Abstract
Long, short and medium chain fatty acids are covalently attached to hundreds of proteins. Each fatty acid confers distinct biochemical properties, enabling fatty acylation to regulate intracellular trafficking, subcellular localization, protein-protein and protein-lipid interactions. Myristate and palmitate represent the most common fatty acid modifying groups. New insights into how fatty acylation reactions are catalyzed, and how fatty acylation regulates protein structure and function continue to emerge. Myristate is typically linked to an N-terminal glycine, but recent studies reveal that lysines can also be myristoylated. Enzymes that remove N-terminal myristoyl-glycine or myristate from lysines have now been identified. DHHC proteins catalyze S-palmitoylation, but the mechanisms that regulate substrate recognition by individual DHHC family members remain to be determined. New studies continue to reveal thioesterases that remove palmitate from S-acylated proteins. Another area of rapid expansion is fatty acylation of the secreted proteins Hedgehog, Wnt and Ghrelin, by Hhat, Porcupine and GOAT, respectively. Understanding how these membrane bound O-acyl transferases recognize their protein and fatty acyl CoA substrates is an active area of investigation, and is punctuated by the finding that these enzymes are potential drug targets in human diseases.
Keywords: myristoylation, myristoyl switch, palmitoylation, depalmitoylation, lipid rafts, MBOAT family
1. Introduction
Proteins can be modified with fatty acids that range in length from eight to over twenty carbons (Table 1). The predominant species acylated to proteins are saturated chain fatty acids, but monounsaturated and polyunsaturated fatty acids can also be attached. There are three major classes of fatty acylation reactions in mammalian cells: N-myristoylation catalyzed by N-myristoyl transferase, S-palmitoylation/acylation catalyzed by DHHC family enzymes, and fatty acylation of secreted proteins catalyzed by MBOAT family enzymes. Each of these fatty acylation reactions utilizes different enzymes, different fatty acyl CoA and protein substrates, and occurs in different intracellular locations. Advances in structural biology analysis have revealed that, besides enhancing membrane binding, fatty acids can form inter- and intramolecular protein-protein interactions by insertion into a hydrophobic binding pocket.
Table 1.
Biochemistry of protein fatty acylation
| Lipid | Structure | Linkage | Modified Residue | Fatty acyl Transferase | Deacylation? |
|---|---|---|---|---|---|
| Octanoate 8:0 |
|
Oxyester | Ser | GOAT | unknown |
| Myristate 14:0 |
|
Amide | Gly | NMT | Removal of Myr-Gly by IpaJ |
| Myristate 14:0 |
|
Amide | Lys | unknown | Sirtuins |
| Palmitate 16:0 |
|
Thioester | Cys | DHHCs | Thioesterases (APT1,2; ABDH17) |
| Palmitate 16:0 |
|
Amide | Cys | Hhat | unknown |
| Palmitoleic 16:1Δ9 |
|
Oxyester | Ser | Porcn | Notum |
| Stearate 18:0 |
|
Thioester | Cys | DHHCs? | Thioesterases? |
| Arach-idonate 20:4 |
|
Thioester | Cys | DHHCs? | Thioesterases? |
The 14-carbon, saturated fatty acid myristate is typically linked to an N-terminal glycine via a stable, amide bond. The acylation reaction occurs cotranslationally in the cytosol. Myristate, along with a second signal (polybasic domain, palmitoylation) can promote membrane binding. Although myristate is generally not physically removed, myristoyl switch mechanisms induce regulated release of the modified protein from membranes, by intramolecular sequestration of the fatty acid. Most palmitoylated proteins contain palmitate linked to the sulfhydryl group of one or more cysteines. These posttranslational reactions are catalyzed by the DHHC family of palmitoyl acyltransferases located on the cytosolic side of intracellular membranes or the plasma membrane. The thioester bond is reversible by thioesterases, allowing some palmitoylated proteins to undergo regulated membrane binding and release. Although the vast majority of fatty acylated proteins are intracellular or transmembrane proteins, fatty acids can also be attached to secreted proteins. The MBOAT (membrane bound O-acyl transferase) family catalyzes attachment of palmitate to hedgehog proteins, palmitoleoylate to Wnt proteins, and octanoate to ghrelin. Each of these protein substrates employs a signal sequence to enter the lumen of the endoplasmic reticulum (ER), where they are then fatty acylated. This review will highlight the similarities and differences among these three types of lipidation reactions.
2. N-Myristoylation
2.1 N-myristoyl transferase (NMT) catalyzes N-myristoylation
Covalent attachment of the 14-carbon fatty acid myristate to an N-terminal glycine has been reported for over 150 proteins in mammalian cells [1]. Nearly all N-myristoylated proteins that are co-translationally modified contain the N-terminal consensus sequence: Met-Gly-X-X-X-Ser/Thr. First, the initiating methionine is removed by methionine aminopeptidase. This reaction is essential to expose Gly as the N-terminal amino acid. Next, Myristoyl-CoA:protein N-myristoyl transferase (NMT) catalyzes transfer of myristate from the myristoyl-CoA donor onto the N-terminal glycine via an amide bond. Glycine is the only residue that can serve as an acceptor substrate for NMT, and Gly-to-Ala (G2A) mutations are typically used to generate non-myristoylated protein mutants. Programs with algorithms for prediction of N-myristoylation sites within proteins are listed in Table 2.
Table 2.
Prediction programs for Protein Fatty Acylation
| Fatty acid modification | Website |
|---|---|
| N-myristoylation | http://mendel.imp.ac.at/myristate/SUPLpredictor.htm |
| N-myristoylation | http://web.expasy.org/myristoylator/ |
| S-palmitoylation |
http://omictools.com/palmitoylation-site-prediction-category (This site has a summary of available prediction programs) |
| S-palmitoylation |
http://swisspalm.epfl.ch/ (This site has a list of published palmitoylome sequences) |
NMT is expressed as a single gene in yeast, flies and worms, where it is required for viability [2]. In vertebrates, two genes, Nmt1 and Nmt2, encode NMT proteins with ~75% sequence identity [3]. NMT1 is essential for development, as knockout of Nmt1 in mice is embryonic lethal [4]. The two isoforms share similar but not identical substrate specificities, and RNAi knockdown experiments reveal functional differences between NMT1 and NMT2 when depleted in cells. For example, depletion of NMT1 reduces cell proliferation, while depletion of both NMT1 and NMT2 with RNAi causes apoptosis [5].
The NMT catalytic reaction proceeds via ordered Bi Bi kinetics [6]. Myristoyl-CoA binds first, with the hydrocarbon chain inserting in a bent conformation into a hydrophobic binding pocket within the enzyme. The size of the binding pocket is only deep enough to accommodate 14 carbons, essentially conferring specificity for myristate over longer fatty acids [7, 8]. Binding of myristoyl-CoA results in formation of a peptide binding site, allowing the protein substrate to bind and form a ternary complex. Myristate is then transferred to the N-terminal glycine, followed by sequential release of CoA and finally the N-myristoylated protein.
Palmitate is too long to fit into the fatty acid binding pocket of NMT. As such, it cannot make the contacts needed for catalysis and thus cannot be transferred to proteins [7]. However, palmitoyl CoA can bind to NMT and competitively inhibit the enzyme in vitro. Given that palmitate and palmitoyl-CoA are far more abundant in cells than myristate and myristoyl-CoA, a mechanism must exist to “protect” NMT from inhibition by palmitoyl-CoA. Most of the intracellular fatty acyl-CoA is bound to Acyl-CoA binding protein (ACBP), a 10 kD cytosolic protein that preferentially binds fatty acyl-CoAs between 14–22 carbons with nM affinity[9]. Binding of long chain fatty acyl-CoAs to membranes further reduces the concentration of “free” fatty acyl-CoA. It is possible that, due to its higher affinity for lipid bilayers[10], binding of palmitoyl CoA to cellular membranes segregates this lipid species away from a pool of cytosolic myristoyl CoA that could serve as a substrate for NMT. However, there is an additional consideration. The Km of NMT for myristoyl CoA is 0.6 μM[11], far above the estimates of free intracellular fatty acyl-CoA concentrations, which range from 5–200 nM[12]. How is sufficient myristoyl CoA delivered to NMT so that it can perform catalysis efficiently? One possibility is that ACBP “donates” the fatty acid as a myristoyl-CoA/ACBP complex to the enzyme. This type of delivery system has been shown to operate for enzymes involved in β-oxidation and glycerolipid synthesis[13] . Another option is illustrated by the recent finding that NMT2 can bind to an Acyl-CoA binding domain containing protein, ACBD6, and this interaction prevents binding of palmitoyl-CoA to NMT2[14]. It has yet to be determined whether this mechanism, which has been demonstrated in vitro, is operative in cells.
Although the majority of N-myristoylated proteins undergo fatty acylation during protein translation, a second class of NMT substrates that are N-myristoylated post-translationally has been uncovered[15, 16]. During apoptosis, caspase-mediated cleavage between Asp-Gly exposes glycine at the N-terminus of the newly cleaved product. If this glycine is contained within a myristoylation consensus sequence, the cleavage product can be N-myristoylated. Examples of caspase-cleaved proteins that are post-translationally N-myristoylated include PAK2, gelsolin, actin, and BID[17, 18] and global profiling reveals that there could be as many as 40 such proteins [19]. Some of the newly myristoylated cleavage products are targeted to mitochondrial or other internal membranes, where they regulate apoptosis[17, 18].
2.2 Role of myristoylation in membrane binding
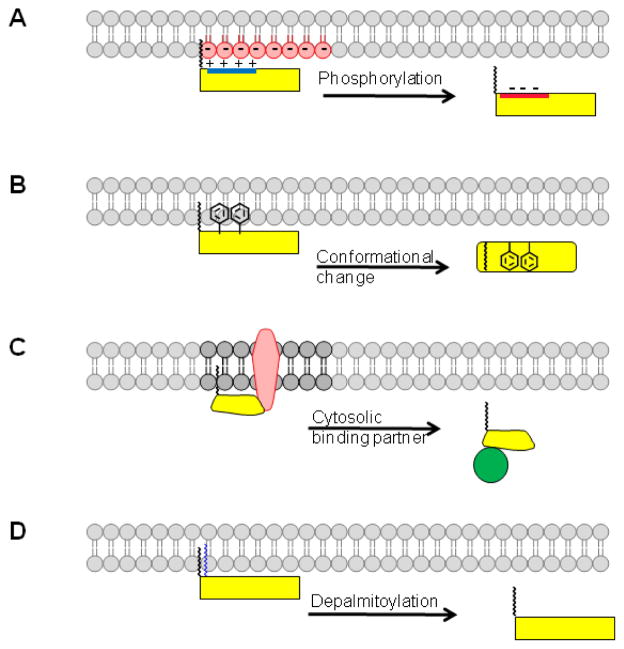
The majority of N-myristoylated proteins are membrane bound. It was initially presumed that attachment of myristate is a de facto signal to direct the modified protein to bind to membranes through hydrophobic interactions. However, this assumption is not correct. In a classic report, Peitzsch and McLaughlin showed that the ΔGo for binding of a model myristoylated peptide to a phospholipid bilayer is 8 kcal/mol, which corresponds to an effective Kd of 10−4 M [10]. This is not sufficient energy to anchor a myristoylated protein stably to a lipid bilayer. In order to achieve stable membrane binding, N-myristoylated proteins employ a second signal[20]. At least four types of second signals have been identified: polybasic regions, hydrophobic residues, another membrane-bound binding partner, or a second lipid modification (Figure 1).
Figure 1. Membrane binding mechanisms for N-myristoylated proteins.
Four different scenarios are illustrated where a second signal, in addition to myristate, contributes to membrane binding. (A) A polybasic cluster of amino acids (blue) enhances membrane binding through electrostatic interactions with head groups of negatively charged phospholipids (red). Phosphorylation of residues within the polybasic cluster reduces or neutralizes the positive charge in the protein, resulting in membrane detachment. (B) Hydrophobic amino acids disposed along the membrane proximal surface of the N-myristoylated protein contribute to membrane binding. Conformational changes can lead to sequestration of the myristate (myristoyl switch) and/or reduced surface accessibility of the hydrophobic residues, leading to release of the protein from the membrane. (C) Interaction with another membrane protein can direct N-myristoylated proteins to the membrane. The protein could detach if another binding partner, with higher affinity, interacts with the N-myristoylated protein and releases it into the cytosol. (D) Palmitoylation of an N-myristoylated protein induces membrane binding, which is reversed upon deacylation.
N-myristoylated proteins such as c-Src, MARCKS (myristoylated alanine-rich C Kinase substrate), hisactophilin, and HIV-1 Gag and Nef contain a polybasic region which synergizes with myristate to provide strong membrane association[21–23]. The positively charged residues form electrostatic interactions with negatively charged phospholipids (phosphatidylserine or phosphatidylinositolphosphates) which are enriched on the cytoplasmic leaflet of the plasma membrane and many intracellular organelle membranes. In the absence of myristate, the binding energy from these electrostatic interactions alone is not sufficient to anchor a protein to a membrane. However, when a myristate + basic motif is present within a protein, hydrophobic and electrostatic forces synergize, resulting in strong membrane binding affinity. Myristoylated proteins that lack an obvious polybasic motif can instead use the presence of nearby hydrophobic amino acids as a second signal. For example, hydrophobic residues (Phe, Leu) from the N-terminal helix provide a hydrophobic face to enhance membrane binding of the small G protein ARF1[24]. An alternative mechanism is to utilize protein-protein interactions with a membrane bound binding partner. For example, the binding of N-myristoylated GRASP65 to Golgi membranes is enhanced by interactions with its Golgi-localized receptor GM130[25].
The second signal for membrane binding of an N-myristoylated protein can also be an additional lipid modification. Src family kinases, α subunits of heterotrimeric G proteins, AKAPs (A-kinase anchoring proteins), and endothelial nitric oxide synthase are examples of proteins that contain N-terminal myristate and thioester linked palmitate attached to one or more nearby cysteine residues[26–28] . N-myristoylation occurs first, and provides increased access to membrane-bound palmitoyl acyltransferases. The additional hydrophobicity provided by two or more fatty acids is sufficient to anchor dually fatty acylated proteins to membrane bilayers.
2.3 Reversible membrane binding: cytosolic states and myristoyl switches
The linkage between myristate and the protein is relatively stable, with a half-life essentially equivalent to that of the peptide bond. There are no known de-myristoylases that cleave myristate from the N-terminal myristoyl-glycine. However, the microbial pathogen Shigella flexneri expresses a cysteine protease, IpaJ, that removes myristoyl-glycine from the N-termini of host N-myristoylated proteins[29]. Although IpaJ is able to remove myristoyl-glycine from a majority of N-myristoylated proteins in vitro, it exhibits apparent specificity for ARF proteins and members of the related ARL (ARF-like) family in Shigella infected cells[30]. Specificity is achieved by binding to the GTPase domain of GTP-bound ARFs localized to the Golgi. This explains why Golgi-associated ARF1 is the major substrate for IpaJ in cells, and provides a molecular explanation for the dramatic alteration in Golgi structure and disruption of the host secretory pathway.
A more generalized mechanism for dissociating N-myristoylated proteins from the membrane is via a myristoyl switch (Figure 1). In this case, the protein exists in two different conformations: a membrane-bound state where the N-terminal myristate is exposed on the surface of the protein and able to promote membrane binding, or a cytosolic state where the fatty acid is buried within a hydrophobic pocket inside the protein. There are multiple ways to switch an N-myristoylated protein from one form to the other. For example, binding of ligand (Ca++ binding to recoverin, GTP binding to ARF1) triggers a myristoyl switch, resulting in reversible association and dissociation from the membrane[31, 32]. A variation on this theme is the myristoyl electrostatic switch which operates for proteins that use a myristate + basic domain for membrane binding. Phosphorylation within the polybasic region by protein kinase C reduces the electrostatic contribution of this domain and releases proteins such as MARCKS and K-Ras4B from the membrane both in vitro and in live cells [33–35]. Another example is the myristoyl entropic switch that operates for the HIV-1 Gag protein[36, 37] . The myristoylated N-terminal matrix (MA) domain of HIV-1 Gag can exist in two states, with myristate either sequestered or exposed. Multimerization drives an entropic switch that promotes myristate exposure and membrane binding. Upon infection of a target cell, the Gag precursor is cleaved, MA shifts to a monomeric form, myristate flips inside, and MA is released from the membrane.
A myristoyl-phosphotyrosine switch regulates the kinase activity of the tyrosine kinase c-Abl. Myristate is bound within a hydrophobic cleft of c-Abl, which holds the kinase in an inactive conformation[38]. Gating by the c-Abl SH2 domain allows the protein to undergo a myristoyl-phosphotyrosine switch, whereupon myristate extrusion leads to kinase activation. Another N-myristoylated tyrosine kinase, c-Src, does not undergo a myristoyl switch. However, N-myristoylation does regulate c-Src kinase activity, but by a different mechanism. The presence of myristate has a positive effect on c-Src tyrosine kinase activity. Wild type N-myristoylated c-Src exhibits increased kinase activity, but is targeted for ubiquitination and has reduced intracellular stability compared to a non-myristoylated c-Src mutant[39]. Thus, in addition to regulating membrane binding, myristoylation regulates steady state levels and kinase activity of c-Src. This prevents activated forms of this tyrosine kinase, which could cause malignant transformation, from building up within the cell.
The myristate + basic motif in some N-myristoylated proteins can confer lipid-binding specificity, promoting binding of these proteins to specific membranes. For example, the HIV-1 MA domain binds specifically to phosphatidylinositol 4,5-bisphosphate (PI(4,5)P(2)). This interaction triggers myristate exposure and promotes targeting to PI(4,5)P(2) enriched regions on the cytosolic face of the plasma membrane[37]. The N-myristoylated, calmodulin binding protein known as NAP-22/CAP-23/BASP1 contains a cluster of N-terminal polybasic residues which function as a nuclear localization sequence. BASP1 specifically binds PI(4,5)P(2) in the nucleus. This leads to binding of the BASP1/ PI(4,5)P(2) complex to the promoter regions of Wilms tumor 1 target genes, recruitment of the histone deacetylase HDAC1, and results in repression of transcription[40].
3. Lysine Acylation: Myristoylation and Demyristoylation
Attachment of myristate to the epsilon amino group of lysine was first reported over 20 years ago, when the membrane-bound precursors of the cytokines interleukin 1α and TNFα were both shown to be myristoylated on two internal lysine residues[41, 42]. More recently, interest has been reignited in lysine myristoylation by the finding that lysines on histone proteins, which are traditionally viewed as being acetylated or methylated, can also be modified with a variety of short and long chain acyl groups. These include propyl, butyryl, and crotonyl as well as myristoyl and palmitoyl moieties[43–46].
The enzyme(s) responsible for lysine acylation has not yet been identified. However, a family of enzymes that removes fatty acyl groups from modified lysines has emerged. Sirtuins were originally characterized as a family of 7 NAD-dependent deacetylases which act to remove acetyl groups from acetylated histones. However, in vitro assays revealed that deacetylase activity of several of the family members is quite weak. Lin and colleagues demonstrated that Sirt2 and Sirt6 actually have deacylase activity, and preferentially hydrolyze long chain fatty acyl groups (myristoyl, palmitoyl) from histone peptides containing acylated lysine[47] . Co-crystallization in complex with a histone H3K9 myristoylated peptide revealed that myristate binds within a large hydrophobic pocket present within both Sirt2 and Sirt6[47, 48].
The physiologic significance of lysine myristoylation/demyristoylation is starting to be understood. Processing of TNFα involves demyristoylation by Sirt6, an event that is required for efficient secretion of the mature form of TNFalpha into the media[47]. For sirtuins, in vitro studies demonstrate reciprocal regulation of deacetylation and deacylation: addition of free fatty acids stimulates deacetylation activity of Sirt6[49] . This suggests that the metabolic state of the cell could regulate histone modification status, ie increased levels of fatty acids would induce histone deacetylation and repression of gene expression.
It remains to be determined whether there are other fatty acylated substrates for sirtuins. Sirt 6, as well as Sirt1 and 7 are localized in the nucleus, whereas Sirt2 is the only sirtuin localized to the cytoplasm. RNAi knockdown or inhibition of Sirt2 was shown to increase the levels of fatty acylation of multiple cellular proteins as well as core histones H1, H2A,H2B, H3 and H4 [50]. Identification of proteins whose lysine fatty acylation levels are altered by sirtuin expression may reveal novel modes of regulation.
4. Protein Palmitoylation
4.1 S-palmitoylation and the palmitoylome
Attachment of the 16-carbon saturated fatty acid palmitate to one or more cysteines via thioester linkage is referred to as S-palmitoylation[26]. The intracellular fatty acid donor is palmitoyl CoA. Several hundred proteins have been reported to be S-palmitoylated. These encompass a wide range of functions, including signaling proteins, receptors, ion channels, and transcriptional regulators. The modified cysteines can be located near the N- or C-terminus, or within internal regions of the protein. Chemical biology approaches combined with mass spectrometry analysis has led to cataloging of “palmitoylomes” in a variety of cell and tissue types[43, 51–55]. Although there is no overall consensus sequence that distinctly characterizes all known S-palmitoylated proteins, algorithms have been developed to predict potential palmitoylation sites (Table 2)[56]. A recent compilation drawn from over 5,000 S-palmitoylated proteins suggests that as much as 10% of the human proteome consists of S-palmitoylated proteins[57].
4.2 DHHC enzymes catalyze S-palmitoylation
S-palmitoylation of proteins is catalyzed by DHHC palmitoyl acyltransferases[58, 59] . Each member of this family of enzymes (5–7 in yeast, 23 in humans) contains a highly conserved Asp-His-His-Cys sequence within a cysteine-rich domain. DHHC proteins are predicted to have four to six transmembrane domains. These enzymes are localized at the plasma membrane or in intracellular membranes (ER, Golgi, endosomes), with the active site exposed to the cytoplasmic side of the membrane. In addition to the 50 amino acid DHHC homology domain, DHHC acyltransferases also contain other domains involved in protein-protein interactions, such as PDZ domains and ankyrin repeats. This may provide a docking function to promote substrate selection. There is a fair amount of functional redundancy built into the mammalian enzyme system: many palmitoylated proteins can be modified by more than one DHHC family member and each DHHC enzyme can apparently palmitoylate more than one protein substrate[60–62]. Moreover, fatty acids other than palmitate can be S-acylated to proteins (see Section 4.3), and evidence for selectivity in fatty acid transfer has been obtained for DHHC2 and DHHC3[63]. Studies of mice with loss of function alleles or knockout of a single DHHC gene have uncovered distinct phenotypes [61]. For example, mice deficient in DHHC13 or DHHC17 exhibit neurological defects similar to Huntington disease [64, 65], while defects in vascular tone and hair follicle differentiation result when mice produce non-functional DHHC21 [66, 67].
DHHC catalyzed S-palmitoylation follows a two step ping-pong enzymatic mechanism[63, 68] . First, palmitate is attached to the cysteine in the DHHC motif[69]. This autoacylation reaction results in formation of an acyl-enzyme intermediate, which then transfers palmitate to a cysteine residue within the acceptor protein substrate. Structural integrity of DHHC enzymes is maintained by coordination of two zinc ions to conserved cysteines within the cysteine rich domain[69]. Although mutation of the DHHC cysteine residue typically generates an inactive enzyme mutant, there are examples of yeast DHHC proteins that can function at reduced efficiency with alanine or arginine in place of the DHHC cysteine[70].
S-palmitoylation can occur on proteins with another lipid modification[20]. For example, N-myristoylated proteins that contain a cysteine residue located at or near an N-terminal glycine (eg Src family kinases, Gα subunits, eNOS) can be S-palmitoylated to produce a dually fatty acylated protein. Prenylated proteins with a cysteine upstream from the C-terminal CAAX-box can be S-palmitoylated (eg H-Ras, N-Ras, K-Ras4A)[71]. In a variation on a theme, proteins that contain a double cysteine (CCAX) in their C-terminal CAAX box have been shown to undergo tandem prenylation and palmitoylation[72]. In the above examples, prior myristoylation or prenylation is required for palmitoylation to occur. The weak hydrophobicity of the myristate or farnesyl group allows the singly modified protein to sample multiple intracellular membranes. In the absence of palmitate, the protein will cycle between membrane and cytosol. When a suitable palmitoyl transferase is encountered, the substrate becomes dually lipidated, and the increase in hydrophobicity reduces the kinetic “off” rate. As a result, the dually modified myristoylated and palmitoylated or farnesylated and palmitoylated protein becomes trapped at the membrane where it was modified. This mechanism, known as kinetic bilayer trapping, enables a particular palmitoyl transferase to dictate the subcellular localization of the lipidated protein to a specific membrane[73].
4.3 Heterogeneous S-acylation
In addition to palmitate, mass spectrometry analyses reveal that longer and shorter fatty acids can be attached to “palmitoylation” sites in S-acylated proteins[74, 75]. These include myristate, palmitoleate, stearate, and oleate, and the polyunsaturated fatty acids arachidonate and eicosapentanoate. In some cases, the same cysteine residue can be acylated with multiple fatty acids (eg rabbit sarcolipin is S-acylated with either palmitate or oleate in a 60/40 ratio[76]). For proteins with multiple palmitoylated cysteines, the heterogeneous fatty acylation pattern is different for each cysteine. For example, the hemagglutinin protein (HA) of influenza virus is S-acylated at three cysteines: two contain only palmitate whereas a cysteine residue near the end of the TMD is modified by stearate[77]. The location of the cysteine relative to the TMD was shown to be critical for stearate attachment.
The presence of different fatty acids on a protein can have functional consequences. Altering the fatty acid composition of the growth media, by feeding cells with an excess of a particular fatty acid, was shown to alter the fatty acylation pattern of S-acylated proteins and regulate signal transduction by Src family kinases [74, 78]. Attachment of saturated fatty acids promotes lipid raft association (see section 4.5) and signaling, while acylation with unsaturated fatty acids has the opposite effect . A recent study reported that attachment of stearate to the transferrin receptor, which is known to be S-palmitoylated[79], inhibits its signaling activity and as a consequence, stimulates mitochondrial fusion[80]. These experiments suggest potential therapeutic utility for dietary intervention by supplementation with specific fatty acids.
4.4 Depalmitoylation
S-palmitoylation differs from N-myristoylation in that the thioester link between the fatty acid and the protein chain is reversible. Two acylprotein thioesterases, APT1 and APT2, that cleave palmitate from S-palmitoylated proteins were initially identified. APT1 and APT2 were reported to catalyze depalmitoylation of membrane-bound substrates such as H-Ras and GAP-43. However, it was unclear how these thioesterases, which are cytosolic enzymes, can access their membrane-bound substrates. It was recently shown that APT1 and APT2 are themselves S-palmitoylated, which is required for membrane association and deacylation of H-Ras and GAP-43[81]. APT1 can also depalmitoylate itself as well as APT2, promoting membrane to cytosol shuttling. An alternative view proposes that the soluble, non-palmitoylated forms of APT are responsible for depalmitoylation of membrane-bound substrates and that this serves to remove non-specifically adsorbed Ras proteins from all internal membranes and enable them to be concentrated on the plasma membrane[82].
APT1 and APT2 are members of a serine hydrolase superfamily which, until recently, had been presumed to be solely responsible for depalmitoylation of all intracellular S-palmitoylated proteins. However, knockdown or inhibition of APT1/APT2 did not alter turnover of palmitate on N-Ras or PSD-95. This finding led to the identification of ABDH17 proteins (isoforms A,B,C), palmitoylated serine hydrolase family members that are capable of catalyzing depalmitoylation of N-Ras and PSD95 and altering palmitate turnover on N-Ras[83]. Using a lipid hydrolase inhibitor and activity based protein profiling, an additional 20 members of the serine hydrolase family have also been implicated as potential depalmitoylating enzymes [52].
4.5 Functions of S-palmitoylation
For proteins that lack a transmembrane domain(s), one of the obvious functions of palmitate is to promote membrane binding of the modified protein. Palmitate is sufficiently hydrophobic so that by itself, or in combination with additional lipids, strong association with a lipid bilayer is achieved. In combination with kinetic trapping, as discussed above, palmitoylation provides not only membrane binding but also membrane targeting. Due to the labile nature of the thioester bond, reversible cycles of palmitoylation/depalmitoylation allow this class of lipidated proteins to dynamically cycle between membranes and the cytosol. The best studied examples of reversible membrane binding are H-Ras and N-Ras, small G proteins that are dually lipidated with farnesyl and palmitate[84, 85]. H- and N-Ras are first prenylated by farnesyl transferase in the cytosol. The C-terminal CAAX box is further processed in the ER by removal of the three C-terminal amino acids and C-terminal carboxymethylation. Upon transport to the Golgi, one (N-Ras) or two (H-Ras) cysteines upstream from the C-terminal farnesyl are palmitoylated by DHHC9/GCP16, a DHHC enzyme complex localized to the Golgi. Vesicles generated from the Golgi are trafficked through the secretory pathway to the plasma membrane, where Ras proteins can be released by depalmitoylation and traffic back to the Golgi by diffusion. The dynamic cycle of acylation/deacylation maintains H-Ras and N-Ras at both the plasma membrane and the Golgi, where they encounter different sets of effector proteins and can activate different signaling pathways[86].
Palmitoylation plays an important role in directing proteins to lipid rafts, membrane subdomains that are highly enriched in cholesterol, glycosphingolipids, and phospholipids containing saturated fatty acid chains. The saturated nature of the fatty acid preferentially drives insertion of palmitoylated proteins into the liquid ordered phase found in raft lipids[74, 78, 87]. For example, Src family tyrosine kinases that are dually acylated with myristate and palmitate are raft associated, whereas substitution of palmitate with mono- or polyunsaturated fatty acids reduces raft localization. For these proteins, which would be soluble in the absence of fatty acylation, palmitate plays a triple role: membrane binding, plasma membrane targeting, and lateral targeting to lipid rafts within the plasma membrane.
There are numerous examples of proteins that contain one or more transmembrane domains (TMDs) that are also S-palmitoylated[26]. The role of palmitate in these cases goes beyond just providing a membrane anchor[88]. For proteins contain TMDs longer than the standard 20 amino acid TMD, hydrophobic mismatch would result from extending a TMD beyond the boundary of the lipid bilayer. S-palmitoylation can address this energetically unfavorable situation in two ways. One option is to promote partitioning of transmembrane proteins into membrane rafts within the ER, as the liquid ordered domain region of the membrane is thicker than the bulk, disordered membrane region. Budding of raft-enriched transport vesicles has been proposed as a sorting signal to direct proteins such as LAT to the plasma membrane[89]. For proteins that have palmitoylation sites adjacent to TMDs (eg LRP6), palmitate may promote tilting of the transmembrane helix to allow proteins with long TMDs to fit within a thinner ER membrane[88]. In both cases, non-palmitoylated mutants would be retained in the ER and exhibit a mislocalization phenotype.
S-palmitoylation can influence protein structure and function by providing an additional anchoring point to the membrane. For example, palmitate-mediated tethering to the bilayer of a region close to a TMD in the γ subunit of the epithelial sodium channel regulates ion channel activity by stabilizing the open state of the channel[90]. When palmitate is present within a long cytoplasmic region, an additional loop will be formed when palmitate inserts into the bilayer. This has been shown to regulate GPCR activity by altering the receptor's ability to bind to different signaling effectors [91].
S-palmitoylation can also influence protein function by altering the accessibility of this region to other post-translational modifications. There are numerous examples of palmitoylation interfering or competing with phosphorylation or ubiquitination at nearby sites [88, 92]. Moreover, analysis of the postsynaptic density protein PSD-95 reveals that modification of the same cysteine residue can be reciprocally regulated by S-nitrosylation vs S-palmitoylation [93].Another mode of regulation is by direct effects on protein-protein interactions. For example, palmitoylation enhances specific interactions between synaptotagmins or integrins and tetraspanins[94, 95]. Finally, at least two proteins have been identified that contain palmitate buried intramolecularly in a deep hydrophobic pocket within the protein. BET3 is a component of the transport protein particle (TRAPP) tethering complex that regulates recruitment and trafficking of of ER-derived vesicles to and through the Golgi. Bet3 exhibits autopalmitoylation activity, which attaches palmitate to an internal cysteine residue. The fatty acid inserts into a hydrophobic groove, where it is required to maintain protein stability[96, 97]. TEAD transcription factors, together with their co-activators YAP and TAX, regulate the Hippo signaling pathway. When purified from E. coli, TEAD proteins were shown to contain covalently bound palmitate, which was attached as a result of autopalmitoylation activity. Mutation of the palmitoylation site in TEAD has no effect on nuclear envelope localization or membrane binding, but reduces interaction with its coactivators and impairs transcriptional activity[98, 99].
5. Fatty acylation of secreted proteins by MBOAT enzymes
5.1 N-palmitoylation of hedgehog proteins
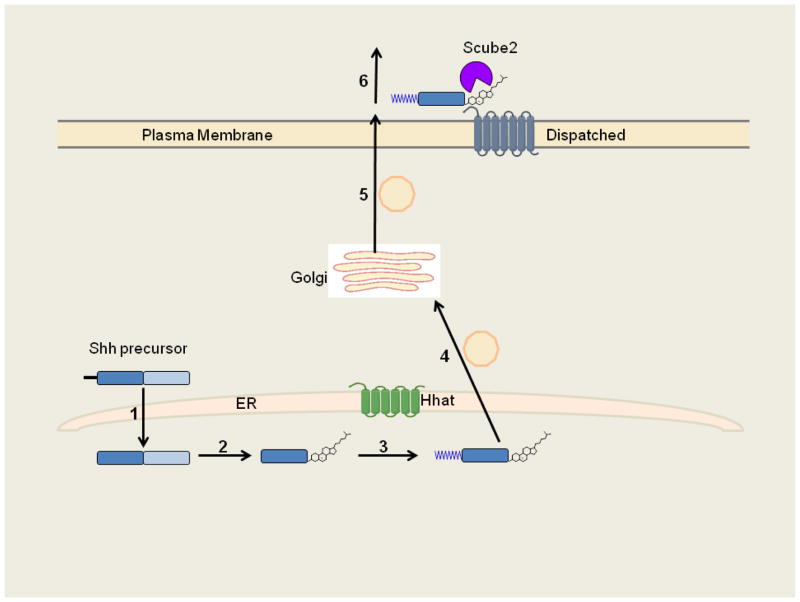
Studies of the hedgehog family of proteins revealed a new mode of attachment for palmitate, via amide linkage to an N-terminal cysteine[100]. Hedgehog proteins (herein referred to as Hh) are secreted from the producing cell and function as morphogens to signal to surrounding cells in a concentration dependent manner. This signaling protein family (hedgehog in flies, Sonic, Indian and Desert hedgehog in mammals,) plays critical roles in embryonic patterning during development and tumorigenesis in adults. Hh is synthesized as a 45kDa precursor protein containing an N-terminal signal sequence[101]. Upon entry into the ER, the N-terminal signal sequence is cleaved by signal peptidase (Figure 2). Hh contains an intein-like autoprocessing domain which promotes autocleavage. The C-terminal 25 kDa fragment is degraded in the lumen of the ER. The remaining 19 kDa N-terminal fragment is further modified by attachment of two lipids. Cholesterol is covalently attached to the C-terminus, and this occurs concomitantly with autocleavage. In a separate, distinct reaction, palmitate is attached through an amide bond formed between the fatty acid and the amino group of the N-terminal cysteine. The end result is a dually lipidated, mature signaling protein.
Figure 2. Fatty acylation and secretion of lipidated Sonic Hedgehog.
Shh undergoes multiple processing steps: 1) The Shh precursor polypeptide enters the lumen of the ER, where the signal peptide is removed. 2) Autocleavage and cholesterol attachment. 3) Hhat catalyzes palmitoylation of the Shh N-terminus. 4,5) Shh undergoes vesicular trafficking from the ER (step 4) through the Golgi (step 5) and is delivered to the cell surface. 6) Shh release is promoted by interaction with Dispatched and Scube2.
N-palmitoylation of Hh is catalyzed by Hhat (Hedgehog acyltransferase) a 57 kDa membrane protein that primarily resides in the ER[102, 103]. The fatty acylation reaction occurs in the lumen of the ER and requires prior removal of the N-terminal signal sequence. N-palmitoylation is independent of C-terminal cholesteroylation; the reaction can occur on the unprocessed Hh precursor in the absence of autocleavage and cholesterol attachment [102]. Hhat has been purified to homogeneity and shown to function as a bona fide palmitoyl acyltransferase with specificity for hedgehog proteins[102]. The N-terminal region of Hh contains the sequence information necessary for substrate recognition: a 10 amino acid peptide can be palmitoylated by Hhat in vitro[102], and fusion of the first 6 amino acids of the mature Hh sequence to GFP is sufficient to confer N-palmitoylation to a GFP fusion protein when expressed in cells[104]. Both Hhat and the fly ortholog Rasp require the presence of a positively charged amino acid within the N-terminal sequence for N-palmitoylation to occur[104].
Deletion of the Hhat gene in mice is embryonic lethal, and recapitulates the phenotype of Shh knockout mice[105] . Similar results were obtained for Hh and Rasp deletion in flies[106, 107]. Notably, a Hh mutant that cannot be palmitoylated cannot rescue the Rasp −/− phenotype. These findings indicate that palmitoylation is required for Hh signaling activity during development. In addition to Hh, the EGF-like ligands Spitz, Keren and Gurken are substrates for N-palmitoylation by Rasp in flies[108]. To date, no mammalian equivalents that are N-palmitoylated have been found. By contrast, cholesterol attachment to Hh helps shape the morphogenetic signaling gradient, but is not absolutely essential for Hh signaling in vivo[109].
The presence of two lipid modifications on a protein leads to the question: how is such a hydrophobic molecule released from the cell? One possibility is that the lipids are removed. Cleavage of both N- and C-terminal residues from Shh by metalloproteases has been observed[110], but it is not clear if this type of “shedding” is universally used. The more likely mechanism is by regulated release (Figure 2). Hh producing cells contain two proteins, Dispatched and Scube2, whose functions are to bind and promote release of lipidated Hh into the surrounding media[111, 112]. In the absence of either protein, Hh remains bound to the cell surface. Dispatched is a plasma membrane-bound protein with a sterol sensing domain that recognizes the aliphatic chain of cholesterol bound to Hh. Scube 2, a secreted protein, recognizes the cholesterol ring structure and its efficiency in promoting Hh release is enhanced by Hh palmitoylation. Once lipidated Hh is released from the cell, it forms multimers in the extracellular milieu, with the hydrophobic groups facing the inside of a micellar structure[113]. These multimeric complexes are presumed to be the active signaling species, but alternative models where Hh travels with lipoprotein particles or is released from exosomes on filopodia-like extensions termed cytonemes have also been proposed[114–116].
5.2 Wnt proteins are palmitoleoylated
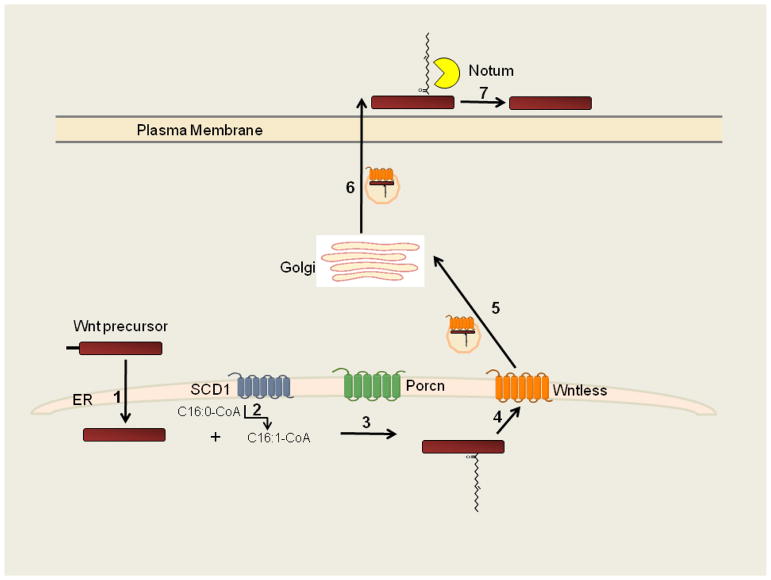
Wnt proteins are modified by a monounsaturated form of palmitate, cis-Δ9-palmitoleate (C16:1”9), which is attached via oxyester linkage to a conserved serine residue (Ser209 in Wnt3a)[117, 118]. Wnts comprise a family of cysteine-rich, secreted glycoproteins that regulate embryonic patterning during development as well as stem cell and tissue renewal in adults. Palmitoleoylation is required for Wnt proteins to bind to Wntless, an intracellular transmembrane protein that promotes Wnt secretion by facilitating Wnt movement from the trans Golgi network to the plasma membrane[119, 120]. In addition, palmitoleate binds within a hydrophobic cleft of the Wnt receptor Frizzled, promoting ligand-receptor interactions[121]. Thus, this monounsaturated palmitate analog is required for Wnt trafficking, secretion, receptor binding and signaling.
The enzyme that catalyzes Wnt protein acylation is Porcupine (Porcn), a multipass membrane protein that is predicted to contain 11 TMDs [117, 122, 123]. The reaction occurs in the lumen of the ER in two steps (Figure 3)[124]. First, stearoyl CoA desaturase (SCD) inserts a cis-double bond at fatty acid position 9 of palmitoyl CoA to form palmitoleoyl-CoA. This monounsaturated fatty acyl CoA is the substrate for Porcn, which transfers the 16:1 fatty acid to the recipient cysteine in Wnt proteins. Porcn can attach monounsaturated fatty acids that are shorter, but not longer than 16 carbons to Wnt proteins[124, 125], and recognizes a consensus sequence surrounding the conserved cysteine in Wnt proteins[126, 127].
Figure 3. Fatty acylation and secretion of palmitoleoylated Wnt proteins.
1) The Wnt precursor enters the lumen of the ER, where the signal peptide is cleaved. 2) SCD1 catalyzes formation of palmitoleoyl-CoA from palmitoyl-CoA. 3) Porcn catalyzes attachment of palmitoleoylate to Wnt. 4) Palmitoleoylated Wnt interacts with Wntless and (5) is packaged into vesicles that transit through the Golgi and then to the plasma membrane (6), where palmitoleoylated Wnt is released. 7) In some cells, Notum releases the fatty acid from Wnt.
The presence of a fatty acid on Wnt poses the same problem as it does for Hh: how is Wnt released from the cell and how does it travel and signal at multiple cell distances? Several new insights into these questions have recently been obtained. Wnt proteins can be detected in the conditioned media from cultured cells, and mechanisms including secretion on exosomes or binding to lipoprotein particles or Wnt interacting proteins have been proposed to carry Wnt proteins through the extracellular milieu[128–130]. However, when Wingless, the major Drosophila Wnt protein, was tethered to the membrane by fusion with a transmembrane protein, embryonic patterning still occurred in a manner similar to that mediated by wild type Wingless[131]. This suggests that short range signaling is critical during development. In organoids formed from mouse intestinal cells, Wnt is apparently transferred from the producing cell to a neighboring cell, where it remains on the cell surface bound to Frizzled[132]. A short range Wnt gradient is then propagated and diluted by cell division.
Wnt palmitoyleolation has recently been shown to be reversible. The fatty acid can be cleaved from secreted Wnt by Notum, a secreted Wnt antagonist that is a member of the α/β hydrolase superfamily[133, 134]. The deacylase activity is specific for Wnt proteins, as Hh is not depalmitoylated by Notum. The shape of the binding pocket in Notum restricts binding to cis-monounsaturated fatty acids, thereby explaining selectivity for Wnt proteins. By removing palmitoleate, which is essential for Wnt binding to Frizzled, deacylation by Notum inactivates Wnt signaling in vitro and in vivo.
5.3 Ghrelin is modified by octanoate
The eight-carbon fatty acid, octanoate, is covalently attached to ghrelin, a secreted 28 amino acid peptide hormone. Ghrelin is synthesized primarily in the stomach. The mature peptide is generated by proteolytic processing from a preprohormone precursor. Fatty acylation is required to produce the active octanoylated form. Only octanoylated ghrelin can bind to the growth hormone secretagogue type 1a receptor, a G protein coupled receptor. The physiologic outputs include growth hormone secretion, appetite stimulation, and increased food uptake and adiposity. Octanoylation of ghrelin is catalyzed by GOAT (ghrelin O-acyltransferase), a multipass membrane protein localized primarily in the gastrointestinal tract[135, 136]. The fatty acid is linked to serine-3 within the N-terminal GSSFL sequence, with glycine-1 and phenylalanine-4 also contributing to enzyme-substrate recognition[137]. Bioinformatics analyses suggest that ghrelin is the only substrate for GOAT in the human proteome.
5.4 How are Fatty Acyl CoA substrates delivered to and recognized by MBOAT enzymes?
Hhat, Porcn and GOAT are members of the MBOAT (Membrane bound O-acyl transferase) family of multipass membrane proteins[138, 139]. Of the 16 MBOAT family members in humans, most catalyze transfer of fatty acids to lipids; only 3 (Hhat, Porcn, GOAT) transfer fatty acids to proteins. The substrates for these three enzymes are secreted proteins, which enter the lumen of the ER via a signal sequence. The fatty acyl donors are the CoA derivatives. In order for fatty acylation to occur, fatty acyl CoA must also be present in the ER lumen. Fatty acids are esterified to fatty acyl CoA in the cytosol, but long chain acyl-CoA esters are not permeable across biological membranes. Thus, a key unanswered question is: how do the long chain fatty acyl CoA substrates access the lumen of the ER? Mitochondria solve this problem with a carnitine-dependent acylation and transfer system: carnitine palmitoyl-transferase1 (CPT1) generates fatty acylcarnitine from fatty acyl CoAs in the cytosol; acylcarnitine traverses the inner membrane bilayer via a carnitine/acylcarnitine exchange carrier (CAC); CPT2 re-forms fatty acyl CoAs in the organelle interior. Carnitine acyltransferase and acylcarnitine exchange activities have been detected and characterized in microsomal membranes[140] but it is not known whether the microsomal enzymes are splice variants of the mitochondrial enzymes or related proteins. Alternatively, it is possible that MBOAT proteins themselves facilitate fatty acyl CoA transport across the ER membrane. Studies of membrane topology reveal that Hhat contains ten TMDs and two reentrant loops[141, 142], with a putative active site histidine disposed near the lumenal surface. Of note, a large fraction of the Hhat sequence is located on the cytosolic side of the ER membrane. Studies of GOAT TMDs reveal a similar membrane topology[143]. The function of the cytosolically disposed loops of these enzymes is unknown, but it is tempting to speculate that these regions may regulate substrate availability and/or selectivity.
MBOAT family members were defined by their O-acyl transferase activity. Porcn and GOAT attach fatty acids via O-acylation to serine in their respective substrates, but threonine has also been shown to serve as an acylation acceptor. At first glance, Hhat is different in that palmitate is attached to cysteine via amide bond. However, in a cell based palmitoylation assay, Shh with an N-terminal serine can also serve as a palmitate acceptor, but at reduced efficiency[104]. This finding may explain why N-terminal cysteine to serine mutants of Shh retain residual signaling activity. Use of a cysteine to alanine mutant is a better choice to mimic fully inactive Shh.
6. Fatty acyl transferases as targets in human diseases
Fatty acylated proteins are critical players in signaling in both normal cells and disease states. Targeted inhibitors that selectively block specific fatty acyl transferases are therefore of great therapeutic interest. The key is to be able to block modification of the protein(s) responsible for the disease state without affecting housekeeping proteins that need to be fatty acylated to maintain normal cellular homeostasis. To date, this approach has been successful for NMT and the MBOAT proteins.
N-myristoylation is necessary for the growth and survival of a number of human pathogens, and many of these organisms encode their own NMTs. Although the myristoyl-CoA binding site is highly conserved between human and pathogen NMTs, the peptide binding sites are quite different. By taking advantage of these differences, NMT has been targeted in Candida albicans, Trypanosoma brucei, and Leishmania major, organisms that cause fungal infections, African sleeping sickness, and leishmaniasis, respectively[144–146].
The Hh signaling pathway is highly active during embryonic development, but is largely shut off in adult tissues. However, aberrant overexpression of Shh has been shown to drive proliferation and metastasis in numerous human tumors, including pancreatic, breast and lung cancers. Thus, Hhat has the potential to provide a tumor-specific target. Using high-throughput screening and an Shh palmitoylation assay as a readout, potent, selective Hhat inhibitors have been identified and shown to block the growth of pancreatic adenocarcinoma cells as well as estrogen receptor positive breast cancer cells[147–149].
Wnt driven cancers provide an attractive system for development of Porcn inhibitors. Proof of concept that small molecule inhibitors of Porcn can selectively inhibit Wnt signaling was first reported in 2009[150]. Since then, orally bioavailable Porcn inhibitors have been developed that show efficacy in Wnt-dependent cancers[151, 152]. Two optimized Porcn inhibitors, LGK974 and ETC-159, are currently in Phase I clinical trials for potential treatment of cancers dependent on Wnt ligand.
Weight gain is stimulated by acyl-ghrelin, making GOAT an obvious target for potential control of obesity and obesity-associated disorders. A bisubstrate analog, GO-CoA-Tat, linking octanoyl CoA and an N-terminal ghrelin peptide, was shown to be a selective GOAT inhibitor in vitro and in vivo and prevented weight gain in mice fed a high fat diet[153]. However, studies in GOAT knockout mice have not mirrored the effects of the GOAT inhibitor. GOAT −/− mice grow at the same rate and achieve the same weight as wild type cohorts, even on a high fat diet[154, 155]. Of note, GOAT −/− mice exhibit reduced food intake and resistance to obesity when placed on a high-fat, high-sucrose diet[155]. Complex interactions with other signaling axes that regulate appetite may complicate the ultimate in vivo use of GOAT inhibitors to control obesity and/or diabetes.
7. Conclusions and perspectives
We have now come to appreciate that protein fatty acylation is not limited to modification by just myristate or palmitate, but encompasses a broad range of saturated and unsaturated fatty acids of varying chain length. We also understand that distinct enzymes are responsible for catalyzing subsets of these fatty acylation reactions. In some cases, there is a dedicated fatty acyltransferase, eg NMT for N-myristoylation, Hhat for Hh, Porcn for Wnts, and GOAT for ghrelin. What we do not understand is why there are so many DHHC enzymes and why there is so much potential redundancy in the S-palmitoylation machinery. We also cannot explain how palmitoylation sites are selected, and how and why palmitate turns over rapidly on some proteins, but not on others. Advances in proteomics have led to identification of even more (potential) fatty acylated proteins, but are we done yet? Are there additional fatty acylated proteins, acyltransferases and/or deacylating enzymes yet to be identified?
Another key concept is that not all fat looks the same to the cell. Myristate and palmitate differ by just two CH2 groups, but have different hydrophobicities and are recognized as substrates by different enzymes at different times and in different places. Palmitate and palmitoleoylate differ by one double bond, but yet only 16:1 is recognized by Porcn and attached to Wnt proteins. The function of heterogeneous fatty acylation is still not clear. What does stearate do to alter the function of the transferrin receptor that is not accomplished by palmitate? Can dietary intervention be successfully used to alter the palette of fatty acids attached to signaling proteins and thereby affect cell signaling?
MBOAT proteins that catalyze fatty acylation of secreted proteins are attractive drug targets in disease, but there is still much to be learned about these enzymes and their acylated substrates. How is the fatty acyl CoA delivered to the lumen of the ER for presentation to the MBOAT enzyme active site? How is release of the lipidated protein into the extracellular milieu regulated and how do lipidated secreted morphogens travel in the aqueous environment? Continued advances in fatty acylation detection methods as well as live imaging promise to deliver answers to some of these questions.
Acknowledgments
Research in the author's laboratory is supported by grants from the National Institutes of Health (CA186957, GM116860), The Geoffrey Beene Cancer Research Center of Memorial Sloan Kettering Cancer Center, the Tri-Institutional Therapeutic Discovery Institute, and the Hirshberg Foundation for Pancreatic Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Resh MD. Covalent lipid modifications of proteins. Curr Biol. 2013;23(10):R431–5. doi: 10.1016/j.cub.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duronio RJ, Towler DA, Heuckeroth RO, Gordon JI. Disruption of the yeast N-myristoyl transferase gene causes recessive lethality. Science. 1989;243(4892):796–800. doi: 10.1126/science.2644694. [DOI] [PubMed] [Google Scholar]
- 3.Giang DK, Cravatt BF. A second mammalian N-myristoyltransferase. J Biol Chem. 1998;273(12):6595–8. doi: 10.1074/jbc.273.12.6595. [DOI] [PubMed] [Google Scholar]
- 4.Yang SH, Shrivastav A, Kosinski C, Sharma RK, Chen MH, Berthiaume LG, Peters LL, Chuang PT, Young SG, Bergo MO. N-myristoyltransferase 1 is essential in early mouse development. J Biol Chem. 2005;280(19):18990–5. doi: 10.1074/jbc.M412917200. [DOI] [PubMed] [Google Scholar]
- 5.Ducker CE, Upson JJ, French KJ, Smith CD. Two N-myristoyltransferase isozymes play unique roles in protein myristoylation, proliferation, and apoptosis. Mol Cancer Res. 2005;3(8):463–76. doi: 10.1158/1541-7786.MCR-05-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudnick DA, McWherter CA, Rocque WJ, Lennon PJ, Getman DP, Gordon JI. Kinetic and structural evidence for a sequential ordered Bi Bi mechanism of catalysis by Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J Biol Chem. 1991;266(15):9732–9. [PubMed] [Google Scholar]
- 7.Bhatnagar RS, Futterer K, Farazi TA, Korolev S, Murray CL, Jackson-Machelski E, Gokel GW, Gordon JI, Waksman G. Structure of N-myristoyltransferase with bound myristoylCoA and peptide substrate analogs. Nat Struct Biol. 1998;5(12):1091–1097. doi: 10.1038/4202. [DOI] [PubMed] [Google Scholar]
- 8.Bhatnagar RS, Futterer K, Waksman G, Gordon JI. The structure of myristoyl-CoA:protein N-myristoyltransferase. Biochim Biophys Acta. 1999;1441(2–3):162–72. doi: 10.1016/s1388-1981(99)00155-9. [DOI] [PubMed] [Google Scholar]
- 9.Rosendal J, Ertbjerg P, Knudsen J. Characterization of ligand binding to acyl-CoA-binding protein. Biochem J. 1993;290( Pt 2):321–6. doi: 10.1042/bj2900321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peitzsch RM, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry (Mosc) 1993;32(39):10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 11.Heuckeroth RO, Glaser L, Gordon JI. Heteroatom-substituted fatty acid analogs as substrates for N-myristoyltransferase: an approach for studying both the enzymology and function of protein acylation. Proc Natl Acad Sci U S A. 1988;85(23):8795–9. doi: 10.1073/pnas.85.23.8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323( Pt 1):1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasmussen JT, Faergeman NJ, Kristiansen K, Knudsen J. Acyl-CoA-binding protein (ACBP) can mediate intermembrane acyl-CoA transport and donate acyl-CoA for beta-oxidation and glycerolipid synthesis. Biochem J. 1994;299( Pt 1):165–70. doi: 10.1042/bj2990165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soupene E, Kao J, Cheng DH, Wang D, Greninger AL, Knudsen GM, DeRisi JL, Kuypers FA. Association of NMT2 with the acyl-CoA carrier ACBD6 protects the N-myristoyltransferase reaction from palmitoyl-CoA. J Lipid Res. 2016;57(2):288–98. doi: 10.1194/jlr.M065003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin DD, Beauchamp E, Berthiaume LG. Post-translational myristoylation: Fat matters in cellular life and death. Biochimie. 2011;93(1):18–31. doi: 10.1016/j.biochi.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Martin DD, Hayden MR. Post-translational myristoylation at the cross roads of cell death, autophagy and neurodegeneration. Biochem Soc Trans. 2015;43(2):229–34. doi: 10.1042/BST20140281. [DOI] [PubMed] [Google Scholar]
- 17.Vilas GL, Corvi MM, Plummer GJ, Seime AM, Lambkin GR, Berthiaume LG. Posttranslational myristoylation of caspase-activated p21-activated protein kinase 2 (PAK2) potentiates late apoptotic events. Proc Natl Acad Sci U S A. 2006;103(17):6542–7. doi: 10.1073/pnas.0600824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 2000;290(5497):1761–5. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 19.Thinon E, Serwa RA, Broncel M, Brannigan JA, Brassat U, Wright MH, Heal WP, Wilkinson AJ, Mann DJ, Tate EW. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat Commun. 2014;5:4919. doi: 10.1038/ncomms5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2(11):584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 21.Sigal CT, Zhou W, Buser CA, McLaughlin S, Resh MD. Amino-terminal basic residues of Src mediate membrane binding through electrostatic interaction with acidic phospholipids. Proc Natl Acad Sci U S A. 1994;91(25):12253–12257. doi: 10.1073/pnas.91.25.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68(4):2556–69. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20(7):272–6. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 24.Harroun TA, Bradshaw JP, Balali-Mood K, Katsaras J. A structural study of the myristoylated N-terminus of ARF1. Biochim Biophys Acta. 2005;1668(1):138–44. doi: 10.1016/j.bbamem.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Bachert C, Linstedt AD. Dual anchoring of the GRASP membrane tether promotes trans pairing. J Biol Chem. 2010;285(21):16294–301. doi: 10.1074/jbc.M110.116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci STKE. 2006;2006(359):re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- 27.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8(1):74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 28.Fraser ID, Tavalin SJ, Lester LB, Langeberg LK, Westphal AM, Dean RA, Marrion NV, Scott JD. A novel lipid-anchored A-kinase Anchoring Protein facilitates cAMP-responsive membrane events. EMBO J. 1998;17(8):2261–72. doi: 10.1093/emboj/17.8.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burnaevskiy N, Fox TG, Plymire DA, Ertelt JM, Weigele BA, Selyunin AS, Way SS, Patrie SM, Alto NM. Proteolytic elimination of N-myristoyl modifications by the Shigella virulence factor IpaJ. Nature. 2013;496(7443):106–9. doi: 10.1038/nature12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnaevskiy N, Peng T, Reddick LE, Hang HC, Alto NM. Myristoylome profiling reveals a concerted mechanism of ARF GTPase deacylation by the bacterial protease IpaJ. Mol Cell. 2015;58(1):110–22. doi: 10.1016/j.molcel.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ames JB, Tanaka T, Stryer L, Ikura M. Portrait of a myristoyl switch protein. Curr Opin Struct Biol. 1996;6(4):432–438. doi: 10.1016/s0959-440x(96)80106-0. [DOI] [PubMed] [Google Scholar]
- 32.Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95(2):237–48. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 33.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, Perez de Castro I, Li C, Thompson CB, Cox AD, Philips MR. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21(4):481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Shishido T, Jiang X, Aderem A, McLaughlin S. Phosphorylation, high ionic strength, and calmodulin reverse the binding of MARCKS to phospholipid vesicles. J Biol Chem. 1994;269(45):28214–9. [PubMed] [Google Scholar]
- 35.Ohmori S, Sakai N, Shirai Y, Yamamoto H, Miyamoto E, Shimizu N, Saito N. Importance of protein kinase C targeting for the phosphorylation of its substrate, myristoylated alanine-rich C-kinase substrate. J Biol Chem. 2000;275(34):26449–57. doi: 10.1074/jbc.M003588200. [DOI] [PubMed] [Google Scholar]
- 36.Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci U S A. 2004;101(2):517–22. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A. 2006;103(30):11364–9. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112(6):859–71. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 39.Patwardhan P, Resh MD. Myristoylation and membrane binding regulate c-Src stability and kinase activity. Mol Cell Biol. 2010;30(17):4094–107. doi: 10.1128/MCB.00246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toska E, Campbell HA, Shandilya J, Goodfellow SJ, Shore P, Medler KF, Roberts SG. Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell reports. 2012;2(3):462–9. doi: 10.1016/j.celrep.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson FT, Bursten SL, Locksley RM, Lovett DH. Myristyl acylation of the tumor necrosis factor alpha precursor on specific lysine residues. J Exp Med. 1992;176(4):1053–62. doi: 10.1084/jem.176.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson FT, Bursten SL, Fanton C, Locksley RM, Lovett DH. The 31-kDa precursor of interleukin 1 alpha is myristoylated on specific lysines within the 16-kDa N-terminal propiece. Proc Natl Acad Sci U S A. 1993;90(15):7245–9. doi: 10.1073/pnas.90.15.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson JP, Raghavan AS, Yang YY, Charron G, Hang HC. Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol Cell Proteomics. 2011;10(3):M110 001198. doi: 10.1074/mcp.M110.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6(5):812–9. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu B, Lin Y, Darwanto A, Song X, Xu G, Zhang K. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J Biol Chem. 2009;284(47):32288–95. doi: 10.1074/jbc.M109.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, Lin H. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496(7443):110–3. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teng YB, Jing H, Aramsangtienchai P, He B, Khan S, Hu J, Lin H, Hao Q. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci Rep. 2015;5:8529. doi: 10.1038/srep08529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288(43):31350–6. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Yang T, Li X, Peng T, Hang HC, Li XD. Integrative chemical biology approaches for identification and characterization of “erasers” for fatty-acid-acylated lysine residues within proteins. Angew Chem Int Ed Engl. 2015;54(4):1149–52. doi: 10.1002/anie.201408763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morrison E, Kuropka B, Kliche S, Brugger B, Krause E, Freund C. Quantitative analysis of the human T cell palmitome. Sci Rep. 2015;5:11598. doi: 10.1038/srep11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9(1):84–9. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dowal L, Yang W, Freeman MR, Steen H, Flaumenhaft R. Proteomic analysis of palmitoylated platelet proteins. Blood. 2011;118(13):e62–73. doi: 10.1182/blood-2011-05-353078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, Lopez CB, Hang HC. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6(8):610–4. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marin EP, Derakhshan B, Lam TT, Davalos A, Sessa WC. Endothelial cell palmitoylproteomic identifies novel lipid-modified targets and potential substrates for protein acyl transferases. Circ Res. 2012;110(10):1336–44. doi: 10.1161/CIRCRESAHA.112.269514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumari B, Kumar R, Kumar M. PalmPred: an SVM based palmitoylation prediction method using sequence profile information. PLoS One. 2014;9(2):e89246. doi: 10.1371/journal.pone.0089246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blanc M, David F, Abrami L, Migliozzi D, Armand F, Burgi J, van der Goot FG. SwissPalm: Protein Palmitoylation database. F1000Res. 2015;4:261. doi: 10.12688/f1000research.6464.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47(6):1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 59.Linder ME, Jennings BC. Mechanism and function of DHHC S-acyltransferases. Biochem Soc Trans. 2013;41(1):29–34. doi: 10.1042/BST20120328. [DOI] [PubMed] [Google Scholar]
- 60.Fernandez-Hernando C, Fukata M, Bernatchez PN, Fukata Y, Lin MI, Bredt DS, Sessa WC. Identification of Golgi-localized acyl transferases that palmitoylate and regulate endothelial nitric oxide synthase. J Cell Biol. 2006;174(3):369–77. doi: 10.1083/jcb.200601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greaves J, Chamberlain LH. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem Sci. 2011;36(5):245–53. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 62.Huang K, Yanai A, Kang R, Arstikaitis P, Singaraja RR, Metzler M, Mullard A, Haigh B, Gauthier-Campbell C, Gutekunst CA, Hayden MR, El-Husseini A. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 2004;44(6):977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 63.Jennings BC, Linder ME. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J Biol Chem. 2012;287(10):7236–45. doi: 10.1074/jbc.M111.337246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singaraja RR, Huang K, Sanders SS, Milnerwood AJ, Hines R, Lerch JP, Franciosi S, Drisdel RC, Vaid K, Young FB, Doty C, Wan J, Bissada N, Henkelman RM, Green WN, Davis NG, Raymond LA, Hayden MR. Altered palmitoylation and neuropathological deficits in mice lacking HIP14. Hum Mol Genet. 2011;20(20):3899–909. doi: 10.1093/hmg/ddr308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sutton LM, Sanders SS, Butland SL, Singaraja RR, Franciosi S, Southwell AL, Doty CN, Schmidt ME, Mui KK, Kovalik V, Young FB, Zhang W, Hayden MR. Hip14l-deficient mice develop neuropathological and behavioural features of Huntington disease. Hum Mol Genet. 2013;22(3):452–65. doi: 10.1093/hmg/dds441. [DOI] [PubMed] [Google Scholar]
- 66.Marin EP, Jozsef L, Di Lorenzo A, Held KF, Luciano AK, Melendez J, Milstone LM, Velazquez H, Sessa WC. The Protein Acyl Transferase ZDHHC21 Modulates alpha1 Adrenergic Receptor Function and Regulates Hemodynamics. Arterioscler Thromb Vac Biol. 2016;36(2):370–9. doi: 10.1161/ATVBAHA.115.306942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mill P, Lee AW, Fukata Y, Tsutsumi R, Fukata M, Keighren M, Porter RM, McKie L, Smyth I, Jackson IJ. Palmitoylation regulates epidermal homeostasis and hair follicle differentiation. PLoS genetics. 2009;5(11):e1000748. doi: 10.1371/journal.pgen.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J Biol Chem. 2010;285(49):38104–14. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gottlieb CD, Zhang S, Linder ME. The Cysteine-rich Domain of the DHHC3 Palmitoyltransferase Is Palmitoylated and Contains Tightly Bound Zinc. J Biol Chem. 2015;290(49):29259–69. doi: 10.1074/jbc.M115.691147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gonzalez Montoro A, Chumpen Ramirez S, Valdez Taubas J. The canonical DHHC motif is not absolutely required for the activity of the yeast S-acyltransferases Swf1 and Pfa4. J Biol Chem. 2015;290(37):22448–59. doi: 10.1074/jbc.M115.651356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai FD, Lopes MS, Zhou M, Court H, Ponce O, Fiordalisi JJ, Gierut JJ, Cox AD, Haigis KM, Philips MR. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci U S A. 2015;112(3):779–84. doi: 10.1073/pnas.1412811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nishimura A, Linder ME. Identification of a novel prenyl and palmitoyl modification at the CaaX motif of Cdc42 that regulates RhoGDI binding. Mol Cell Biol. 2013;33(7):1417–29. doi: 10.1128/MCB.01398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schroeder H, Leventis R, Rex S, Schelhaas M, Nagele E, Waldmann H, Silvius JR. S-Acylation and plasma membrane targeting of the farnesylated carboxyl-terminal peptide of N-ras in mammalian fibroblasts. Biochemistry (Mosc) 1997;36(42):13102–13109. doi: 10.1021/bi9709497. [DOI] [PubMed] [Google Scholar]
- 74.Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P, Resh MD. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem. 2001;276(33):30987–30994. doi: 10.1074/jbc.M104018200. [DOI] [PubMed] [Google Scholar]
- 75.Liang X, Lu Y, Neubert TA, Resh MD. Mass spectrometric analysis of GAP-43/neuromodulin reveals the presence of a variety of fatty acylated species. J Biol Chem. 2002;277(36):33032–33040. doi: 10.1074/jbc.M204607200. [DOI] [PubMed] [Google Scholar]
- 76.Montigny C, Decottignies P, Le Marechal P, Capy P, Bublitz M, Olesen C, Moller JV, Nissen P, le Maire M. S-palmitoylation and s-oleoylation of rabbit and pig sarcolipin. J Biol Chem. 2014;289(49):33850–61. doi: 10.1074/jbc.M114.590307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brett K, Kordyukova LV, Serebryakova MV, Mintaev RR, Alexeevski AV, Veit M. Site-specific S-acylation of influenza virus hemagglutinin: the location of the acylation site relative to the membrane border is the decisive factor for attachment of stearate. J Biol Chem. 2014;289(50):34978–89. doi: 10.1074/jbc.M114.586180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem. 2000;275(1):261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 79.Alvarez E, Girones N, Davis RJ. Inhibition of the receptor-mediated endocytosis of diferric transferrin is associated with the covalent modification of the transferrin receptor with palmitic acid. J Biol Chem. 1990;265(27):16644–55. [PubMed] [Google Scholar]
- 80.Senyilmaz D, Virtue S, Xu X, Tan CY, Griffin JL, Miller AK, Vidal-Puig A, Teleman AA. Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature. 2015;525(7567):124–8. doi: 10.1038/nature14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kong E, Peng S, Chandra G, Sarkar C, Zhang Z, Bagh MB, Mukherjee AB. Dynamic palmitoylation links cytosol-membrane shuttling of acyl-protein thioesterase-1 and acyl-protein thioesterase-2 with that of proto-oncogene H-ras product and growth-associated protein-43. J Biol Chem. 2013;288(13):9112–25. doi: 10.1074/jbc.M112.421073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vartak N, Papke B, Grecco HE, Rossmannek L, Waldmann H, Hedberg C, Bastiaens PI. The autodepalmitoylating activity of APT maintains the spatial organization of palmitoylated membrane proteins. Biophys J. 2014;106(1):93–105. doi: 10.1016/j.bpj.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin DT, Conibear E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. Elife. 2015;4 doi: 10.7554/eLife.11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307(5716):1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 85.Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, Kenworthy AK. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol. 2005;170(2):261–272. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bivona TG, Perez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, Cullen PJ, Pellicer A, Cox AD, Philips MR. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424(6949):694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 87.Lorent JH, Levental I. Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem Phys Lipids. 2015;192:23–32. doi: 10.1016/j.chemphyslip.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 88.Blaskovic S, Blanc M, van der Goot FG. What does S-palmitoylation do to membrane proteins? The FEBS journal. 2013;280(12):2766–74. doi: 10.1111/febs.12263. [DOI] [PubMed] [Google Scholar]
- 89.Diaz-Rohrer BB, Levental KR, Simons K, Levental I. Membrane raft association is a determinant of plasma membrane localization. Proc Natl Acad Sci U S A. 2014;111(23):8500–5. doi: 10.1073/pnas.1404582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mukherjee A, Mueller GM, Kinlough CL, Sheng N, Wang Z, Mustafa SA, Kashlan OB, Kleyman TR, Hughey RP. Cysteine palmitoylation of the gamma subunit has a dominant role in modulating activity of the epithelial sodium channel. J Biol Chem. 2014;289(20):14351–9. doi: 10.1074/jbc.M113.526020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miggin SM, Lawler OA, Kinsella BT. Palmitoylation of the human prostacyclin receptor. Functional implications of palmitoylation and isoprenylation. J Biol Chem. 2003;278(9):6947–58. doi: 10.1074/jbc.M210637200. [DOI] [PubMed] [Google Scholar]
- 92.Shipston MJ. Ion channel regulation by protein palmitoylation. J Biol Chem. 2011;286(11):8709–16. doi: 10.1074/jbc.R110.210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ho GP, Selvakumar B, Mukai J, Hester LD, Wang Y, Gogos JA, Snyder SH. S-nitrosylation and S-palmitoylation reciprocally regulate synaptic targeting of PSD-95. Neuron. 2011;71(1):131–41. doi: 10.1016/j.neuron.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang X, Kovalenko OV, Tang W, Claas C, Stipp CS, Hemler ME. Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J Cell Biol. 2004;167(6):1231–1240. doi: 10.1083/jcb.200404100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Flannery AR, Czibener C, Andrews NW. Palmitoylation-dependent association with CD63 targets the Ca2+ sensor synaptotagmin VII to lysosomes. J Cell Biol. 2010;191(3):599–613. doi: 10.1083/jcb.201003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kummel D, Heinemann U, Veit M. Unique self-palmitoylation activity of the transport protein particle component Bet3: a mechanism required for protein stability. Proc Natl Acad Sci U S A. 2006;103(34):12701–6. doi: 10.1073/pnas.0603513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Turnbull AP, Kummel D, Prinz B, Holz C, Schultchen J, Lang C, Niesen FH, Hofmann KP, Delbruck H, Behlke J, Muller EC, Jarosch E, Sommer T, Heinemann U. Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization. EMBO J. 2005;24(5):875–884. doi: 10.1038/sj.emboj.7600565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan P, Han X, Zheng B, DeRan M, Yu J, Jarugumilli GK, Deng H, Pan D, Luo X, Wu X. Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat Chem Biol. 2016;12(4):282–9. doi: 10.1038/nchembio.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Noland CL, Gierke S, Schnier PD, Murray J, Sandoval WN, Sagolla M, Dey A, Hannoush RN, Fairbrother WJ, Cunningham CN. Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling. Structure. 2016;24(1):179–86. doi: 10.1016/j.str.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 100.Pepinsky RB, Zeng C, Wen D, Rayhorn P, Baker DP, Williams KP, Bixler SA, Ambrose CM, Garber EA, Miatkowski K, Taylor FR, Wang EA, Galdes A. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem. 1998;273(22):14037–14045. doi: 10.1074/jbc.273.22.14037. [DOI] [PubMed] [Google Scholar]
- 101.Mann RK, Beachy PA. Novel lipid modifications of secreted protein signals. Annu Rev Biochem. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- 102.Buglino JA, Resh MD. Hhat is a palmitoyl acyltransferase with specificity for N-palmitoylation of sonic hedgehog. J Biol Chem. 2008;283:22076–22088. doi: 10.1074/jbc.M803901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chamoun Z, Mann RK, Nellen D, von Kessler DP, Bellotto M, Beachy PA, Basler K. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science. 2001;293(5537):2080–2084. doi: 10.1126/science.1064437. [DOI] [PubMed] [Google Scholar]
- 104.Hardy RY, Resh MD. Identification of N-terminal Residues of Sonic Hedgehog Important for Palmitoylation by Hedgehog Acyltransferase. J Biol Chem. 2012;287(51):42881–9. doi: 10.1074/jbc.M112.426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004;18(6):641–659. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee JD, Kraus P, Gaiano N, Nery S, Kohtz J, Fishell G, Loomis CA, Treisman JE. An acylatable residue of Hedgehog is differentially required in Drosophila and mouse limb development. Dev Biol. 2001;233(1):122–136. doi: 10.1006/dbio.2001.0218. [DOI] [PubMed] [Google Scholar]
- 107.Lee JD, Treisman JE. Sightless has homology to transmembrane acyltransferases and is required to generate active Hedgehog protein. Curr Biol. 2001;11(14):1147–1152. doi: 10.1016/s0960-9822(01)00323-2. [DOI] [PubMed] [Google Scholar]
- 108.Miura GI, Buglino J, Alvarado D, Lemmon MA, Resh MD, Treisman JE. Palmitoylation of the EGFR ligand Spitz by Rasp increases Spitz activity by restricting its diffusion. Dev Cell. 2006;10(2):167–176. doi: 10.1016/j.devcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 109.Ciepla P, Magee AI, Tate EW. Cholesterylation: a tail of hedgehog. Biochem Soc Trans. 2015;43(2):262–7. doi: 10.1042/BST20150032. [DOI] [PubMed] [Google Scholar]
- 110.Jakobs P, Exner S, Schurmann S, Pickhinke U, Bandari S, Ortmann C, Kupich S, Schulz P, Hansen U, Seidler DG, Grobe K. Scube2 enhances proteolytic Shh processing from the surface of Shh-producing cells. J Cell Sci. 2014;127(Pt 8):1726–37. doi: 10.1242/jcs.137695. [DOI] [PubMed] [Google Scholar]
- 111.Creanga A, Glenn TD, Mann RK, Saunders AM, Talbot WS, Beachy PA. Scube/You activity mediates release of dually lipid-modified Hedgehog signal in soluble form. Genes Dev. 2012;26(12):1312–25. doi: 10.1101/gad.191866.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tukachinsky H, Kuzmickas RP, Jao CY, Liu J, Salic A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell reports. 2012;2(2):308–20. doi: 10.1016/j.celrep.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goetz JA, Singh S, Suber LM, Kull FJ, Robbins DJ. A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. J Biol Chem. 2006;281(7):4087–4093. doi: 10.1074/jbc.M511427200. [DOI] [PubMed] [Google Scholar]
- 114.Bischoff M, Gradilla AC, Seijo I, Andres G, Rodriguez-Navas C, Gonzalez-Mendez L, Guerrero I. Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nat Cell Biol. 2013;15(11):1269–81. doi: 10.1038/ncb2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eugster C, Panakova D, Mahmoud A, Eaton S. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev Cell. 2007;13(1):57–71. doi: 10.1016/j.devcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 116.Gradilla AC, Gonzalez E, Seijo I, Andres G, Bischoff M, Gonzalez-Mendez L, Sanchez V, Callejo A, Ibanez C, Guerra M, Ortigao-Farias JR, Sutherland JD, Gonzalez M, Barrio R, Falcon-Perez JM, Guerrero I. Exosomes as Hedgehog carriers in cytoneme-mediated transport and secretion. Nat Commun. 2014;5:5649. doi: 10.1038/ncomms6649. [DOI] [PubMed] [Google Scholar]
- 117.Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, Takao T, Takada S. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev Cell. 2006;11(6):791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 118.Nile AH, Hannoush RN. Fatty acylation of Wnt proteins. Nat Chem Biol. 2016;12(2):60–9. doi: 10.1038/nchembio.2005. [DOI] [PubMed] [Google Scholar]
- 119.Coombs GS, Yu J, Canning CA, Veltri CA, Covey TM, Cheong JK, Utomo V, Banerjee N, Zhang ZH, Jadulco RC, Concepcion GP, Bugni TS, Harper MK, Mihalek I, Jones CM, Ireland CM, Virshup DM. WLS-dependent secretion of WNT3A requires Ser209 acylation and vacuolar acidification. J Cell Sci. 2010;123(Pt 19):3357–67. doi: 10.1242/jcs.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Herr P, Basler K. Porcupine-mediated lipidation is required for Wnt recognition by Wls. Dev Biol. 2011 doi: 10.1016/j.ydbio.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 121.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337(6090):59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kadowaki T, Wilder E, Klingensmith J, Zachary K, Perrimon N. The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes Dev. 1996;10(24):3116–28. doi: 10.1101/gad.10.24.3116. [DOI] [PubMed] [Google Scholar]
- 123.van den Heuvel M, Harryman-Samos C, Klingensmith J, Perrimon N, Nusse R. Mutations in the segment polarity genes wingless and porcupine impair secretion of the wingless protein. EMBO J. 1993;12(13):5293–302. doi: 10.1002/j.1460-2075.1993.tb06225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rios-Esteves J, Resh MD. Stearoyl CoA desaturase is required to produce active, lipid-modified Wnt proteins. Cell reports. 2013;4(6):1072–81. doi: 10.1016/j.celrep.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gao X, Hannoush RN. Single-cell imaging of Wnt palmitoylation by the acyltransferase porcupine. Nat Chem Biol. 2014;10(1):61–8. doi: 10.1038/nchembio.1392. [DOI] [PubMed] [Google Scholar]
- 126.Rios-Esteves J, Haugen B, Resh MD. Identification of key residues and regions important for porcupine-mediated Wnt acylation. J Biol Chem. 2014;289(24):17009–19. doi: 10.1074/jbc.M114.561209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miranda M, Galli LM, Enriquez M, Szabo LA, Gao X, Hannoush RN, Burrus LW. Identification of the WNT1 residues required for palmitoylation by Porcupine. FEBS Lett. 2014;588(24):4815–24. doi: 10.1016/j.febslet.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Neumann S, Coudreuse DY, van der Westhuyzen DR, Eckhardt ER, Korswagen HC, Schmitz G, Sprong H. Mammalian Wnt3a is released on lipoprotein particles. Traffic. 2009;10(3):334–43. doi: 10.1111/j.1600-0854.2008.00872.x. [DOI] [PubMed] [Google Scholar]
- 129.Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol. 2012;14(10):1036–45. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- 130.Mulligan KA, Fuerer C, Ching W, Fish M, Willert K, Nusse R. Secreted Wingless-interacting molecule (Swim) promotes long-range signaling by maintaining Wingless solubility. Proc Natl Acad Sci U S A. 2012;109(2):370–7. doi: 10.1073/pnas.1119197109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Alexandre C, Baena-Lopez A, Vincent JP. Patterning and growth control by membrane-tethered Wingless. Nature. 2014;505(7482):180–5. doi: 10.1038/nature12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DV, de Punder K, Angers S, Peters PJ, Maurice MM, Clevers H. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530(7590):340–3. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 133.Zhang X, Cheong SM, Amado NG, Reis AH, MacDonald BT, Zebisch M, Jones EY, Abreu JG, He X. Notum is required for neural and head induction via Wnt deacylation, oxidation, and inactivation. Dev Cell. 2015;32(6):719–30. doi: 10.1016/j.devcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kakugawa S, Langton PF, Zebisch M, Howell SA, Chang TH, Liu Y, Feizi T, Bineva G, O'Reilly N, Snijders AP, Jones EY, Vincent JP. Notum deacylates Wnt proteins to suppress signalling activity. Nature. 2015;519(7542):187–92. doi: 10.1038/nature14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell. 2008;132(3):387–96. doi: 10.1016/j.cell.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 136.Gutierrez JA, Solenberg PJ, Perkins DR, Willency JA, Knierman MD, Jin Z, Witcher DR, Luo S, Onyia JE, Hale JE. Ghrelin octanoylation mediated by an orphan lipid transferase. Proc Natl Acad Sci U S A. 2008;105(17):6320–5. doi: 10.1073/pnas.0800708105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Darling JE, Zhao F, Loftus RJ, Patton LM, Gibbs RA, Hougland JL. Structure-activity analysis of human ghrelin O-acyltransferase reveals chemical determinants of ghrelin selectivity and acyl group recognition. Biochemistry (Mosc) 2015;54(4):1100–10. doi: 10.1021/bi5010359. [DOI] [PubMed] [Google Scholar]
- 138.Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci. 2000;25(3):111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- 139.Shindou H, Eto M, Morimoto R, Shimizu T. Identification of membrane O-acyltransferase family motifs. Biochem Biophys Res Commun. 2009;383(3):320–5. doi: 10.1016/j.bbrc.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 140.Gooding JM, Shayeghi M, Saggerson ED. Membrane transport of fatty acylcarnitine and free L-carnitine by rat liver microsomes. Eur J Biochem. 2004;271(5):954–61. doi: 10.1111/j.1432-1033.2004.03997.x. [DOI] [PubMed] [Google Scholar]
- 141.Matevossian A, Resh MD. Membrane topology of hedgehog acyltransferase. J Biol Chem. 2015;290(4):2235–43. doi: 10.1074/jbc.M114.625764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Konitsiotis AD, Jovanovic B, Ciepla P, Spitaler M, Lanyon-Hogg T, Tate EW, Magee AI. Topological analysis of Hedgehog acyltransferase, a multipalmitoylated transmembrane protein. J Biol Chem. 2015;290(6):3293–307. doi: 10.1074/jbc.M114.614578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Taylor MS, Ruch TR, Hsiao PY, Hwang Y, Zhang P, Dai L, Huang CR, Berndsen CE, Kim MS, Pandey A, Wolberger C, Marmorstein R, Machamer C, Boeke JD, Cole PA. Architectural organization of the metabolic regulatory enzyme ghrelin O-acyltransferase. J Biol Chem. 2013;288(45):32211–28. doi: 10.1074/jbc.M113.510313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Brannigan JA, Smith BA, Yu Z, Brzozowski AM, Hodgkinson MR, Maroof A, Price HP, Meier F, Leatherbarrow RJ, Tate EW, Smith DF, Wilkinson AJ. N-myristoyltransferase from Leishmania donovani: structural and functional characterisation of a potential drug target for visceral leishmaniasis. J Mol Biol. 2010;396(4):985–99. doi: 10.1016/j.jmb.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Frearson JA, Brand S, McElroy SP, Cleghorn LA, Smid O, Stojanovski L, Price HP, Guther ML, Torrie LS, Robinson DA, Hallyburton I, Mpamhanga CP, Brannigan JA, Wilkinson AJ, Hodgkinson M, Hui R, Qiu W, Raimi OG, van Aalten DM, Brenk R, Gilbert IH, Read KD, Fairlamb AH, Ferguson MA, Smith DF, Wyatt PG. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature. 2010;464(7289):728–32. doi: 10.1038/nature08893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hutton JA, Goncalves V, Brannigan JA, Paape D, Wright MH, Waugh TM, Roberts SM, Bell AS, Wilkinson AJ, Smith DF, Leatherbarrow RJ, Tate EW. Structure-based design of potent and selective Leishmania N-myristoyltransferase inhibitors. J Med Chem. 2014;57(20):8664–70. doi: 10.1021/jm5011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Petrova E, Matevossian A, Resh MD. Hedgehog acyltransferase as a target in pancreatic ductal adenocarcinoma. Oncogene. 2015;34(2):263–8. doi: 10.1038/onc.2013.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Petrova E, Rios-Esteves J, Ouerfelli O, Glickman JF, Resh MD. Inhibitors of Hedgehog acyltransferase block Sonic Hedgehog signaling. Nat Chem Biol. 2013;9:247–249. doi: 10.1038/nchembio.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Matevossian A, Resh MD. Hedgehog Acyltransferase as a target in estrogen receptor positive, HER2 amplified, and tamoxifen resistant breast cancer cells. Molecular cancer. 2015;14:72. doi: 10.1186/s12943-015-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, Roth MG, Amatruda JF, Chen C, Lum L. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5(2):100–7. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Proffitt KD, Madan B, Ke Z, Pendharkar V, Ding L, Lee MA, Hannoush RN, Virshup DM. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 2013;73(2):502–7. doi: 10.1158/0008-5472.CAN-12-2258. [DOI] [PubMed] [Google Scholar]
- 152.Madan B, Ke Z, Harmston N, Ho SY, Frois AO, Alam J, Jeyaraj DA, Pendharkar V, Ghosh K, Virshup IH, Manoharan V, Ong EH, Sangthongpitag K, Hill J, Petretto E, Keller TH, Lee MA, Matter A, Virshup DM. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. 2015 doi: 10.1038/onc.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Barnett BP, Hwang Y, Taylor MS, Kirchner H, Pfluger PT, Bernard V, Lin YY, Bowers EM, Mukherjee C, Song WJ, Longo PA, Leahy DJ, Hussain MA, Tschop MH, Boeke JD, Cole PA. Glucose and weight control in mice with a designed ghrelin O-acyltransferase inhibitor. Science. 2010;330(6011):1689–92. doi: 10.1126/science.1196154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhao TJ, Liang G, Li RL, Xie X, Sleeman MW, Murphy AJ, Valenzuela DM, Yancopoulos GD, Goldstein JL, Brown MS. Ghrelin O-acyltransferase (GOAT) is essential for growth hormone-mediated survival of calorie-restricted mice. Proc Natl Acad Sci U S A. 2010;107(16):7467–72. doi: 10.1073/pnas.1002271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kouno T, Akiyama N, Ito T, Okuda T, Nanchi I, Notoya M, Oka S, Yukioka H. Ghrelin O-acyltransferase knockout mice show resistance to obesity when fed high-sucrose diet. J Endocrinol. 2016;228(2):115–25. doi: 10.1530/JOE-15-0330. [DOI] [PubMed] [Google Scholar]