Abstract
The accuracy of antifungal susceptibility tests is important for accurate resistance surveillance and for the clinical management of patients with serious infections. Our main objective was to compare the results of fluconazole disk diffusion testing of Candida spp. performed by ARTEMIS participating centers with disk diffusion and MIC results obtained by the central reference laboratory. A total of 2,949 isolates of Candida spp. were tested by NCCLS disk diffusion and reference broth microdilution methods in the central reference laboratory. These results were compared to the results of disk diffusion testing performed in the 54 participating centers. All tests were performed and interpreted following NCCLS recommendations. Overall categorical agreement between participant disk diffusion test results and reference laboratory MIC results was 87.4%, with 0.2% very major errors (VME) and 3.3% major errors (ME). The categorical agreement between the disk diffusion test results obtained in the reference laboratory with the MIC test results was similar: 92.8%. Likewise, good agreement was observed between participant disk diffusion test results and reference laboratory disk diffusion test results: 90.4%, 0.4% VME, and 3.4% ME. The disk diffusion test was especially reliable in detecting those isolates of Candida spp. that were characterized as resistant by reference MIC testing. External quality assurance data obtained by surveillance programs such as the ARTEMIS Global Antifungal Surveillance Program ensure the generation of useful surveillance data and result in the continued improvement of antifungal susceptibility testing practices.
Disk diffusion testing of fluconazole against Candida spp. was developed in order to provide a simple inexpensive method for monitoring fluconazole susceptibility in a variety of laboratory settings (2, 4, 6-8, 12, 13). Barry and colleagues (2) demonstrated that the accuracy and precision of fluconazole disk diffusion testing, using a 25-μg disk and Mueller-Hinton agar supplemented with 2% glucose and 0.5 μg of methylene blue per ml (MH-MB), were comparable to those of reference broth microdilution (BMD) MIC testing. Recently, the National Committee for Clinical Laboratory Standards (NCCLS) Subcommittee on Antifungal Testing has approved this agar disk diffusion method, M44-A, for testing fluconazole against yeasts (11).
The M44 fluconazole disk diffusion test with MH-MB has also been used for more than 4 years as part of the ARTEMIS Global Antifungal Surveillance Program (4, 8, 12). In this program, fluconazole disk diffusion testing is performed in more than 80 different laboratories in 35 countries, and the data are used to follow trends in fluconazole susceptibility patterns worldwide (4, 8). Isolates of Candida spp. collected by ARTEMIS participants are also sent to a central reference laboratory at the University of Iowa for additional testing using NCCLS disk diffusion and BMD methods (12-14). Data generated from the ARTEMIS Program have been useful in documenting the sustained activity of fluconazole versus clinical isolates of Candida (4, 8, 12, 14).
In addition to assessing the scale of the resistance problem at the local, national, or international level, antimicrobial resistance surveillance may also provide an opportunity for improving the quality of susceptibility testing among those taking part in the surveillance (5). The ARTEMIS Program demonstrates two different, and complementary, approaches to the performance of antifungal resistance surveillance. The central reference laboratory provides a means to overcome differences between methods, or differences in the same method performed in different laboratories. The centralized approach ensures that standardized, internationally recognized quantitative methods are used and that isolates are available for further studies of resistance mechanisms and for epidemiological typing (5, 15, 16). Alternatively, fluconazole disk diffusion testing performed on-site in the participant laboratories results in the generation of large amounts of data in a short period of time, using a standard protocol and with results submitted to a central database (4, 8). Despite the use of a standard protocol, it is recognized that any surveillance system based on susceptibility tests performed by the participating laboratories needs to include some measure of quality assurance, beyond simple quality control (QC) testing, in order to provide an independent assessment of laboratory performance and validation of results generated from the various laboratories (5, 18). The ARTEMIS DISK Surveillance Study (4) generated massive amounts of data but was limited by a lack of validation and comparison with reference MIC results. In the present study, we assess the accuracy of the ARTEMIS participant disk diffusion test results by comparison to those obtained for the same isolates tested in the central reference laboratory by disk diffusion and BMD methods recommended by the NCCLS.
MATERIALS AND METHODS
Study design.
The ARTEMIS Program was established to monitor the species and antifungal susceptibility patterns of Candida spp. isolated from clinically significant sites of infection (e.g., blood and normally sterile sites [NSSI]) via a broad network of sentinel hospitals in North America, Latin America, Europe, Africa, and Asia. Candida spp. causing invasive disease (in blood and NSSI) and the accompanying fluconazole disk zone diameters were reported from 54 different medical centers in 2001 and 2002.
Each participant medical center contributed results (organism identification and fluconazole disk zone diameter) for consecutive blood and NSSI culture isolates (one isolate per patient) of Candida spp. judged to be clinically significant by local criteria and detected in each calendar month during the study. All isolates were saved on agar slants and were sent to the University of Iowa College of Medicine (Iowa City) for storage and further characterization by reference identification methods and susceptibility testing (3, 10, 11). In the central reference laboratory, isolates were tested by both NCCLS disk diffusion (11) and BMD (10) methods.
Organism identification.
All Candida spp. isolates were identified at participating institutions by the routine method used in each laboratory. Upon receipt at the University of Iowa, the isolates were subcultured onto potato dextrose agar (Remel, Lenexa, Kans.) and CHROMagar Candida medium (Hardy Laboratories, Santa Maria, Calif.) to ensure viability and purity. Confirmation of species identification was performed with Vitek and API products (bioMerieux, St. Louis, Mo.), as recommended by the manufacturer, or by conventional methods, as required (3). Isolates were stored as suspensions in water or on agar slants at ambient temperature until needed.
Susceptibility testing.
Disk diffusion testing of fluconazole was performed in both participant laboratories and the central reference laboratory according to the methods described in NCCLS document M44-A (11). MIC testing was performed by the NCCLS BMD reference method M27-A2 (10). Standard fluconazole reference powder was obtained from Pfizer Pharmaceuticals (Groton, Conn.), and 25-μg fluconazole disks were obtained from Becton Dickinson (Sparks, Md.).
Disk diffusion testing of fluconazole was performed as described by Hazen et al. (4) and in NCCLS document M44-A (11). Agar plates (150-mm diameter) containing MH-MB at a depth of 4.0 mm were used. The agar surface was inoculated by using a swab dipped in a cell suspension adjusted to the turbidity of a 0.5 McFarland standard. The plates were incubated in air at 35 to 37°C and read at 18 to 24 h. Zone diameter endpoints were read at 80% growth inhibition by using the BIOMIC image analysis plate reader system (version 5.9; Giles Scientific, Santa Barbara, Calif.) (4, 12).
MIC interpretive criteria for fluconazole were those published by Rex et al. (17) and the NCCLS (10) and were as follows: susceptible, MIC of ≤8 μg/ml; susceptible dose dependent, MIC of 16 to 32 μg/ml; resistant, MIC of ≥64 μg/ml. The interpretive criteria for the fluconazole disk diffusion test were those published by Barry et al. (2) and the NCCLS (11): susceptible, zone diameter of ≥19 mm; susceptible dose dependent, zone diameter of 15 to 18 mm; resistant, zone diameter of ≤14 mm.
QC.
QC was performed for BMD in accordance with NCCLS document M27-A2 (10) by using Candida krusei ATCC 6258 and Candida parapsilosis ATCC 22019 (1, 10). QC for disk diffusion was performed using Candida albicans ATCC 90028 and C. parapsilosis ATCC 22019 (2, 4).
Analysis of results.
The diameters of the zones of inhibition (in millimeters) surrounding the fluconazole disks at 24 h of incubation obtained in the participant laboratories and in the reference laboratory were plotted against their respective BMD MICs read at 48 h (see Fig. 1 and 2, below) (2, 12, 13). Similarly, the diameters of the zones of inhibition surrounding the fluconazole disks at 24 h of incubation obtained in the participant laboratories were plotted against their respective zones obtained in the reference laboratory read at 24 h. The interpretive breakpoints described by the NCCLS (10, 11) were used to determine the categorical agreement between disk diffusion and BMD results and between participant and reference disk diffusion results. Major errors (ME) were classified as resistant by disk diffusion (participant or central laboratory) and susceptible by BMD or resistant by the participant disk diffusion test and susceptible by the central laboratory disk diffusion test. Very major errors (VME) were classified as susceptible by the participant or central laboratory disk diffusion test and resistant by BMD or susceptible by the participant disk diffusion test and resistant by the central laboratory disk diffusion test. Minor errors (M) occurred when the result of one of the tests was susceptible or resistant and that of the other test was susceptible dose dependent.
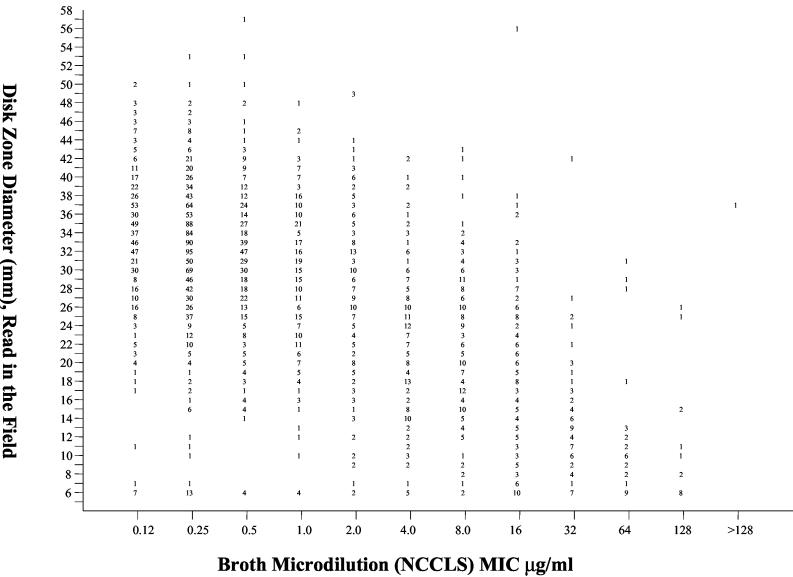
FIG. 1.
Zones of inhibition around 25-μg fluconazole disks tested in ARTEMIS participant laboratories plotted against the MICs determined at 48 h by using the reference BMD method for 2,949 isolates of Candida spp. The numbers represent the number of results (isolates) for each data point.
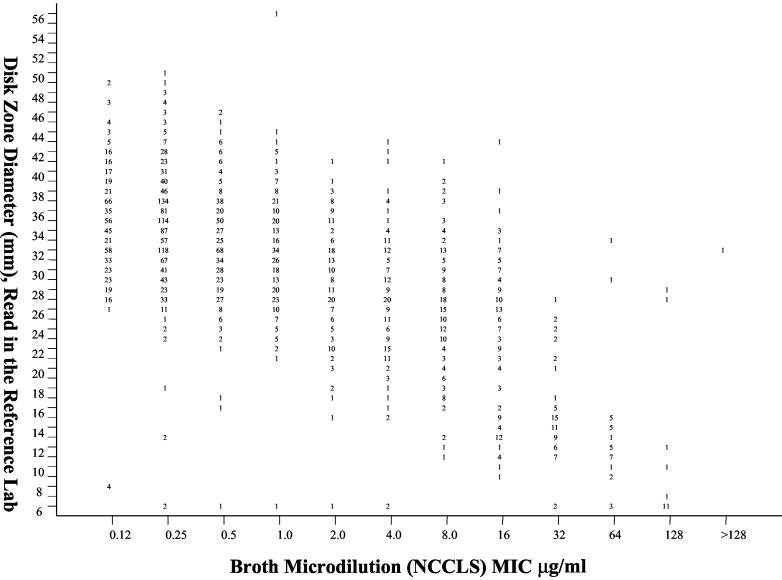
FIG. 2.
Zones of inhibition around 25-μg fluconazole disks tested on MH agar supplemented with 2% glucose and methylene blue (0.5 μg/ml) in the central reference laboratory plotted against the MICs at 48 h determined by using the reference BMD method for 2,949 isolates of Candida spp. The numbers represent the number of results (isolates) for each data point.
RESULTS AND DISCUSSION
During the 2001-2002 study period, a total of 2,949 isolates of Candida from blood and other NSSI and their accompanying fluconazole disk diffusion zone diameters were submitted from 54 participating centers in North America (8 centers), Latin America (12 centers), Europe (20 centers), and the Asia-Pacific region (14 centers). The frequencies of infections due to the various species of Candida identified in the central reference laboratory are presented in Table 1 and represent a total of 14 different species.
TABLE 1.
Isolates of Candida spp. tested by participant and reference laboratories in the ARTEMIS Global Antifungal Surveillance Program
| Organism | No. tested (%) |
|---|---|
| C. albicans | 1,635 (55.4) |
| C. glabrata | 403 (13.7) |
| C. parapsilosis | 402 (13.6) |
| C. tropicalis | 328 (11.2) |
| C. krusei | 87 (3.0) |
| C. guilliermondii | 34 (1.1) |
| C. lusitaniae | 27 (0.9) |
| C. pelliculosa | 12 (0.4) |
| Candida spp.a | 21 (0.7) |
| Total | 2,949 (100) |
Includes C. kefyr (n = 9), C. rugosa (n = 5), C. dubliniensis (n = 2), C. lipolytica (n = 2), C. zeylanoides (n = 2), and C. famata (n = 1).
In vitro susceptibility testing performed in both participant laboratories (disk diffusion) (Fig. 1) and the reference laboratory (disk diffusion and MIC test) (Fig. 2) indicated that resistance to fluconazole was uncommon among isolates of Candida from blood and NSSI (Table 2). Notably, more isolates of all species appeared resistant to fluconazole by disk diffusion testing in both participant and reference laboratories, with the greatest resistance observed with disk diffusion testing performed in the participant laboratories. Overall, the level of categorical agreement between the disk diffusion test results and the reference MIC results was quite good in both participant (87.4%) and reference (92.8%) laboratories (Table 2). Importantly, there were very few VME, indicating that the disk diffusion test was reliable in detecting isolates resistant to fluconazole by MIC testing. The level of agreement between disk diffusion and MIC results was highest with C. albicans and lowest with Candida glabrata. The low level of agreement between disk diffusion and MIC results with C. glabrata was similar to that observed previously (9, 12) and was due almost entirely to M-type errors. Interestingly, the M-type errors observed with C. glabrata involved shifts between the susceptible and susceptible-dose-dependent categories in the reference laboratory and between the susceptible-dose-dependent and resistant categories in the participant laboratories. This was likely the result of the distribution of both zones and MICs around the respective breakpoints and a more conservative reading of zone diameters in the participant laboratories. Importantly, the number of VME remained low for C. glabrata disk diffusion tests in both participant and reference laboratory settings.
TABLE 2.
Interpretive agreement between results of fluconazole disk diffusion tests and standard 48-h BMDa
| Organism (no. tested) | Test methodd | % of results in each categoryb
|
% Agreementc | % errors
|
||||
|---|---|---|---|---|---|---|---|---|
| S | SDD | R | VME | ME | M | |||
| C. albicans (1,631) | Ref.-MIC | 99.8 | 0.2 | 0.0 | ||||
| Ref.-disk | 99.5 | 0.4 | 0.1 | 99.6 | 0.0 | 0.1 | 0.3 | |
| Part.-disk | 97.6 | 0.7 | 1.7 | 97.7 | 0.0 | 1.4 | 0.9 | |
| C. glabrata (403) | Ref.-MIC | 69.5 | 26.3 | 4.2 | ||||
| Ref.-disk | 93.6 | 2.2 | 4.2 | 71.7 | 0.8 | 0.5 | 27.0 | |
| Part.-disk | 71.5 | 14.4 | 14.1 | 60.6 | 0.7 | 7.4 | 31.3 | |
| C. parapsilosis (400) | Ref.-MIC | 93.3 | 6.2 | 0.5 | ||||
| Ref.-disk | 91.0 | 3.0 | 6.0 | 93.3 | 0.0 | 1.0 | 5.7 | |
| Part.-disk | 85.5 | 4.0 | 10.5 | 85.5 | 0.0 | 4.8 | 9.7 | |
| C. tropicalis (327) | Ref.-MIC | 99.1 | 0.3 | 0.6 | ||||
| Ref.-disk | 98.8 | 0.6 | 0.6 | 97.9 | 0.6 | 0.6 | 0.9 | |
| Part.-disk | 89.6 | 7.0 | 3.4 | 88.7 | 0.6 | 3.4 | 7.3 | |
| All Candida spp. (2,949) | Ref.-MIC | 91.6 | 6.7 | 1.7 | ||||
| Ref.-disk | 94.1 | 2.2 | 3.7 | 92.8 | 0.2 | 0.4 | 6.6 | |
| Part.-disk | 87.9 | 4.5 | 7.6 | 87.4 | 0.2 | 3.3 | 9.1 | |
Fluconazole disk diffusion testing was performed according to NCCLS method M44-A, and fluconazole BMD MIC testing was performed according to NCCLS method M27-A2.
Fluconazole susceptibility categories: S, susceptible, MIC of ≤8 μg/ml (≥19 mm); SDD, susceptible dose dependent, MIC of 16 to 32 μg/ml (15 to 18 mm); R, resistant, MIC of ≥64 μg/ml (≤14 mm).
Percent categorical agreement between disk diffusion and MIC test results.
Ref.-MIC, MIC testing performed by ARTEMIS reference laboratory; Ref.-disk, disk testing performed by ARTEMIS reference laboratory; Part.-disk, disk testing performed by ARTEMIS participants.
A similar level of agreement was seen when disk diffusion results obtained in the participant laboratories were compared with those obtained in the reference laboratory (Table 3). The overall categorical agreement was excellent at 90.4%, and the number of VME, ME, and M errors was quite low. As with the comparison to BMD testing, agreement was highest with C. albicans and lowest with C. glabrata, with most of the errors in the M category.
TABLE 3.
Overall interpretive agreement between results of fluconazole disk diffusion tests performed in participant and reference laboratoriesa
| Organism | No. testedb | No. (%) of discrepant results
|
% Agree- mentc | ||
|---|---|---|---|---|---|
| Minor | Major | Very major | |||
| C. albicans | 1,631 | 16 (1.0) | 23 (1.4) | 1 (0.1) | 97.5 |
| C. glabrata | 403 | 65 (16.1) | 40 (10.0) | 2 (0.5) | 73.4 |
| C. parapsilosis | 400 | 26 (6.5) | 19 (4.8) | 5 (1.2) | 87.5 |
| C. tropicalis | 327 | 21 (6.4) | 11 (3.4) | 1 (0.3) | 89.9 |
| All Candida spp. | 2,949 | 170 (5.8) | 100 (3.4) | 13 (0.4) | 90.4 |
The fluconazole disk diffusion method was performed according to NCCLS method M44-A.
Total number of isolates tested in 54 laboratories participating in the ARTEMIS program and also in the ARTEMIS reference laboratory.
Percent total agreement, the categorical agreement between participant and reference laboratory results.
This study is the largest one of its kind validating the performance of disk diffusion testing in surveillance program participant laboratories by testing the same isolates in a central reference laboratory. We demonstrate that fluconazole disk diffusion testing using the NCCLS M44-A method can be performed with a high level of accuracy in more than 50 different laboratories worldwide. The participant results were not only validated by fluconazole disk diffusion testing performed in the central reference laboratory, but also by reference BMD MIC testing. Importantly, the participant laboratories tended to err on the side of calling isolates more resistant than the reference laboratory. This was most pronounced with C. glabrata and other non-C. albicans species. False susceptible results were uncommon. Similar results have also been reported by Morace et al. (9).
Previously, Barry et al. (2) demonstrated that when replicate fluconazole disk diffusion tests were performed in three different laboratories, 94 to 97% of zone diameters fell within a range of the median zone ±6 to 8 mm. Although a formal precision analysis was not conducted in the present study, we found that among replicate zone diameters generated in both reference and participant laboratories, 80% fell within ±8 mm (data not shown).
In summary, we have shown that fluconazole disk diffusion testing performed in accordance with the NCCLS M44-A method can be performed with an acceptable degree of accuracy in a wide range of laboratory settings throughout the world. The test was especially reliable in identifying those strains of Candida characterized as resistant by reference BMD MIC testing. False resistant results may be an issue with disk diffusion, especially with C. glabrata and other non-C. albicans species. Further assessment of such isolates by BMD should be performed if clinically indicated. Continued collaboration and external validation of antifungal susceptibility testing results such as that presented here for the ARTEMIS Global Antifungal Surveillance Program will ensure the generation of useful antifungal surveillance data and result in continued improvement of antifungal susceptibility testing practices.
Acknowledgments
This study was supported in part by research and educational grants from Pfizer Inc., Pfizer Global Pharmaceuticals, New York, N.Y.
We thank Linda Elliott and Shanna Duffy for secretarial assistance in the preparation of the manuscript. We appreciate the participation of all ARTEMIS site participants. A list of ARTEMIS participants can be found on the following website: http://www.medicine.uiowa.edu/pathology/path_folder/research/acknowledgments/artemis_participants.pdf.
REFERENCES
- 1.Barry, A. L., M. A. Pfaller, S. D. Brown, A. Espinel-Ingroff, M. A. Ghannoum, C. Knapp, R. P. Rennie, J. H. Rex, and M. G. Rinaldi. 2000. Quality control limits for broth microdilution susceptibility tests of ten antifungal agents. J. Clin. Microbiol. 38:3457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, A. L., M. A. Pfaller, R. P. Rennie, P. C. Fuchs, and S. D. Brown. 2002. Precision and accuracy of fluconazole susceptibility tests by broth microdilution, Etest, and disk diffusion methods. Antimicrob. Agents Chemother. 46:1781-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazen, K. C., and S. A. Howell. 2003. Candida, Cryptococcus, and other yeasts of medical importance, p. 1693-1711. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 4.Hazen, K. C., E. J. Baron, A. L. Colombo, C. Girmenia, A. Sanchez-Sousa, A. del Palacio, C. de Bedout, D. L. Gibbs, and The Global Antifungal Surveillance Group. 2003. Comparison of the susceptibilities of Candida spp. to fluconazole and voriconazole in a 4-year global evaluation using disk diffusion. J. Clin. Microbiol. 41:5623-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahlmeter, G., and D. F. J. Brown. 2002. Resistance surveillance studies—comparability of results and quality assurance of methods. J. Antimicrob. Chemother. 50:775-777. [DOI] [PubMed] [Google Scholar]
- 6.Kirkpatrick, W. R., T. M. Turner, A. W. Fothergill, D. I. McCarthy, S. W. Redding, M. G. Rinaldi, and T. F. Patterson. 1998. Fluconazole disk diffusion susceptibility testing of Candida species. J. Clin. Microbiol. 36:3429-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, S.-C., C.-P. Fung, N. Lee, L.-C. See, J.-S. Huang, C.-J. Tsai, K.-S. Chen, and W.-B. Shieh. 2001. Fluconazole disk diffusion test with methylene blue- and glucose-enriched Mueller-Hinton agar for determining susceptibility of Candida species. J. Clin. Microbiol. 39:1615-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meis, J., M. Petrou, J. Bille, D. Ellis, D. Gibbs, and The Global Antifungal Surveillance Group. 2000. A global evaluation of the susceptibility of Candida species to fluconazole by disk diffusion. Diagn. Microbiol. Infect. Dis. 36:215-223. [DOI] [PubMed] [Google Scholar]
- 9.Morace, G., G. Amato, F. Bistoni, G. Fadda, P. Marone, M. T. Montagna, S. Oliveri, L. Polonelli, R. Rigoli, I. Mancuson, S. La Face, L. Masucci, L. Romano, C. Napoli, D. Tato, M. G. Buscema, C. M. C. Belli, M. M. Piccirillo, S. Conti, S. Covan, F. Fanti, C. Cavanna, F. D'Alo, and L. Pitzurra. 2002. Multicenter comparative evaluation of six commercial systems and the National Committee for Clinical Laboratory Standards M27-A broth microdilution method for fluconazole susceptibility testing of Candida species. J. Clin. Microbiol. 40:2953-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution testing of yeasts. Approved standard, 2nd ed., M27-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.National Committee for Clinical Laboratory Standards. 2004. Method for antifungal disk diffusion susceptibility testing of yeasts: approved guideline M44-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Pfaller, M. A., D. J. Diekema, S. A. Messer, L. Boyken, and R. J. Hollis. 2003. Activities of fluconazole and voriconazole against 1,586 recent clinical isolates of Candida species determined by broth microdilution, disk diffusion, and Etest methods: report from the ARTEMIS Global Antifungal Susceptibility Program, 2001. J. Clin. Microbiol. 41:1440-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaller, M. A., D. J. Diekema, L. Boyken, S. A. Messer, S. Tendolkar, and R. J. Hollis. 2003. Evaluation of the Etest and disk diffusion methods for determining susceptibilities of 235 bloodstream isolates of Candida glabrata to fluconazole and voriconazole. J. Clin. Microbiol. 41:1875-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaller, M. A., S. A. Messer, L. Boyken, R. J. Hollis, C. Rice, S. Tendolkar, and D. J. Diekema. 2004. In vitro activities of voriconazole, posaconazole, and fluconazole against 4,169 clinical isolates of Candida spp. and Cryptococcus neoformans collected during 2001 and 2002 in the ARTEMIS Global Antifungal Surveillance Program. Diagn. Microbiol. Infect. Dis. 48:201-205. [DOI] [PubMed] [Google Scholar]
- 15.Pujol, C., M. Pfaller, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans bloodstream isolates from the United States, Canada, South America, and Europe reveals a European clade. J. Clin. Microbiol. 40:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pujol, C., M. A. Pfaller, and D. R. Soll. 2004. Flucytosine resistance is restricted to a single genetic clade of Candida albicans. Antimicrob. Agents Chemother. 48:262-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rex, J. H., M. A. Pfaller, J. N. Galgiani, M. S. Bartlett, A. Espinel-Ingroff, M. A. Ghannoum, M. Lancaster, M. G. Rinaldi, T. J. Walsh, and A. L. Barry. 1997. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin. Infect. Dis. 24:235-247. [DOI] [PubMed] [Google Scholar]
- 18.Tenover, F. C., M. J. Mohammed, J. Stelling, T. O'Brien, and R. Williams. 2001. Ability of laboratories to detect emerging antimicrobial resistance: proficiency testing and quality control results from the World Health Organizations' external quality assurance system for antimicrobial susceptibility testing. J. Clin. Microbiol. 39:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]