Abstract
The assumption that Mycobacterium tuberculosis infections should be considered clonally homogeneous has been weakened in the last few years. Recent studies have shown (i) the isolation of different M. tuberculosis strains from sequential episodes, (ii) mixed infections by two M. tuberculosis strains, and (iii) genetic variations in M. tuberculosis subpopulations due to microevolution events. Nevertheless, it is unknown whether clonal heterogeneity could be found in the initial steps of M. tuberculosis infection, i.e., the primary infection. In the present study we analyzed the clonal composition of the M. tuberculosis isolates causing primary infections in children. Cultures were clonally homogeneous in most cases (11 of 12). In 1 of the 12 cases (8.3%), clonal heterogeneity among the M. tuberculosis isolates was found by spoligotyping and IS6110-based restriction fragment length polymorphism analysis. This case occurred in a 2-year-old child in whom microevolution events were unlikely and who had no risk factors for overexposure to M. tuberculosis. Clonal heterogeneity should also be considered in primary M. tuberculosis infections, including circumstances in which it is usually unexpected.
In recent years, clonal heterogeneity in the Mycobacterium tuberculosis populations causing infections has been found more frequently than was initially expected. Different studies have shown that a relevant proportion of tuberculosis (TB) recurrences are caused by strains other than those involved in the first episode (1, 3, 4, 7, 9, 11, 13). Furthermore, mixed infections by more than one M. tuberculosis strain during the same episode have been reported (2, 4, 10, 14, 15), and we have found (8) that in some cases this polyclonality leads to compartmentalization of the infection, with different clones infecting respiratory and extrarespiratory sites. In addition, clonal heterogeneity has also been proposed to be the result of the acquisition of subtle genetic changes in an M. tuberculosis subpopulation due to microevolution events during the course of the infection (6).
In general, clonal heterogeneity in an M. tuberculosis isolate is assumed to be the result of a superinfection in a previous TB episode by a new M. tuberculosis strain. This would lead to the simultaneous or sequential presence of two different strains in the same patient. In some cases, a simultaneous coinfection with more than one strain has been proved, but only in high-risk circumstances (2, 4). As far as the microevolution events causing clonal heterogeneity in the M. tuberculosis isolates are concerned, it has been proposed that a minimum time of progression of the infection is required to allow mutations and rearrangements to occur (5).
Therefore, the circumstances in which clonal heterogeneity has been explored in TB are mainly restricted to (i) recurrences, (ii) superinfections of uncured cases, and (iii) cases in high-incidence settings. Our study aimed to analyze the clonal composition of M. tuberculosis isolates in a circumstance, primary infection, that has received little attention from a molecular point of view.
(This study was presented in part at the 41st Eur. Congr. Clin. Microbiol. Infect. Dis., Glasgow, Scotland, May 2003 [M. Marín, D. García de Viedma, G. Lorenzo, M. J. Ruiz, and E. Bouza, Abstr. 41st Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P-1648, 2003].)
MATERIALS AND METHODS
Patients.
Over a 2-year period, 12 children with TB (Table 1) were selected as candidates for primary M. tuberculosis infection. Cultures of respiratory samples taken from all patients close to the time of appearance of clinical symptoms were positive for M. tuberculosis.
TABLE 1.
Clinical and microbiological features of the study patientsa
| Patient no. | Age/sexb | Type of TB at diagnosis | HIV infection status | Sample tested | Predisposing risk factor |
|---|---|---|---|---|---|
| 1 | 16 yr/M | Lymphatic | Unknown | Sputum | None |
| 2 | 13 yr/F | Pulmonary | Unknown | Sputum | None |
| 3 | 10 mo/M | Pulmonary | − | Gastric aspirate | Parent(s) IVDUc |
| 4 | 13 mo/M | Pulmonary | − | Gastric aspirate | Parent(s) IVDU |
| 5 | 3 yr/M | Meningitis | − | Gastric aspirate | Unknown |
| 6 | 13 yr/F | Pulmonary | − | Sputum | Unknown |
| 7 | 15 yr/F | Pulmonary | Unknown | Bronchial aspirate | None |
| 8 | 2 yr/F | Meningitis | − | Gastric aspirate | None |
| 9 | 5 mo/F | Meningitis | + | Gastric aspirate | HIV infection |
| 10 | 14 yr/M | Pulmonary | Unknown | Sputum | None |
| 11 | 4 yr/M | Pulmonary | − | Gastric aspirate | None |
| 12 | 7 mo/M | Lymphatic | − | Gastric aspirate | Parent(s) IVDU |
None of the isolates was drug resistant.
M, male; F, female.
IVDU, intravenous drug user.
Microbiological methods.
Clinical specimens were processed by standard methods and inoculated on Lowenstein-Jensen slants and in mycobacterial growth indicator tube (MGIT; Becton Dickinson, Sparks, Md.) liquid medium. Testing for susceptibility to isoniazid, rifampin, streptomycin, and ethambutol was performed for all the strains with the MB/BacT system (Organon Teknika, Durham, N.C.).
The MTB cultures from the clinical samples were totally harvested from the solid media (in order to guarantee that the different M. tuberculosis clones potentially present in the culture were selected for analysis) and were stored frozen at −70°C until analysis.
Molecular typing.
To analyze the clonal composition of the M. tuberculosis isolates, molecular typing was performed with a random selection of single colonies from the M. tuberculosis cultures. These single colonies were obtained by plating the positive cultures onto Middlebrook 7H11 agar plates to obtain single colonies. Thirty independent colonies from each plate were picked and subcultured in MGIT medium for molecular analysis. All the cultures that grew from these single colonies were fingerprinted by spoligotyping. When different spoligotypes were observed for colonies from the same clinical culture, spoligotyping was repeated to confirm the results. Molecular typing by IS6110-based restriction fragment length polymorphism analysis (RFLP) was also performed as a second-line assay to confirm polyclonality.
Laboratory cross-contamination was ruled out as a cause of misassignment of the typing patterns by checking that the typing patterns were not shared by any other sample handled on the same day.
Spoligotyping.
Spoligotyping, which explores polymorphisms in the direct repeat (DR) locus of M. tuberculosis, was performed as follows. For chromosome extractions, 1 ml of the bacteria cultured in MGIT (BACTEC; Becton Dickinson) was centrifuged. The cells were resuspended in a dilution of lysis reagent from the Accuprobe kit (Gene Probe) and boiled for 5 min. Glass beads (Sigma) were added, and the suspension was sonicated for 5 min. The PCR for amplification of the DR region was performed with the primers in the spoligotyping kit (Isogen, Maarsen, The Netherlands) and according to the instructions of the manufacturer. All spoligotypes were confirmed by repeating the assay. When a spoligotype showed ambiguities in any of the boxes, the assay was repeated until definite hybridization signals were obtained. Spoligotypes were considered the same when all spacers showed identical hybridization patterns between strains.
IS6110-based RFLP typing.
RFLP was used as a confirmatory typing method when different spoligotyping patterns were obtained. The procedure was performed according to international standardization guidelines (12). In the RFLP assay, apart from the standard assay in which the DNA was digested with PvuII, an independent assay in which the DNA was digested with SmaI was also performed to confirm clonal heterogeneity. A marker strain (M. tuberculosis 14323) was included as a reference, and the molecular sizes of the bands were calculated with the ChemiDoc system (Bio-Rad, Hercules, Calif.) and Diversity database software (Bio-Rad).
RESULTS
The M. tuberculosis isolates from the respiratory samples of 12 children were available for study (Table 1). Spoligotyping was performed with 360 independent colonies (30 colonies per patient).
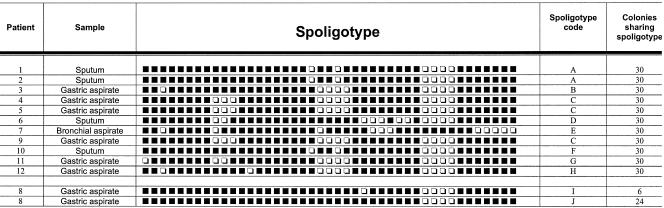
For 11 patients, all 30 colonies analyzed shared the same spoligotype (Fig. 1), indicating that a single M. tuberculosis clone was causing the primary infection.
FIG. 1.
Spoligotypes for the analysis of multiple independent single colonies of the clinical isolates. Black boxes, hybridization with the corresponding spacer in the DR region; white boxes, absence of hybridization. The patient number, sample origin, a letter representing each genotype, and the number of colonies sharing each of the spoligotypes are shown in the columns. The spoligotypes (I and J) of the isolates from patient 8 (with clonal heterogeneity during primary infection) are shown at the bottom. Patient 7’s spoligotype corresponded to M. bovis.
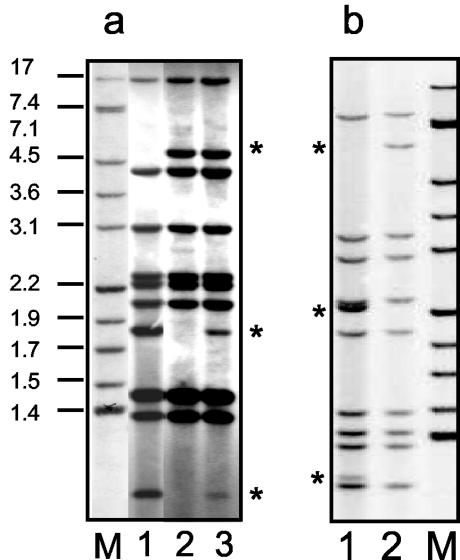
For the remaining patient (patient 8), two different spoligotypes (genotypes I and J) were obtained (proportion, 6:24) (Fig. 1). Molecular typing by IS6110-based RFLP analysis was also performed with all single colonies corresponding to spoligotypes I and J (Fig. 2a, lanes 1 and 2), and the differences between the genetic profiles detected by spoligotyping were confirmed, with RFLP types differing at three of nine bands (Fig. 2b, lanes 1 and 2). The sizes of the bands which differed between the patterns were 4.94, 1.83, and 1.07 kb, thus ruling out the possibility that the two small bands were derived from the long band by loss of a restriction site. RFLP analysis was also performed directly with a sample from the culture of a respiratory sample from this patient, and the typing pattern corresponded to the addition of the two profiles observed in the assay with single colonies (Fig. 2a, lane 3). Lastly, RFLP analysis was performed after digestion of the DNA with a different enzyme (SmaI) (Fig. 2b, lanes 1 and 2), and clonal differences were again observed. These data together confirm the presence of clonal heterogeneity during the primary infection.
FIG. 2.
(a) RFLP patterns, after digestion of the DNA with PvuII, corresponding to the analysis of representatives of single colonies of genotypes I and J (lanes 1 and 2, respectively) and the clinical isolate, which includes simultaneously isolates of both genotype I and genotype J (lane 3) from patient 8. The differential bands between the genotypes are indicated with asterisks. (b) RFLP patterns, after digestion of the DNA with SmaI, corresponding to the analysis of representatives of single colonies of genotypes I and J (lanes 1 and 2, respectively). The differential bands between the genotypes are indicated with asterisks. Lanes M, reference strain 14323 (band sizes are indicated in kilobases).
The child infected with isolates with clonal heterogeneity during primary infection (patient 8) was a 2-year-old girl with tuberculous meningitis (Table 1) and no risk factors for TB. She was human immunodeficiency virus (HIV) negative, and high-risk, socioepidemiological circumstances to justify exposure to TB were not present. The epidemiological research covered her household and school background and did not find an index case.
DISCUSSION
During recent years, different studies have led us to reconsider the assumption that infection by M. tuberculosis is clonally homogeneous, and we are beginning to understand that the clonal composition of M. tuberculosis infection is much more complex than was initially thought.
It has been proved that recurrences are caused, at a greater rate than expected, by strains other than those involved in the first episode. This has been demonstrated not only in high-prevalence settings (13) but also in contexts with moderate incidences of the disease (1, 3, 7). In addition to sequential episodes of TB, the simultaneous isolation of different M. tuberculosis strains from single episodes has been explored (4, 10, 14, 15), and we have found that polyclonality could lead to compartmentalization of the infection, with one of the strains being selected to disseminate, whereas the other remains at the respiratory level (8).
In general, infections in patients infected with M. tuberculosis isolates with clonal heterogeneity are considered to correspond to polyclonality caused by a superinfection of an uncured previous episode. Clonal heterogeneity due to polyclonality has also been thought to result from simultaneous coinfection by multiple strains (2, 4, 9), and other investigators consider clonal heterogeneity in M. tuberculosis isolates to be due to microevolution events during the course of the infection.
Recently, an ambitious study performed in The Netherlands (6) examined the presence of clonal heterogeneity in 1,277 cases of TB by analyzing the presence of low-intensity bands (LIBs) by RFLP analysis. The investigators ruled out the presence of multiple infections and assumed that the proportion of cases in which LIBs are detected is a consequence of genetic variations caused by microevolution events, probably as a result of adaptation of the bacterial population to latency or reactivation status. This hypothesis is consistent with the observation that LIBs are more frequently observed in older patients (6).
Our study analyzes the clonal composition of M. tuberculosis isolates in a context in which clonal heterogeneity is usually unexpected: primary infections in a general setting. The main limitation of studying primary infection is that it is difficult to be certain that in adults, cases are not reactivations of latent infections. Therefore, we selected children (median age, 3.5 years) with TB as candidates for primary infection because in this group the possibility of reactivation of latent infections is highly minimized. Unfortunately, it is not easy to compile cases of TB in children with an M. tuberculosis-positive culture, a requirement for isolate typing, and we were able to study only 12 patients over a 2-year period.
Our data indicate that the clinical isolates from most patients analyzed showed clonal homogeneity. Nevertheless, in 1 of our 12 patients (8.3%) we detected the coexistence of two different typing patterns when independent colonies from the clinical culture were analyzed. The potential role of laboratory cross-contamination in misassignment of a typing pattern was ruled out after analysis of all the typing profiles for the strains handled in the laboratory at that time and by checking that they were not shared by the analysis group.
As explained above, the clonal heterogeneity of strains in patients with TB can be due to (i) genetic variations in the bacterial population due to intrapatient microevolution or (ii) sequential or simultaneous dual infections by two different strains.
Microevolution events were unlikely in our study, when it is considered that the course of the infection in our patients was not long enough to allow microevolution to occur. In children, the time between exposure and the appearance of clinical symptoms is very short. In addition, it has been reported that the rate of change in RFLP patterns as a result of microevolution is 3.2 years (5). These aspects, together with the age of our patient (2 years), minimize the possibility that the bacterial population could have acquired genetic variations either randomly or as a result of adaptations to dormancy and reactivation states. Finally, in our patient whose isolates showed clonal heterogeneity, the RFLP patterns between the isolates differed by three bands (one large band disappeared and two new smaller bands appeared), and it can be considered that these differences were due to the loss of a restriction site. Nevertheless, the sum of the sizes of the two smaller bands is not equivalent to the size of the large band. This is not consistent with the hypothesis that subtle genetic changes due to microevolution are responsible for the clonal heterogeneity detected.
With regard to the alternative explanation for clonal heterogeneity, i.e., dual infection by two different strains, the profile of the case did not fit with a risk for overexposure. The patient in whom clonal heterogeneity was detected during primary infection did not have risk factors for TB, was HIV negative, and did not live under high-risk socioepidemiological circumstances associated with overexposure. On the contrary, in our study other patients who had risk factors for TB or who came from environments with conflicting high-risk socioepidemiological circumstances for TB (Table 1) were infected with clonally homogeneous populations. Therefore, in the analysis group, there did not seem to be a direct correlation between clonal heterogeneity and the socioepidemiological circumstances usually associated with overexposure. The possibility that the patient in whom clonal heterogeneity was detected was infected by two variants of a single parental clone which had microevolved in the source patient should also be considered.
It is difficult to understand the real cause of clonal heterogeneity in the primary infection found in our study, and the limitations of our sample size make it difficult to draw definitive conclusions. Additional studies with patients with primary infections are therefore needed to further evaluate clonality. Nevertheless, we believe that the relevant issue is that clonal heterogeneity can also be detected in circumstances in which neither microevolution nor overexposure is expected.
Our data suggest that the factors involved in the clonal composition of TB isolates are much more complex than generally expected. Our study extends the contexts in which clonal heterogeneity in TB can be found and looks closely at cases in which it is usually disregarded.
Acknowledgments
We are indebted to Thomas O'Boyle for revision of the English in the manuscript and Sandra Andrés for excellent technical assistance with the typing analysis.
This study was partially financed by grants from the Comunidad de Madrid (grant 08.2/0029.1/2001) and Fondo de Investigaciones Sanitarias (grant PI020882). M.M. has a contract from Fondo de Investigaciones Sanitarias (contract FIS 01/F048).
We do not have a commercial or other association that might pose a conflict of interest.
REFERENCES
- 1.Bandera, A., A. Gori, L. Catozzi, A. Degli Esposti, G. Marchetti, C. Molteni, G. Ferrario, L. Codecasa, V. Penati, A. Matteelli, and F. Franzetti. 2001. Molecular epidemiology study of exogenous reinfection in an area with a low incidence of tuberculosis. J. Clin. Microbiol. 39:2213-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braden, C. R., G. P. Morlock, C. L. Woodley, K. R. Johnson, A. C. Colombel, M. D. Cave, Z. Yang, S. E. Valway, I. M. Onorato, and J. T. Crawford. 2001. Simultaneous infection with multiple strains of Mycobacterium tuberculosis. Clin. Infect. Dis. 33:e42-e47. [Online.] [DOI] [PubMed] [Google Scholar]
- 3.Caminero, J. A., M. J. Pena, M. I. Campos-Herrero, J. C. Rodriguez, O. Afonso, C. Martin, J. M. Pavon, M. J. Torres, M. Burgos, P. Cabrera, P. M. Small, and D. A. Enarson. 2001. Exogenous reinfection with tuberculosis on a European island with a moderate incidence of disease. Am. J. Respir. Crit. Care Med. 163(Pt. 1):717-720. [DOI] [PubMed] [Google Scholar]
- 4.Chaves, F., F. Dronda, M. Alonso-Sanz, and A. R. Noriega. 1999. Evidence of exogenous reinfection and mixed infection with more than one strain of Mycobacterium tuberculosis among Spanish HIV-infected inmates. AIDS 13:615-620. [DOI] [PubMed] [Google Scholar]
- 5.de Boer, A. S., M. W. Borgdorff, P. E. de Haas, N. J. Nagelkerke, J. D. van Embden, and D. van Soolingen. 1999. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J. Infect. Dis. 180:1238-1244. [DOI] [PubMed] [Google Scholar]
- 6.de Boer, A. S., K. Kremer, M. W. Borgdorff, P. E. de Haas, H. F. Heersma, and D. van Soolingen. 2000. Genetic heterogeneity in Mycobacterium tuberculosis isolates reflected in IS6110 restriction fragment length polymorphism patterns as low-intensity bands. J. Clin. Microbiol. 38:4478-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García de Viedma, D., M. Marin, S. Hernangomez, M. Diaz, M. J. Ruiz Serrano, L. Alcala, and E. Bouza. 2002. Tuberculosis recurrences: reinfection plays a role in a population whose clinical/epidemiological characteristics do not favor reinfection. Arch. Intern. Med. 162:1873-1879. [DOI] [PubMed] [Google Scholar]
- 8.García de Viedma, D., M. Marín, M. Ruiz Serrano, L. Alcalá, and E. Bouza. 2003. Polyclonal and compartmentalized infection by Mycobacterium tuberculosis in patients with both respiratory and extrarespiratory involvement. J. Infect. Dis. 187:695-699. [DOI] [PubMed] [Google Scholar]
- 9.Nardell, E., B. McInnis, B. Thomas, and S. Weidhaas. 1986. Exogenous reinfection with tuberculosis in a shelter for the homeless. N. Engl. J. Med. 315:1570-1575. [DOI] [PubMed] [Google Scholar]
- 10.Raleigh, J. W. W. R., T. A. Rado, and J. H. Bates. 1975. Evidence for infection by two distinct strains of Mycobacterium tuberculosis in pulmonary tuberculosis: report of 9 cases. Am. Rev. Respir. Dis. 112:497-503. [DOI] [PubMed] [Google Scholar]
- 11.Small, P. M., R. W. Shafer, P. C. Hopewell, S. P. Singh, M. J. Murphy, E. Desmond, M. F. Sierra, and G. K. Schoolnik. 1993. Exogenous reinfection with multidrug-resistant Mycobacterium tuberculosis in patients with advanced HIV infection. N. Engl. J. Med. 328:1137-1144. [DOI] [PubMed] [Google Scholar]
- 12.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Rie, A., R. Warren, M. Richardson, T. C. Victor, R. P. Gie, D. A. Enarson, N. Beyers, and P. D. van Helden. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N. Engl. J. Med. 341:1174-1179. [DOI] [PubMed] [Google Scholar]
- 14.Warren, R. M., T. C. Victor, E. M. Streicher, M. Richardson, N. Beyers, N. C. van Pittius, and P. D. van Helden. 2004. Patients with active tuberculosis often had different strains in the same sputum specimen. Am. J. Respir. Crit. Care Med. 169:610-614. [DOI] [PubMed] [Google Scholar]
- 15.Yeh, R. W., P. C. Hopewell, and C. L. Daley. 1999. Simultaneous infection with two strains of Mycobacterium tuberculosis identified by restriction fragment length polymorphism analysis. Int. J. Tuberc. Lung Dis. 3:537-539. [PubMed] [Google Scholar]