in 3 subgroups according to the level of Bacteroides and Ruminococcus did not lead to significantly different rates of fibrosis F≥2 patients (Figure 4b). Of the 46 patients with a metabolic syndrome, 12 had fibrosis F0/1 without NASH, 10 had fibrosis F0/1 with NASH, and 24 had fibrosis F≥2. The stratification of these 46 patients according to Bacteroides and Ruminococcus abundance led to subgroups with significantly increased severity of NAFLD lesions (Figure 4b). The same pattern was observed when diabetes or the various components of the metabolic syndrome were considered instead of the metabolic syndrome (Table s3).
Inferred metagenome content of stool samples
The functional potential of bacterial assemblages associated to each stool sample was predicted with PICRUSt using level 3 of KEGG orthologs. As assessed with LEfSE at a p value <0.05, the gut microbiome of NASH patients was significantly enriched in 6 functional categories compared to patients without NASH. These enriched functional categories (Table 4) were related to carbohydrate metabolism (e.g. glyoxylate/dicarboxylate metabolism, starch/sucrose metabolism), lipid metabolism (e.g. sphingolipid metabolism), amino acid metabolism (e.g. cyanoamino acid metabolism) and secondary metabolism (e.g. phenylpropanoid biosynthesis). Compared to F0/1 patients, the gut microbiome of patients with significant F≥2 fibrosis was significantly enriched in 6 functional categories. These enriched categories (Table 4) were also related to carbohydrate metabolism (e.g. glyoxylate/dicarboxylate metabolism, pentose and glucuronate interconversions, pentose phosphate pathway) and lipid metabolism (e.g. fatty acid biosynthesis, lipid biosynthesis proteins).
Table 4. Functional profile of the gut microbiota from NASH or F ≥2 patients.
KEGG pathways were inferred from 16s rRNA gene sequences using PICRUSt. Functional categories significantly enriched or depleted in NASH patients or patients with F ≥2 fibrosis were assessed with LEfSE (p ≤0.05, LDA >2). NASH and F ≥2 fibrosis were associated with microbiota enrichment in KEGG categories related to metabolic functions.
| KO functional categories |
NASH vs no NASH |
F ≥2 vs F0/1 |
|||
|---|---|---|---|---|---|
| Level 2 | Level 3 | LDA | p-value | LDA | p-value |
| Biosynthesis of Other Secondary Metabolites | Phenylpropanoid biosynthesis | 2.34 | 0.03 | - | - |
| Carbohydrate metabolism | Glyoxylate and dicarboxylate metabolism | 2.28 | 0.03 | 2.28 | 0.02 |
| Carbohydrate metabolism | Pentose and glucuronate interconversions | - | - | 2.74 | 0.02 |
| Carbohydrate metabolism | Pentose phosphate pathway | - | - | 2.44 | 0.01 |
| Carbohydrate metabolism | Starch and sucrose metabolism | 2.68 | 0.01 | - | - |
| Carbohydrate metabolism | Unclassified | 2.22 | 0.02 | 2.27 | 0.01 |
| Lipid Metabolism | Fatty acid biosynthesis | - | - | 2.10 | 0.03 |
| Lipid Metabolism | Lipid biosynthesis proteins | - | - | 2.07 | 0.03 |
| Lipid Metabolism | Sphingolipid metabolism | 2.45 | 0.04 | - | - |
| Metabolism of Other Amino Acids | Cyanoamino acid metabolism | 2.27 | 0.03 | - | - |
| Translation | Translation factors | - | - | −2.25 | 0.05 |
| Replication and repair | DNA replication proteins | - | - | −2.55 | 0.04 |
DISCUSSION
Recent evidence has linked gut microbiota to the pathogenesis of NAFLD (14, 15). Indeed, by manipulating the gut microbiota, animal studies have demonstrated direct roles for gut microbiota in each of the liver lesions observed in NAFLD: steatosis (28), NASH (29), fibrosis (30), and liver cancer (31). However, human data in this field remain scarce in the literature. Our work has several strengths compared to previously published human studies that have evaluated gut dysbiosis in NAFLD (16-18). First, our population was well phenotyped for liver lesions since each patient enrolled underwent a diagnostic liver biopsy. Because NASH is poorly diagnosed by usual clinical and serological parameters, studies that have compared individuals with biopsy-proven NASH to an un-biopsied obese control group might have been biased by un-diagnosed NASH in controls. In addition, our population encompassed the entire spectrum of non-malignant liver lesions observed in NAFLD, i.e. steatosis, NASH, and fibrosis. This permitted us to determine if particular gut microbiota profiles associate with different liver phenotypes of NAFLD, and to evaluate for the first time in human NAFLD the association between gut dysbiosis and liver fibrosis. Finally, this is the first work to study gut dysbiosis in adult NAFLD patients from the European continent. Our results show that the more serious NAFLD lesions (i.e., NASH and significant fibrosis) associate with gut dysbiosis. More specifically, we found that an increased abundance of Bacteroides genus independently associated with NASH, and that increased abundance in Ruminococcus was independently associated with fibrosis. As discussed below, this dysbiosis shifted the metabolic potential of the gut microbiota, thereby potentially altering host exposure to various factors that have been linked to NAFLD pathogenesis
NAFLD lesions are more severe in patients with the metabolic syndrome than those without it (32). Indeed, the latest AASLD guidelines state that the presence of the metabolic syndrome can be used to identify NAFLD patients in whom liver biopsy is particularly justified (27). Despite the relatively small sample size, our study revealed an association between Bacteroides abundance in the gut and NASH, and this relationship was independent of metabolic factors (metabolic syndrome, BMI, diabetes, elevated blood pressure, elevated triglycerides, reduced HDL-cholesterol). Further, within the patients who had a metabolic syndrome, the severity of NAFLD lesions significantly increased as a function of the 3 subgroups defined by Bacteroides and Ruminococcus abundance. These results show that gut microbiota analysis adds prognostic information to the classical risk factors for NAFLD severity, and strongly suggests that the gut microbiota has a significant role in the pathogenesis of human NAFLD. As NAFLD is a complex disease resulting from the interaction of several factors, further studies are required to determine how members of the gut microbiota, environmental factors such as nutrition, and host genetics interact to modulate NAFLD pathogenesis. This knowledge will enable more precise profiling of NAFLD patients who are at risk for progressive liver damage.
Published literature suggests several mechanisms that may explain why increased Bacteroides abundance in the gut promotes NASH. Bacteroides abundance displays strong positive correlations with the fecal content of deoxycholic acid, D-pinitol, choline, raffinose and stachyose (the two last contain glucose and fructose). Conversely, negative correlations between fecal Bacteroides and fecal short chain fatty acids (SCFA) and amino acids have been reported (33). Most of these compounds influence the pathogenesis of NASH. For example, deoxycholic acid induces apoptosis in rat liver, and is increased in the livers of NASH patients (34, 35). Fructose has been associated with increased liver inflammation and fibrosis in NAFLD patient (36). Hence, Bacteroides-associated increases in deoxycholic acid, raffinose, and stachyose are predicted to promote NASH, while decreased SCFAs might be detrimental for NAFLD (37, 38).
We also observed that increases in fecal Bacteroides abundance were paralleled by decreases in Prevotella. This finding is consistent with evidence that Bacteroides and Prevotella are competitors. Dietary composition is known to influence the balance between Bacteroides and Prevotella in the gut: Western diets rich in fat, animal proteins and sugar favor Bacteroides; while agrarian society diets that are rich in fiber, starch and plant polysaccharides promote Prevotella abundance (26, 39-41). Western-type diets high in fructose and saturated fats have been associated with NASH. Thus, evidence of increased Bacteroides and decreased Prevotella in our NASH patients is in line with previously published information about the relationship between diet and human NASH (6). A mechanism that might explain this relationship was identified by a recent study that evaluated rapid and diet-specific alterations of gut microbial communities under a ‘plant-based’ or an ‘animal-based’ diet (42). Namely, the animal-based diet rapidly induced a shift of gut microbial community to favor Bacteroides abundance. Bacteroides accumulation correlated with an accumulation of branched-chain fatty acids that are produced by amino acid fermentation. The latter are known to promote insulin-resistance (43), and insulin resistance increases the risk for NASH.
Our results also demonstrated an independent, positive correlation between Ruminococcus abundance and significant (F≥2) liver fibrosis. There is little published information to guide hypotheses generation about this association. Ruminococcus are able to ferment complex carbohydrates such as cellulose, pectine, resistant starch (44, 45), and are acetate and propionate producers (46, 47). The Ruminococcus genus is quite heterogeneous, including both beneficial and deleterious bacteria. For example, Ruminococcus bromii is known to have beneficial effects on health (48), while other Ruminococcus species have been shown to be pro-inflammatory (49, 50). In a recent study, Ruminococcus abundance increased in formula-fed infant rhesus monkeys, and this was accompanied by increased branched-chain amino acids, hyperinsulinemia, and an inflammatory state (51). Ruminococcus are also able to produce alcohol (46) and this might have detrimental actions on intestinal permeability and hepatic inflammation. As with Bacteroides, Ruminococcus abundance in the gut seems to be influenced by diet composition, but the effects of diet appear to be complex. For example, animal based diets increase Ruminococcus gnavus but decrease Ruminococcus bromii and Ruminococcus callidus (42).
To further explore hypotheses linking Bacteroides and Ruminoccocus to the severity of NAFLD, we used PICRUSt to estimate the metagenomic profile of the gut microbiota from our patients (24). Interestingly, this functional approach showed that more serious NAFLD lesions (i.e., NASH and significant fibrosis) associated with significant shifts in the metabolic function of the gut microbiota, mainly impacting KEGG pathways that relate to metabolism of carbohydrates, lipids, and amino acids. These results provide exciting, new insights about potential roles of gut microbiota in NAFLD pathogenesis. They must be confirmed by further ‘classical’ metagenomics studies to precisely identify which metabolic pathways of the gut microbiota associate with NASH and/or fibrosis and thus, might promote NAFLD progression.
One limitation of our study is the relative small sample size which didn't allow us to demonstrate that small variations in the bacterial counts were statistically significant. However, we may assume that the bigger the difference between the subgroups studied, the stronger the potential effect of the bacteria on the phenotype should be. Consequently, the present work provides relevant information about the potential role of gut microbiota in the processes that drive NAFLD to the severe form of the disease, i.e. NASH and fibrosis.
In conclusion, histologic subtypes of NAFLD that increase the risk for liver-related morbidity and mortality associate with gut dysbiosis and altered metabolic functions of the gut microbiota. Enrichment of the fecal microbiome with Bacteroides independently and positively associates with NASH, and Ruminococcus accumulation similarly correlates with fibrosis stage, showing that gut microbiota analysis adds information to classical predictors of NAFLD severity. Further studies will have to decipher how metabolic functions of the gut microbiota might contribute to NASH and fibrosis in NAFLD.
Supplementary Material
Acknowledgments
Financial support: This research is supported by NIH R01-DK053792 and R01-DK106633 (Diehl AM). JB received grants from the French association for the study of the liver (AFEF), and the French national society of hepato-gastroenterology (SNFGE).
Abbreviations
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- OTU
operational taxonomic unit
- SCFA
short chain fatty acid
Footnotes
Conflict of interest: none
REFERENCES
- 1.de Alwis NM, Day CP. Non-alcoholic fatty liver disease: the mist gradually clears. J Hepatol. 2008;48(Suppl 1):S104–112. doi: 10.1016/j.jhep.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Singh S, Allen AM, Wang Z, Prokop LJ, Murad MH, Loomba R. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin Gastroenterol Hepatol. 2015;13:643–654. e641–649. doi: 10.1016/j.cgh.2014.04.014. quiz e639-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 4.Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang JD, Abdelmalek MF, Pang H, Guy CD, Smith AD, Diehl AM, et al. Gender and menopause impact severity of fibrosis among patients with nonalcoholic steatohepatitis. Hepatology. 2014;59:1406–1414. doi: 10.1002/hep.26761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Wit NJ, Afman LA, Mensink M, Muller M. Phenotyping the effect of diet on non-alcoholic fatty liver disease. J Hepatol. 2012;57:1370–1373. doi: 10.1016/j.jhep.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 9.Murphy EF, Cotter PD, Hogan A, O'Sullivan O, Joyce A, Fouhy F, et al. Divergent metabolic outcomes arising from targeted manipulation of the gut microbiota in diet-induced obesity. Gut. 2013;62:220–226. doi: 10.1136/gutjnl-2011-300705. [DOI] [PubMed] [Google Scholar]
- 10.Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916. e917. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 11.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boursier J, Diehl AM. Implication of gut microbiota in nonalcoholic fatty liver disease. PLoS Pathog. 2015;11:e1004559. doi: 10.1371/journal.ppat.1004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnabl B, Brenner DA. Interactions Between the Intestinal Microbiome and Liver Diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–127. doi: 10.1002/hep.26319. [DOI] [PubMed] [Google Scholar]
- 17.Wong VW, Tse CH, Lam TT, Wong GL, Chim AM, Chu WC, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis--a longitudinal study. PLoS One. 2013;8:e62885. doi: 10.1371/journal.pone.0062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 19.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 20.Sanyal AJ, Brunt EM, Kleiner DE, Kowdley KV, Chalasani N, Lavine JE, et al. Endpoints and clinical trial design for nonalcoholic steatohepatitis. Hepatology. 2011;54:344–353. doi: 10.1002/hep.24376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 24.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology. 2012;142:1592–1609. doi: 10.1053/j.gastro.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787–1794. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 29.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Minicis S, Rychlicki C, Agostinelli L, Saccomanno S, Candelaresi C, Trozzi L, et al. Dysbiosis contributes to fibrogenesis in the course of chronic liver injury. Hepatology. 2014;59:1738–1749. doi: 10.1002/hep.26695. [DOI] [PubMed] [Google Scholar]
- 31.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 32.Marchesini G, Bugianesi E, Forlani G, Cerrelli F, Lenzi M, Manini R, et al. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 2003;37:917–923. doi: 10.1053/jhep.2003.50161. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Wu J, Li JV, Zhou NY, Tang H, Wang Y. Gut microbiota composition modifies fecal metabolic profiles in mice. J Proteome Res. 2013;12:2987–2999. doi: 10.1021/pr400263n. [DOI] [PubMed] [Google Scholar]
- 34.Aranha MM, Cortez-Pinto H, Costa A, da Silva IB, Camilo ME, de Moura MC, et al. Bile acid levels are increased in the liver of patients with steatohepatitis. Eur J Gastroenterol Hepatol. 2008;20:519–525. doi: 10.1097/MEG.0b013e3282f4710a. [DOI] [PubMed] [Google Scholar]
- 35.Ferreira DM, Afonso MB, Rodrigues PM, Simao AL, Pereira DM, Borralho PM, et al. c-Jun N-terminal kinase 1/c-Jun activation of the p53/microRNA 34a/sirtuin 1 pathway contributes to apoptosis induced by deoxycholic acid in rat liver. Mol Cell Biol. 2014;34:1100–1120. doi: 10.1128/MCB.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdelmalek MF, Suzuki A, Guy C, Unalp-Arida A, Colvin R, Johnson RJ, et al. Increased fructose consumption is associated with fibrosis severity in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1961–1971. doi: 10.1002/hep.23535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puertollano E, Kolida S, Yaqoob P. Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Curr Opin Clin Nutr Metab Care. 2014;17:139–144. doi: 10.1097/MCO.0000000000000025. [DOI] [PubMed] [Google Scholar]
- 39.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98:111–120. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christopherson MR, Dawson JA, Stevenson DM, Cunningham AC, Bramhacharya S, Weimer PJ, et al. Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis. BMC Genomics. 2014;15:1066. doi: 10.1186/1471-2164-15-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One. 2013;8:e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott KP, Antoine JM, Midtvedt T, van Hemert S. Manipulating the gut microbiota to maintain health and treat disease. Microb Ecol Health Dis. 2015;26:25877. doi: 10.3402/mehd.v26.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 50.Sartor RB. Key questions to guide a better understanding of host-commensal microbiota interactions in intestinal inflammation. Mucosal Immunol. 2011;4:127–132. doi: 10.1038/mi.2010.87. [DOI] [PubMed] [Google Scholar]
- 51.O'Sullivan A, He X, McNiven EM, Haggarty NW, Lonnerdal B, Slupsky CM. Early diet impacts infant rhesus gut microbiome, immunity, and metabolism. J Proteome Res. 2013;12:2833–2845. doi: 10.1021/pr4001702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.