Abstract
Autophagy is a cellular degradative pathway that involves the delivery of cytoplasmic components, including proteins and organelles, to the lysosome for degradation. Autophagy is implicated in the maintenance of skeletal muscle; increased autophagy leads to muscle atrophy while decreased autophagy leads to degeneration and weakness. A growing body of work suggests that reactive oxygen species (ROS) are important cellular signal transducers controlling autophagy. Nicotinamide adenine dinucleotide phosphate (NADPH) oxidases and mitochondria are major sources of ROS generation in skeletal muscle that are likely regulating autophagy through different signaling cascades based on localization of the ROS signals. This review aims to provide insight into the redox control of autophagy in skeletal muscle. Understanding the mechanisms by which ROS regulate autophagy will provide novel therapeutic targets for skeletal muscle diseases.
Keywords: reactive oxygen species, free radicals, autophagy, skeletal muscle
Introduction
Cellular homeostasis, essential for tissue development and cell survival, is maintained by a balance of protein synthesis and degradation. Skeletal muscle is a highly plastic tissue, effectively adapting to changes in metabolic demand. There are three major pathways regulating proteolysis is skeletal muscle: 1) the ubiquitin proteasome pathway (UPP); 2) the caspase-3 and calpain (calcium dependent protease) pathway; and 3) the autophagy-lysosomal pathway. Recently, mitochondrial specific proteases (i.e. Lon protease) have been shown to be upregulated in skeletal muscle in response to acute oxidative stress (1); however, its role in regulation of autophagy has not been investigated. Oxidative stress has been shown to increase protein breakdown through increased gene expression of key atrophy related protein such as atrogins and MuRF-1 (2, 3), as well as increase the activity of calpain and caspase-3 (4, 5). Oxidative modification of proteins also causes partial unfolding, promoting the exposure of hidden recognition sequences that facilitate their proteolytic degradation. Oxidation of myofibrillar proteins promotes proteolytic cleavage by calpain and caspase-3 (4, 5), which is required to facilitate degradation by UPP (6–8). While oxidized proteins are cleared by UPP and the calpain/caspase-3 pathways, large protein aggregates and damaged organelles are degraded by the autophagy-lysosomal pathway.
Autophagy is a homeostatic process that clears protein aggregates and damaged organelles through the autophagosome-lysosome system. Autophagy has recently gained immense attention for its role in metabolic homeostasis and disease progression of skeletal muscle. Alterations in autophagic flux are commonly observed in response to stress and have been shown to increase in skeletal muscle in response to starvation, denervation, disuse atrophy, hypoxia, and exercise (9–12). A number of factors and signaling pathways have been shown to contribute to the regulation of autophagic flux. Among them, reactive oxygen species (ROS) have been implicated in the control of autophagic flux.
Oxidative stress may occur through an increase in ROS levels or a decrease in the cellular antioxidant capacity. While a certain level of ROS is essential for the regulation of cell growth and various biological functions, a disrupted ROS balance has negative implications. For example, oxidative stress has been associated with a number of pathological conditions, including neurodegenerative disorders (13–18), skeletal muscle disorders (19–23), lysosomal storage disorders (24, 25), cardiomyopathy (26, 27), carcinogenesis (28, 29), atherosclerosis (30, 31), diabetes (32, 33), and aging (34, 35). While the involvement of oxidative stress is firmly demonstrated in these pathological conditions, the specific source of ROS generation and the mechanisms by which each disease is regulated by ROS has yet to be elucidated. While ROS and autophagy were first described a number of years ago; the precise mechanisms of ROS-regulated autophagy and effective therapeutic strategies still remain to be discovered. Due to the compelling recent evidence associating autophagy with skeletal muscle homeostasis, we focus this review on summarizing the identified molecular mechanisms of ROS-regulated autophagy and their relevance to skeletal muscle health and disease.
Overview of Autophagy Signaling
Autophagy is an evolutionarily conserved cellular degradation pathway that involves breakdown of cytoplasmic components by the lysosome. In general, autophagy is categorized by three main types: microautophagy, chaperone-mediated autophagy (CMA), and macroautophagy (36, 37).
Microautophagy
Microautophagy is a non-selective lysosomal degradative process which directly engulfs the cytoplasmic cargo and eliminates them by both invagination and vesicle scission (38, 39). While microautophagy is unresponsive to amino-acid deprivation (39), little else is known regarding the mechanisms regulating miroautophagy in mammalian cells.
Chaperone-mediate autophagy (CMA)
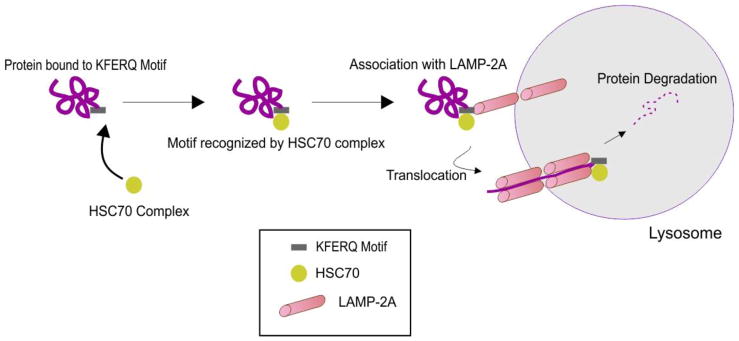
During CMA, cytoplasmic cargo is targeted to the lysosome, where it is degraded by lysosomal enzymes (40–42) (Figure 1). The pentapeptide motif of CMA substrates contains a glutamine (Q) residue at the beginning or end of the sequence, one or two of the positive charged amino acids lysine (K) or arginine (R), one of the hydrophobic amino acids, phenylalanine (F), valine (V), leucine (L) or isoleucine (I) and one of the negatively charged amino acids, glutamic (E) or aspartic acid (D) (40, 43, 44). In the cytosol, a constitutive chaperone, heat shock cognate protein of 70 kDa (Hsc70), along with other co-chaperones (Bag1, Hip, Hop and Hsp40), bind to the substrate on the pentapeptide motif KFERQ, which is present in the amino acid sequence of all CMA substrates, consequently transporting it to the surface of the lysosomal membrane (40, 43, 45).
Figure 1. Schematic diagram of chaperone mediated autophagy.
Chaperone mediated autophagy is involved in the breakdown of damaged cytosolic proteins. Chaperones recognize a KFERQ motif on the targeted protein and deliver the protein to LAMP-2A on the lysosomal membrane for degradation. See text for details.
Once the substrate complex is targeted to the lysosomal surface, it interacts with the cytosolic tail of lysosomal-associated membrane protein type 2A (LAMP-2A). The monomeric LAMP-2A forms multi-protein complex structures, along with many other proteins, promoting the translocation of CMA substrates. CMA substrates can be introduced to this multi-protein complex in the folded or unfolded state; however, translocation of the substrates can only be carried out in the unfolded form (46, 47). The folding and unfolding of CMA substrates are tightly regulated by Hsc70 and the other co-chaperones. Once the CMA substrates are internalized into the lysosomes, they are degraded by lysosomal hydrolases. Subsequently, LAMP-2A dissociates from the multi-protein complex to form monomers, where another CMA substrate can bind, and thus this dynamic process maintains the homeostasis of CMA (48). Alterations in redox balance and subsequent oxidative stress is one of the major factors that regulate the levels of LAMP-2A (40, 48–50). The role of CMA in skeletal muscle has not been widely studied. Increased LAMP2A has been reported in mouse skeletal muscle after a single bout of exercise (51). Additionally, abnormalities of CMA have been observed in sporadic inclusion-body myositis muscle fibers (52).
Macroautophagy
Macroautophagy, referred to here as autophagy, is the most investigated form of autophagy and is characterized by the formation of double-membrane structures, called autophagosomes, which sequester cytoplasmic substrates and fuse with lysosomes to eliminate damaged components or recycle end products for production of energy that regulates cellular homeostasis (53, 54) (Figure 2). Substrates of autophagy include damaged proteins, organelles, inclusion bodies, and superfluous and invasive bacteria (36, 53, 55). Precise regulation of autophagy is a highly selective process, as it critically depends on engulfment of specific substrates within autophagosomes, while preventing engulfment of undamaged cytoplasmic contents (55). Due to the vast range of substrate selectivity, autophagic pathways can be impaired through a wide range of mechanisms that vary in each disease. Therefore, understanding the key regulators of autophagy in mammalian cells and how they are altered under different pathological conditions has gained immense attention in recent years.
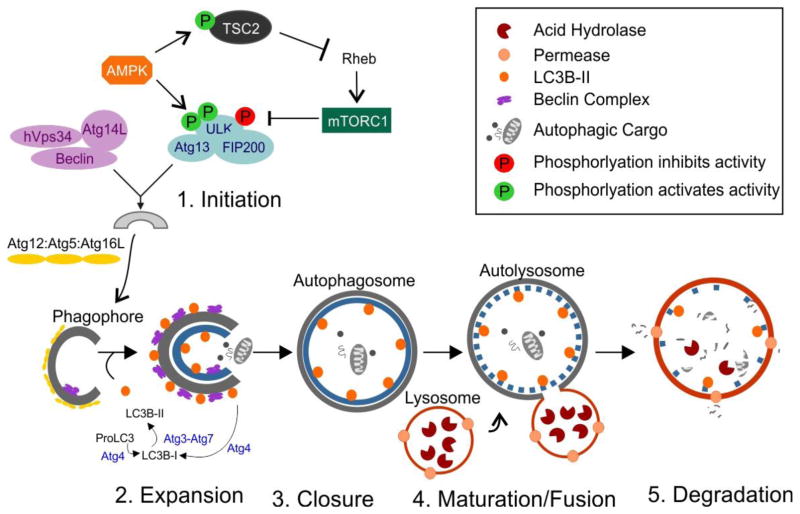
Figure 2. Schematic diagram of macroautophagy.
Macroautophagy involves the formation of distinct complexes during five sequential stages: (1) initiation, (2) expansion and elongation, (3) closure, (4) maturation and fusion of autophagosomes with lysosomes, and (5) degradation of the autophagic cargo. See text for details.
Control and regulation of autophagy
Low basal levels of autophagy allow cells to break down long-lived and large cytosolic protein aggregates and organelles, which has been shown to be necessary for cell survival. The regulatory process of autophagy is divided into two distinct forms, selective and nonselective autophagy. Selective autophagy is mainly regulated under homeostatic conditions; while, nonselective autophagy is induced upon starvation or in response to external or internal stress related conditions (56). Both selective and nonselective autophagy are regulated by core autophagic machinery structured by a number of autophagy-related (ATG) genes that have been identified by large-scale genetic screening in yeast almost three decades ago (57). Most of the ATGs identified in yeast have mammalian counterparts, where they are actively involved in regulating autophagy by highly-conserved mechanisms in the initiation of double-membrane autophagosome formation (54, 56–58).
Initiation of autophagosome formation
In mammalian cells, formation of the autophagosome is initiated by a complex consisting of the serine/threonine protein kinase unc-51-like kinase-1 (ULK1) and -2 (ULK2), FIP200, and Atg13 (53, 56–58). The classical paradigm of autophagosome formation is controlled by two master regulators of ULK, mammalian target of rapamycin (mTOR) complex1 and AMP-activated protein kinase (AMPK) (59–63). mTOR is a serine/threonine kinase which acts as a central inhibitor of autophagy by inhibiting ULK1 activity through phosphorylation at S757, thereby disrupting its interaction with AMPK. Under nutrient starvation, mTORC1 is inhibited, resulting in its dissociation from the ULK1 complex with subsequent dephosphorylation and activation of ULK1. Recent studies have identified the association between AMPK and ULK1. Bioinformatics approaches have screened several possible AMPK-phosphorylation sites in ULK1 (S555, T574, S637, and S467). However, all the phosphorylation sites have not been confirmed in vivo. Recently, using systematic mutagenesis, two major AMPK-phosphorylation sites in ULK1 (S317 and S777) were identified and later confirmed by cell based assays (62, 63). AMPK-dependent phosphorylation of ULK1 increases ULK1 activity and promotes autophagy (63). In addition to ULK1 phosphorylation, AMPK can directly phosphorylate the Tuberous sclerosis complex2 (TSC2), leading to the inactivation of the GTPase Rheb, which directly binds to and activates mTORC1 kinase activity (62). AMPK-mediated inactivation of mTORC1 increases ULK1 activity and promotes autophagy.
Nucleation of the phagophore
The nucleation and assembly of the initial phagophore membrane is a major determinant of mature autophagosome formation. This process is centrally regulated by a complex which consists of class III phosphatidylinositol 3-kinase (PI3K or hVps34), its regulatory subunits p150 or hVps15, Beclin1, and Atg14L, the relatively recent discovered mammalian homology of Atg14 (56, 57, 64, 65). The activity of this complex is tightly controlled by several positive and negative regulators, and is often dysregulated in various pathological conditions. Recent studies have implicated an mTORC1/AMPK-independent regulation of autophagy, which is directly dependent on the interaction between Beclin1 and hVps34 (66). However, the precise mechanisms by which ULK:Atg13:FIP200 complex connects with beclin1:hVps34:Atg14L complex has not been clearly established.
Elongation of the phagophore, autophagosome formation, and fusion
Elongation of the autophagophore is critical for the completion of the autophagosome. Atg12 conjugates with Atg5, and the conjugated Atg12:Atg5 complex interacts with Atg16L to form a multimeric complex Atg12:Atg5:Atg16L. This multimeric complex associates with the microtubule-associated protein-light chain 3 (LC3) conjugation system, which is recruited by the beclin1:hVps34:Atg14L complex to form the mature autophagosome (56, 57, 67–69). While mammalian cells express three variants of LC3 (LC3A, LC3B, LC3C), LC3B is expressed in nearly all tissues and is the most widely used marker of autophagic flux (70). Conjugation of phosphatidylethanolamine (PE) to soluble LC3B (LC3B-I) is mediated by the protease Atg4 followed by Atg3 and Atg7. The lipidated form of LC3B (LC3B-II) is associated with the outer and inner membranes of the autophagosome for the induction and maturation of the autophagosome (71–73). Atg4 also acts to delipidate LC3B-II present on the cytoplasmic face of the autophagosome, recycling it back to LC3B-I, thereby ensuring elongation of the autophagosome. Autophagosomes move along microtubules in a dynein/kinesin dependent manner to the lysosome and fuse with the lysosome to form the autolysosome. Lysosomal acid hydrolases then degrade the autohagic cargo.
Redox balance, oxidative stress and redox control of autophagy in skeletal muscle
ROS are produced at relatively low rates under physiological conditions in skeletal muscle fibers and exert positive effects on gene expression, regulation of cell signaling, and modulation of contractile force. In contrast, high levels of ROS result in damage to cellular components such as proteins and organelles, leading to muscle dysfunction. The role of ROS and oxidative stress in the regulation of skeletal muscle has been extensively reviewed elsewhere (74–81) as well as in this Special Issue.
Indeed, autophagic flux has been shown to participate in pro-atrophic stimuli (11, 12, 82–90), fasting (91, 92), high fat diet/insulin resistance (93, 94), hypoxia (95), and exercise (9, 96–101). Conversely, impaired autophagy has been reported in several myopathies (23, 102–108). While autophagy and oxidative stress have been studied individually, little is known about the molecular regulation of autophagy by ROS. A number of studies, as reviewed below, report that ROS induces autophagy and, vice-versa, autophagy serves to reduce oxidative stress.
Much of the work that has reported on autophagy and oxidative stress in skeletal muscle merely hypothesize that ROS are crucial for induction of autophagy (11, 82–84, 86, 87, 91, 95), as direct cause and effect was not established. More direct evidence that ROS regulates autophagic flux comes from Dobrowolny et al (109), who have shown that skeletal muscle expressing a mutant form of superoxide dismutase 1 (SOD1G93A) increases oxidative stress and triggers activation of autophagy, leading to muscle atrophy and weakness. In addition, pharmacological application of H2O2 has been shown to induce autophagy in C2C12 myotubes (85, 110, 111). Concomitantly, antioxidant treatment has been shown to inhibit the induction of autophagy (92, 94, 110, 112). In a mouse model of muscular dystrophy, we have shown that blocking Nox2-dependent ROS production relieves inhibition of autophagy and improves muscle function (23). On the contrary, skeletal muscle specific genetic knockout of Atg7 showed an altered metabolic profile, defective mitochondrial respiration, and increased steady state ROS production (113–115), suggesting that decreased autophagy results in increased production of ROS. It is clear that ROS and autophagy play a role in skeletal muscle homeostasis; however, it is unclear how up or down regulation of these processes induce a negative or beneficial response. The emerging theme is that the amount of ROS generated and its sub-cellular localization are major determinants of ROS-mediated autophagy regulation in skeletal muscle (Figure 3).
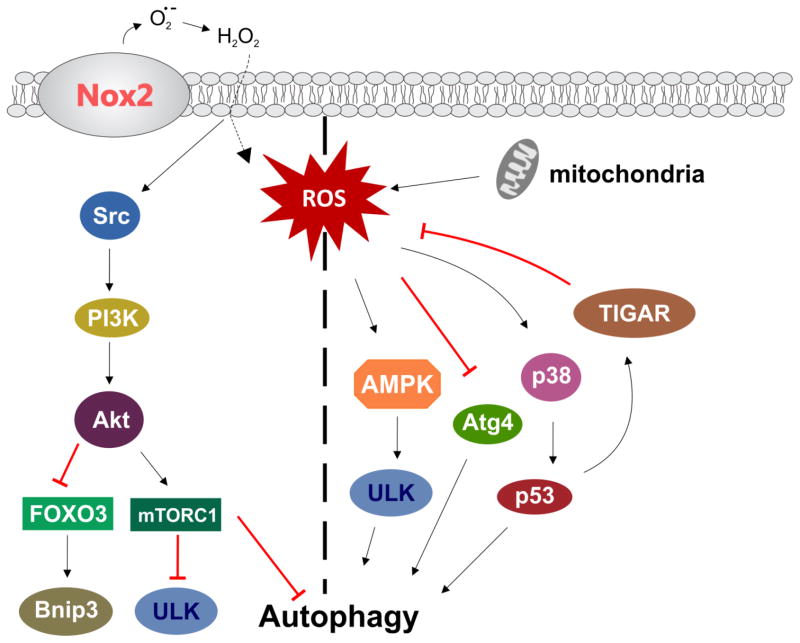
Figure 3. Regulation of autophagy by ROS.
Nox2 and mitochondria are the main sources of ROS in skeletal muscle. Activation or inhibition of autophagy is likely governed by micro-domain redox signaling and the amount of ROS produced. See text for details.
The source of ROS in regulation of not only autophagy, but many skeletal muscle cell signaling cascades is an area of active research. There are a number of potential sources of ROS production in skeletal muscle. These include mitochondria, NADPH oxidase (Nox), xanthine oxidase, and phospholipase A2 (reviewed in (74, 75)). However, mitochondria and Nox isoform 2 (Nox2) have emerged as the two main sources of ROS production. Mitochondria have been proposed as the primary source of ROS to regulate autophagy in many cell types (116–120), including skeletal muscle (86, 91, 109, 110). The role of Nox2 in regulation of autophagy is less clear. In macrophages, Nox2-dependent ROS has been shown to induce autophagy upon bacterial infection (121, 122), functioning as an innate immune-defense mechanism. In a cellular model of neurodegeneration we have shown that rotenone, a prototypical mitochondrial complex I inhibitor, increases Nox2-dependent ROS production (18), resulting in inhibition of autophagy. To our knowledge, we are the only group to show that Nox2-generated ROS regulates autophagy in skeletal muscle. We found that exuberant Nox2-dependent ROS production impairs autophagic flux in skeletal muscle (23).
While many studies show that ROS are a signal to induce or impair autophagy, we still know very little about the mechanisms of action. Some studies have shown that ROS activate autophagy by regulating the activation of the PI3K/Akt/mTORC1-signaling pathway. In malignant glioma, ROS promoted autophagy by inhibiting Akt/mTOR signaling (123, 124). In a hindlimb casting model of disuse atrophy, Talbert et al (125) have shown that mitochondrial ROS promote inhibition of Akt/mTOR and subsequent induction of autophagy. In our recent study, we have demonstrated that ROS generated from Nox2 induces activation of mTORC1 through activation of a Src/PI3K/Akt pathway, and thus ROS-mediated activation of mTORC1 inhibits autophagy in a dystrophic mouse model (23).
Another potential pathway for ROS dependent regulation of autophagy is through p38 MAPK/p53. Mitochondrial ROS was shown to induce autophagy through a p38/p53 dependent path in A375 cells (126). In skeletal muscle, ROS induced autophagy in a p38 MAPK dependent manner (85). Interestingly, Yuan et al (127) have shown that the p38/p53 pathway appears to not only activate autophagy, but to be involved in a positive feed-back response, as both p38 and p53 were shown to increase ROS production in cardiomyocytes. Inhibition of the p53-target gene TIGAR (TP53-induced glycolysis and apoptosis regular) results in increased ROS production and activation of autophagy (mitophagy), (128) while TIGAR overexpression results in decreased ROS levels and inhibition of autophagy (129).
AMPK, a widely established sensor of cellular energy levels, is an essential regulator of muscle metabolism during exercise, as well as in skeletal muscle adaptation to exercise training (reviewed in (130)). Alterations in redox balance have been shown to regulate AMPK activity. Exposure of C2C12 cells to pharmacological H2O2 concentrations resulted in activation of AMPK (110, 111), with a subsequent increase in autophagy (110).
The tumor suppressor gene PTEN (phosphatase and tensin homolog deleted on chromosome 10) is inhibited by oxidative stress (131–135). Inactivation of PTEN results in an increase in cellular PIP3 levels, activation of PI3K/Akt, and subsequent activation of autophagy. PTEN can also regulate ROS production, providing a feedback loop in which Nox may be intimately poised to regulate this signaling (131). While ROS has been shown to activate Akt through inhibition of PTEN in C2C12 myotubules (135), its role in regulating autophagy in skeletal muscle has not been directly assessed.
In Chinese Hamster Ovary (CHO) cells, nutrient deprivation resulted in increased ROS production, specifically mitochondrial H2O2, oxidation and inactivation of Atg4, thus preventing its delipidation of LC3B-II and ensuring elongation of the autophagosome (120). REDD1 (regulated in development and DNA damage responses 1), is a hypoxia-inducible factor-1 target gene and plays a crucial role in inhibiting mTORC1 (136). Ellisen and colleagues (137) have shown that hypoxia and exercise increase ROS production through a REDD1/TXNIP pro-oxidant complex, inhibiting Atg4 activity and promoting autophagy. Mice lacking REDD1 displayed impaired oxidative phosphorylation and reduced exercise capacity, presumably due to altered Atg4 activity and decreased mitophagy.
The Forkhead box O (FoxO) transcription factors play essential roles in regulation of muscle physiology (10, 12). They are phosphorylated and inactivated by Akt/PKB and predominantly localize in the cytosol. However, in response to Akt supression, FoxO translocates to the nucleus, inducing transcription of atrophy related genes (atrogin 1 and MuRF-1) and the autophagy related genes cathepsin L, Bnip3, and LC3B (10). While basal autophagy is essential for the maintenance of metabolic homeostasis, oxidative stress-dependent activation of FoxO3 and subsequent up-regulation of FoxO3-mediated autophagy have been shown to promote muscle atrophy and weakness (10, 138). Other studies have also demonstrated that ROS generation induces activation of Akt (139–142), an event that negatively regulates transcriptional activity of FoxO3. Therefore, the precise role of oxidative stress on FoxO-mediated autophagy in skeletal muscle remains unclear.
Regulation of redox balance by autophagy
Dysregulation in the homeostasis of autophagy promotes ROS generation and subsequent alterations in redox balance. Impairment of autophagy leads to cytosolic-accumulation of ubiquitinated proteins that induces mitochondrial damage and promotes ROS generation (143). In squamous cell carcinoma cells autophagy was shown to increases ROS generation from xanthine oxidase, leading to mitochondrial damage and exacerbating oxidative stress (144). Keap1 (Kelch-like ECH- associated protein 1) binds Nrf2 (nuclear factor erythroid 2-related factor 2), sequestering Nrf2 in the cytoplasm and preventing transcriptional regulation of antioxidant genes. Autophagic degradation of Keap1 allows Nrf2 to translocate to the nucleus and bind the antioxidant-responsive elements (ARE) in the promoter region of antioxidant genes (145). Skeletal muscle specific knockout of Atg7 leads to dysfunctional mitochondria, oxidative stress, myofiber atrophy, and muscle weakness (102, 113–115). Although it is not clearly established whether autophagy is a major checkpoint for the control of redox balance, together, these studies suggest that alterations in redox balance coordinate with changes in autophagy.
Autophagy is differentially regulated in fast-twitch glycolytic and slow-twitch oxidative muscle
Several lines of evidence suggest that autophagy signaling is differentially regulated between muscles with distinct fiber type distribution and metabolic characteristics. Analysis of autophagsome formation in skeletal muscle in response to starvation has indicated that there is a significantly greater increase in glycolytic muscle (tibialis anterior, TA) compared to oxidative muscle like the diaphragm (105, 108, 146). After spontaneous wheel running for 3 months, TA muscle from mice, did not show any evidence of autophagy induction (LC3 lipidation) (9). Conversely, in plantaris muscle, composed of glycolic and oxidative fibers, from mice subjected to 4 weeks of voluntary running displayed increased LC3 lipidation, decreased p62 protein content, and increased expression of several autophagy-related proteins (i.e., Atg6, LC3, and Bnip3) (101, 147). Inactivation of mTORC1 signaling resulted in greater atrophy in glycolytic muscle fibers compared to oxidative muscle fibers (148, 149). In this regard, muscle specific ATG5 knockout mice showed increased p62 protein content and accumulation of cytoplasmic ubiquitinated proteins in glycolytic muscle fibers but not oxidative muscle fibers (150).
Autophagic flux also shows fiber-type variability in myopathic conditions. Autophagy is severely compromised in glycolytic TA compared to the more oxidative diaphragm muscle in both Collagen VI (105) and Duchenne muscular dystrophies (108, 146). Deficiency of the glycogen-degrading lysosomal enzyme acid-alpha glucosidase (Pompe disease) resulted in accumulation of p62, LAMP-1, and ubiquitinated proteins in fast glycolytic fiber of the gastrocnemius but not slow oxidative fibers of the soleus (150). In a mouse model of sepsis, LPS induced upregulation of autophagy was greater in the TA than either the diaphragm or soleus (86). Pessin and colleagues (151) have shown that a Fyn/STAT3/Vps34 pathway upregulates macroautophagy in glycolytic muscle, with less effect on oxidative muscle. Finally, in a rat model of myocardial infarction, autophagy-related genes (MAP1LC3B, GABARAPL1, BNIP3 and CTSL1) were upregulated in plantaris (glycolytic) muscle fibers but not soleus muscle fibers; even though both fiber types showed marked atrophy (152).
Taken together, there is sufficient data indicating that autophagy is differentially regulated in a fiber-type specific manner. However, the basis for the selectivity under different physiological or pathophysiological conditions and what role oxidative stress may play are important yet unresolved issues.
Conclusions
ROS play an important role in controlling a wide range of cell signal transduction pathways and modulation of skeletal muscle force. Autophagy is an important cell survival mechanism that is now recognized to be crucial in skeletal muscle health. Both ROS and autophagy likely have either beneficial or detrimental effects, depending on their balance. While ROS have been shown to promote autophagy in skeletal muscle, ROS have also been shown to impair autophagy. An important question is how ROS crosstalk with autophagic signaling. While we have begun to uncover the role of redox signaling in regulation of specific signaling cascades in autophagy, intricate details of this process have yet to be elucidated. It is likely that the amount of ROS generated and the specific sub-cellular localization of ROS are major determinants of ROS-mediated autophagy regulation in skeletal muscle. Future studies aimed at understanding the control of autophagy through micro-domain redox signaling will aid in our understanding of the control of autophagy, providing valuable information for the development of selective therapies for skeletal muscle dysfunction.
Highlights.
Autophagy regulates skeletal muscle homeostasis.
Skeletal muscle atrophy occurs due to increased autophagy.
Impaired autophagy leads skeletal muscle degeneration due to accumulation of damaged proteins and organelles.
Regulation of skeletal muscle autophagy by reactive oxygen species is due to sub-cellular production of ROS.
Acknowledgments
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R01 AR061370, the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under Award Number R21 NS085208, and a Clifford Elder White Graham Endowed Fellowship to G.G.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ngo JK, Pomatto LC, Davies KJ. Upregulation of the mitochondrial Lon Protease allows adaptation to acute oxidative stress but dysregulation is associated with chronic stress, disease, and aging. Redox Biol. 2013;1:258–264. doi: 10.1016/j.redox.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrigo J, Rivera JC, Simon F, Cabrera D, Cabello-Verrugio C. Transforming growth factor type beta (TGF-beta) requires reactive oxygen species to induce skeletal muscle atrophy. Cell Signal. 2016;28:366–376. doi: 10.1016/j.cellsig.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Li YP, Chen Y, Li AS, Reid MB. Hydrogen peroxide stimulates ubiquitin-conjugating activity and expression of genes for specific E2 and E3 proteins in skeletal muscle myotubes. Am J Physiol Cell Physiol. 2003;285:C806–812. doi: 10.1152/ajpcell.00129.2003. [DOI] [PubMed] [Google Scholar]
- 4.Powers SK, Kavazis AN, DeRuisseau KC. Mechanisms of disuse muscle atrophy: role of oxidative stress. Am J Physiol Regul Integr Comp Physiol. 2005;288:R337–344. doi: 10.1152/ajpregu.00469.2004. [DOI] [PubMed] [Google Scholar]
- 5.Smuder AJ, Kavazis AN, Hudson MB, Nelson WB, Powers SK. Oxidation enhances myofibrillar protein degradation via calpain and caspase-3. Free Radical Biology and Medicine. 2010;49:1152–1160. doi: 10.1016/j.freeradbiomed.2010.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du J, Wang X, Miereles C, Bailey JL, Debigare R, Zheng B, Price SR, Mitch WE. Activation of caspase-3 is an initial step triggering accelerated muscle proteolysis in catabolic conditions. The Journal of Clinical Investigation. 2004;113:115–123. doi: 10.1172/JCI200418330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecker SH, Goldberg AL, Mitch WE. Protein Degradation by the–Ubiquitin Proteasome Pathway in Normal and Disease States. Journal of the American Society of Nephrology. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 8.Murton AJ, Constantin D, Greenhaff PL. The involvement of the ubiquitin proteasome system in human skeletal muscle remodelling and atrophy. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2008;1782:730–743. doi: 10.1016/j.bbadis.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Grumati P, Coletto L, Schiavinato A, Castagnaro S, Bertaggia E, Sandri M, Bonaldo P. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy. 2011;7:1415–1423. doi: 10.4161/auto.7.12.17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 Controls Autophagy in Skeletal Muscle In Vivo. Cell metabolism. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 11.O’Leary MF, Vainshtein A, Carter HN, Zhang Y, Hood DA. Denervation-induced mitochondrial dysfunction and autophagy in skeletal muscle of apoptosis-deficient animals. Am J Physiol Cell Physiol. 2012;303:C447–454. doi: 10.1152/ajpcell.00451.2011. [DOI] [PubMed] [Google Scholar]
- 12.Sandri M. Autophagy in skeletal muscle. FEBS Lett. 2010;584:1411–1416. doi: 10.1016/j.febslet.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 13.Pal R, Cristan EA, Schnittker K, Narayan M. Rescue of ER oxidoreductase function through polyphenolic phytochemical intervention: Implications for subcellular traffic and neurodegenerative disorders. Biochemical and Biophysical Research Communications. 2010;392:567–571. doi: 10.1016/j.bbrc.2010.01.071. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez V, Pal R, Narayan M. The Oxidoreductase Behavior of Protein Disulfide Isomerase Impedes Fold Maturation of Endoplasmic Reticulum-Processed Proteins in the Pivotal Structure-Coupled Step of Oxidative Folding: Implications for Subcellular Protein Trafficking. Biochemistry. 2010;49:6282–6289. doi: 10.1021/bi100753s. [DOI] [PubMed] [Google Scholar]
- 15.Uttara B, Singh AV, Zamboni P, Mahajan RT. Oxidative Stress and Neurodegenerative Diseases: A Review of Upstream and Downstream Antioxidant Therapeutic Options. Current Neuropharmacology. 2009;7:65–74. doi: 10.2174/157015909787602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 17.Pal R, Miranda M, Narayan M. Nitrosative stress-induced Parkinsonian Lewy-like aggregates prevented through polyphenolic phytochemical analog intervention. Biochemical and biophysical research communications. 2011;404:324–329. doi: 10.1016/j.bbrc.2010.11.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pal R, Monroe TO, Palmieri M, Sardiello M, Rodney GG. Rotenone induces neurotoxicity through Rac1-dependent activation of NADPH oxidase in SHSY-5Y cells. FEBS letters. 2014;588:472–481. doi: 10.1016/j.febslet.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loehr JA, Abo-Zahrah R, Pal R, Rodney GG. Sphingomyelinase promotes oxidant production and skeletal muscle contractile dysfunction through activation of NADPH oxidase. Frontiers in Physiology. 2014;5:530. doi: 10.3389/fphys.2014.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moylan JS, Reid MB. Oxidative stress, chronic disease, and muscle wasting. Muscle & Nerve. 2007;35:411–429. doi: 10.1002/mus.20743. [DOI] [PubMed] [Google Scholar]
- 21.Powers SK, Smuder A, Judge A. Oxidative stress and disuse muscle atrophy: cause or consequence? Current opinion in clinical nutrition and metabolic care. 2012;15:240–245. doi: 10.1097/MCO.0b013e328352b4c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powers SK, Smuder AJ, Criswell DS. Mechanistic Links Between Oxidative Stress and Disuse Muscle Atrophy. Antioxidants & Redox Signaling. 2011;15:2519–2528. doi: 10.1089/ars.2011.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pal R, Palmieri M, Loehr JA, Li S, Abo-Zahrah R, Monroe TO, Thakur PB, Sardiello M, Rodney GG. Src-dependent impairment of autophagy by oxidative stress in a mouse model of Duchenne muscular dystrophy. Nature communications. 2014;5:4425–4425. doi: 10.1038/ncomms5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mello AS, da Silva Garcia C, de Souza Machado F, da Silva Medeiros N, Wohlenberg MF, Marinho JP, Dani C, Funchal C, Coelho JC. Oxidative stress parameters of Gaucher disease type I patients. Molecular Genetics and Metabolism Reports. 2015;4:1–5. doi: 10.1016/j.ymgmr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitner EB, Platt FM, Futerman AH. Common and Uncommon Pathogenic Cascades in Lysosomal Storage Diseases. The Journal of Biological Chemistry. 2010;285:20423–20427. doi: 10.1074/jbc.R110.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimitrow PPPP. Enhanced oxidative stress in hypertrophic cardiomyopathy. Pharmacological reports. 61:491. doi: 10.1016/s1734-1140(09)70091-x. [DOI] [PubMed] [Google Scholar]
- 27.Cai L, Kang YJ. Oxidative stress and diabetic cardiomyopathy. Cardiovasc Toxicol. 2001;1:181–193. doi: 10.1385/ct:1:3:181. [DOI] [PubMed] [Google Scholar]
- 28.Klaunig JE, Kamendulis LM, Hocevar BA. Oxidative Stress and Oxidative Damage in Carcinogenesis. Toxicologic Pathology. 2010;38:96–109. doi: 10.1177/0192623309356453. [DOI] [PubMed] [Google Scholar]
- 29.Klaunig JE, Kamendulis LM. THE ROLE OF OXIDATIVE STRESS IN CARCINOGENESIS. Annual Review of Pharmacology & Toxicology. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 30.Bonomini FF. Atherosclerosis and oxidative stress. Histology and histopathology. 23:381. doi: 10.14670/HH-23.381. [DOI] [PubMed] [Google Scholar]
- 31.Harrison D, Griendling KK, Landmesser U, Hornig B, Drexler H. Role of oxidative stress in atherosclerosis. The American Journal of Cardiology. 2003;91:7–11. doi: 10.1016/s0002-9149(02)03144-2. [DOI] [PubMed] [Google Scholar]
- 32.Maritim AC, Sanders RA, Watkins JB. Diabetes, oxidative stress, and antioxidants: A review. Journal of Biochemical and Molecular Toxicology. 2003;17:24–38. doi: 10.1002/jbt.10058. [DOI] [PubMed] [Google Scholar]
- 33.Wright E, Scism-Bacon JL, Glass LC. Oxidative stress in type 2 diabetes: the role of fasting and postprandial glycaemia. International Journal of Clinical Practice. 2006;60:308–314. doi: 10.1111/j.1368-5031.2006.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan LJ. Positive oxidative stress in aging and aging-related disease tolerance. Redox Biology. 2014;2:165–169. doi: 10.1016/j.redox.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 36.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 37.Price FD, Kuroda K, Rudnicki MA. Stem cell based therapies to treat muscular dystrophy. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2007;1772:272–283. doi: 10.1016/j.bbadis.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Li W-w, Li J, Bao J-k. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 40.Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaushik S, Cuervo AM. Chaperone-Mediated Autophagy. Methods in molecular biology (Clifton, NJ) 2008;445:227–244. doi: 10.1007/978-1-59745-157-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dice JF. Chaperone-Mediated Autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 43.Chiang HL, Terlecky SR, Plant CP, Dice JF. A Role for a 70-Kilodaton Heat Shock Protein in Lysosomal Degradation of Intracellular Proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 44.Fred Dice J. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends in biochemical sciences. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 45.Kaushik S, Bandyopadhyay U, Sridhar S, Kiffin R, Martinez-Vicente M, Kon M, Orenstein SJ, Wong E, Cuervo AM. Chaperone-mediated autophagy at a glance. Journal of Cell Science. 2011;124:495–499. doi: 10.1242/jcs.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The Chaperone-Mediated Autophagy Receptor Organizes in Dynamic Protein Complexes at the Lysosomal Membrane. Molecular and Cellular Biology. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salvador N, Aguado C, Horst M, Knecht E. Import of a Cytosolic Protein into Lysosomes by Chaperone-Mediated Autophagy depends on its Folding State. Journal of Biological Chemistry. 2000 doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- 48.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of Chaperone-mediated Autophagy during Oxidative Stress. Molecular Biology of the Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. Journal of Cell Science. 2000;113:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 50.Cuervo AM, Dice JF. Regulation of Lamp2a Levels in the Lysosomal Membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 51.Kim Y, Kim Y, Song W. Autophagic response to a single bout of moderate exercise in murine skeletal muscle. J Physiol Biochem. 2012;68:229–235. doi: 10.1007/s13105-011-0135-x. [DOI] [PubMed] [Google Scholar]
- 52.Cacciottolo M, Nogalska A, D’Agostino C, Engel WK, Askanas V. Chaperone-mediated autophagy components are upregulated in sporadic inclusion-body myositis muscle fibres. Neuropathology and Applied Neurobiology. 2013;39:750–761. doi: 10.1111/nan.12038. [DOI] [PubMed] [Google Scholar]
- 53.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nature cell biology. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong ASL, Cheung ZH, Ip NY. Molecular machinery of macroautophagy and its deregulation in diseases. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2011;1812:1490–1497. doi: 10.1016/j.bbadis.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 56.Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive, ubiquitin-like proteins. Nature structural & molecular biology. 2014;21:336–345. doi: 10.1038/nsmb.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mehrpour M, Esclatine A, Beau I, Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- 58.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi RK, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NYO, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai X-Y, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ETW, Juhász G, Bartholomew CR, Bassham DC, Bast RC, Batoko H, Bay B-H, Beau I, Béchet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin J-P, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahová M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac V, Carneiro LAM, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Chae H-J, Chai C-Y, Chan DC, Chan EY, Chang RC-C, Che C-M, Chen C-C, Chen G-C, Chen G-Q, Chen M, Chen Q, Chen SSL, Chen W, Chen X, Chen X, Chen X, Chen Y-G, Chen Y, Chen Y, Chen Y-J, Chen Z, Cheng A, Cheng CHK, Cheng Y, Cheong H, Cheong J-H, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang H-L, Chiarelli R, Chiba T, Chin L-S, Chiou S-H, Chisari FV, Cho CH, Cho D-H, Choi AMK, Choi D, Choi KS, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Chuang T-H, Chueh S-H, Chun T, Chwae Y-J, Chye M-L, Ciarcia R, Ciriolo MR, Clague MJ, Clark RSB, Clarke PGH, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronjé MJ, Cuervo AM, Cullen JJ, Czaja MJ, D’Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, de Groot JF, de Haan CAM, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LMD, Demarchi F, Deng YZ, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Ding W-X, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff KE, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Eissa NT, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Eng KE, Engelbrecht A-M, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Eskelinen E-L, Espert L, Espina V, Fan H, Fan J, Fan Q-W, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Farre J-C, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fésüs L, Feuer R, Figueiredo-Pereira ME, Fimia GM, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Gao F-B, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Ginsberg HN, Golab J, Goligorsky MS, Golstein P, Gomez-Manzano C, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, González-Estévez C, González-Polo RA, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Granato GE, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Guan J-L, Guan K-L, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson ÅB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Harper JW, Harris AL, Harris J, Harris SD, Hashimoto M, Haspel JA, Hayashi S-i, Hazelhurst LA, He C, He Y-W, Hébert M-J, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Höhfeld J, Holyoake TL, Hong M-H, Hood DA, Hotamisligil GS, Houwerzijl EJ, H⊘yer-Hansen M, Hu B, Hu C-aA, Hu H-M, Hua Y, Huang C, Huang J, Huang S, Huang W-P, Huber TB, Huh W-K, Hung T-H, Hupp TR, Hur GM, Hurley JB, Hussain SNA, Hussey PJ, Hwang JJ, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Isobe K-i, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jäättelä M, Jackson GR, Jackson WT, Janji B, Jendrach M, Jeon J-H, Jeung E-B, Jiang H, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jiang X, Jiménez A, Jin M, Jin SV, Joe CO, Johansen T, Johnson DE, Johnson GVW, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhász G, Juillerat-Jeanneret L, Jung CH, Jung Y-K, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kågedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Kang KB, Kang KI, Kang R, Kang Y-A, Kanki T, Kanneganti T-D, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Ke P-Y, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JAKW, Kiger AA, Kihara A, Kim DR, Kim D-H, Kim D-H, Kim E-K, Kim H-R, Kim J-S, Kim JH, Kim JC, Kim JK, Kim PK, Kim SW, Kim Y-S, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BCB, Koch JC, Koga H, Koh J-Y, Koh YH, Koike M, Komatsu M, Kominami E, Kong HJ, Kong W-J, Korolchuk VI, Kotake Y, Koukourakis MI, Flores JBK, Kovács AL, Kraft C, Krainc D, Krämer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Krüger R, Krut O, Ktistakis NT, Kuan C-Y, Kucharczyk R, Kumar A, Kumar R, Kumar S, Kundu M, Kung H-J, Kurz T, Kwon HJ, La Spada AR, Lafont F, Lamark T, Landry J, Lane JD, Lapaquette P, Laporte JF, László L, Lavandero S, Lavoie JN, Layfield R, Lazo PA, Le W, Le Cam L, Ledbetter DJ, Lee AJX, Lee B-W, Lee GM, Lee J, lee J-h, Lee M, Lee M-S, Lee SH, Leeuwenburgh C, Legembre P, Legouis R, Lehmann M, Lei H-Y, Lei Q-Y, Leib DA, Leiro J, Lemasters JJ, Lemoine A, Lesniak MS, Lev D, Levenson VV, Levine B, Levy E, Li F, Li J-L, Li L, Li S, Li W, Li X-J, Li Y-B, Li Y-P, Liang C, Liang Q, Liao Y-F, Liberski PP, Lieberman A, Lim HJ, Lim K-L, Lim K, Lin C-F, Lin F-C, Lin J, Lin JD, Lin K, Lin W-W, Lin W-C, Lin Y-L, Linden R, Lingor P, Lippincott-Schwartz J, Lisanti MP, Liton PB, Liu B, Liu C-F, Liu K, Liu L, Liu QA, Liu W, Liu Y-C, Liu Y, Lockshin RA, Lok C-N, Lonial S, Loos B, Lopez-Berestein G, López-Otín C, Lossi L, Lotze MT, Low P, Lu B, Lu B, Lu B, Lu Z, Luciano F, Lukacs NW, Lund AH, Lynch-Day MA, Ma Y, Macian F, MacKeigan JP, Macleod KF, Madeo F, Maiuri L, Maiuri MC, Malagoli D, Malicdan MCV, Malorni W, Man N, Mandelkow E-M, Manon S, Manov I, Mao K, Mao X, Mao Z, Marambaud P, Marazziti D, Marcel YL, Marchbank K, Marchetti P, Marciniak SJ, Marcondes M, Mardi M, Marfe G, Mariño G, Markaki M, Marten MR, Martin SJ, Martinand-Mari C, Martinet W, Martinez-Vicente M, Masini M, Matarrese P, Matsuo S, Matteoni R, Mayer A, Mazure NM, McConkey DJ, McConnell MJ, McDermott C, McDonald C, McInerney GM, McKenna SL, McLaughlin B, McLean PJ, McMaster CR, McQuibban GA, Meijer AJ, Meisler MH, Meléndez A, Melia TJ, Melino G, Mena MA, Menendez JA, Menna-Barreto RFS, Menon MB, Menzies FM, Mercer CA, Merighi A, Merry DE, Meschini S, Meyer CG, Meyer TF, Miao C-Y, Miao J-Y, Michels PAM, Michiels C, Mijaljica D, Milojkovic A, Minucci S, Miracco C, Miranti CK, Mitroulis I, Miyazawa K, Mizushima N, Mograbi B, Mohseni S, Molero X, Mollereau B, Mollinedo F, Momoi T, Monastyrska I, Monick MM, Monteiro MJ, Moore MN, Mora R, Moreau K, Moreira PI, Moriyasu Y, Moscat J, Mostowy S, Mottram JC, Motyl T, Moussa CEH, Müller S, Muller S, Münger K, Münz C, Murphy LO, Murphy ME, Musarò A, Mysorekar I, Nagata E, Nagata K, Nahimana A, Nair U, Nakagawa T, Nakahira K, Nakano H, Nakatogawa H, Nanjundan M, Naqvi NI, Narendra DP, Narita M, Navarro M, Nawrocki ST, Nazarko TY, Nemchenko A, Netea MG, Neufeld TP, Ney PA, Nezis IP, Nguyen HP, Nie D, Nishino I, Nislow C, Nixon RA, Noda T, Noegel AA, Nogalska A, Noguchi S, Notterpek L, Novak I, Nozaki T, Nukina N, Nürnberger T, Nyfeler B, Obara K, Oberley TD, Oddo S, Ogawa M, Ohashi T, Okamoto K, Oleinick NL, Oliver FJ, Olsen LJ, Olsson S, Opota O, Osborne TF, Ostrander GK, Otsu K, Ou J-hJ, Ouimet M, Overholtzer M, Ozpolat B, Paganetti P, Pagnini U, Pallet N, Palmer GE, Palumbo C, Pan T, Panaretakis T, Pandey UB, Papackova Z, Papassideri I, Paris I, Park J, Park OK, Parys JB, Parzych KR, Patschan S, Patterson C, Pattingre S, Pawelek JM, Peng J, Perlmutter DH, Perrotta I, Perry G, Pervaiz S, Peter M, Peters GJ, Petersen M, Petrovski G, Phang JM, Piacentini M, Pierre P, Pierrefite-Carle V, Pierron G, Pinkas-Kramarski R, Piras A, Piri N, Platanias LC, Pöggeler S, Poirot M, Poletti A, Poüs C, Pozuelo-Rubio M, Prætorius-Ibba M, Prasad A, Prescott M, Priault M, Produit-Zengaffinen N, Progulske-Fox A, Proikas-Cezanne T, Przedborski S, Przyklenk K, Puertollano R, Puyal J, Qian S-B, Qin L, Qin Z-H, Quaggin SE, Raben N, Rabinowich H, Rabkin SW, Rahman I, Rami A, Ramm G, Randall G, Randow F, Rao VA, Rathmell JC, Ravikumar B, Ray SK, Reed BH, Reed JC, Reggiori F, Régnier-Vigouroux A, Reichert AS, Reiners JJ, Reiter RJ, Ren J, Revuelta JL, Rhodes CJ, Ritis K, Rizzo E, Robbins J, Roberge M, Roca H, Roccheri MC, Rocchi S, Rodemann HP, Rodríguez de Córdoba S, Rohrer B, Roninson IB, Rosen K, Rost-Roszkowska MM, Rouis M, Rouschop KMA, Rovetta F, Rubin BP, Rubinsztein DC, Ruckdeschel K, Rucker EB, Rudich A, Rudolf E, Ruiz-Opazo N, Russo R, Rusten TE, Ryan KM, Ryter SW, Sabatini DM, Sadoshima J, Saha T, Saitoh T, Sakagami H, Sakai Y, Salekdeh GH, Salomoni P, Salvaterra PM, Salvesen G, Salvioli R, Sanchez AMJ, Sánchez-Alcázar JA, Sánchez-Prieto R, Sandri M, Sankar U, Sansanwal P, Santambrogio L, Saran S, Sarkar S, Sarwal M, Sasakawa C, Sasnauskiene A, Sass M, Sato K, Sato M, Schapira AHV, Scharl M, Schätzl HM, Scheper W, Schiaffino S, Schneider C, Schneider ME, Schneider-Stock R, Schoenlein PV, Schorderet DF, Schüller C, Schwartz GK, Scorrano L, Sealy L, Seglen PO, Segura-Aguilar J, Seiliez I, Seleverstov O, Sell C, Seo JB, Separovic D, Setaluri V, Setoguchi T, Settembre C, Shacka JJ, Shanmugam M, Shapiro IM, Shaulian E, Shaw RJ, Shelhamer JH, Shen H-M, Shen W-C, Sheng Z-H, Shi Y, Shibuya K, Shidoji Y, Shieh J-J, Shih C-M, Shimada Y, Shimizu S, Shintani T, Shirihai OS, Shore GC, Sibirny AA, Sidhu SB, Sikorska B, Silva-Zacarin ECM, Simmons A, Simon AK, Simon H-U, Simone C, Simonsen A, Sinclair DA, Singh R, Sinha D, Sinicrope FA, Sirko A, Siu PM, Sivridis E, Skop V, Skulachev VP, Slack RS, Smaili SS, Smith DR, Soengas MS, Soldati T, Song X, Sood AK, Soong TW, Sotgia F, Spector SA, Spies CD, Springer W, Srinivasula SM, Stefanis L, Steffan JS, Stendel R, Stenmark H, Stephanou A, Stern ST, Sternberg C, Stork B, Strålfors P, Subauste CS, Sui X, Sulzer D, Sun J, Sun S-Y, Sun Z-J, Sung JJY, Suzuki K, Suzuki T, Swanson MS, Swanton C, Sweeney ST, Sy L-K, Szabadkai G, Tabas I, Taegtmeyer H, Tafani M, Takács-Vellai K, Takano Y, Takegawa K, Takemura G, Takeshita F, Talbot NJ, Tan KSW, Tanaka K, Tanaka K, Tang D, Tang D, Tanida I, Tannous BA, Tavernarakis N, Taylor GS, Taylor GA, Taylor JP, Terada LS, Terman A, Tettamanti G, Thevissen K, Thompson CB, Thorburn A, Thumm M, Tian F, Tian Y, Tocchini-Valentini G, Tolkovsky AM, Tomino Y, Tönges L, Tooze SA, Tournier C, Tower J, Towns R, Trajkovic V, Travassos LH, Tsai T-F, Tschan MP, Tsubata T, Tsung A, Turk B, Turner LS, Tyagi SC, Uchiyama Y, Ueno T, Umekawa M, Umemiya-Shirafuji R, Unni VK, Vaccaro MI, Valente EM, Van den Berghe G, van der Klei IJ, van Doorn WG, van Dyk LF, van Egmond M, van Grunsven LA, Vandenabeele P, Vandenberghe WP, Vanhorebeek I, Vaquero EC, Velasco G, Vellai T, Vicencio JM, Vierstra RD, Vila M, Vindis C, Viola G, Viscomi MT, Voitsekhovskaja OV, von Haefen C, Votruba M, Wada K, Wade-Martins R, Walker CL, Walsh CM, Walter J, Wan X-B, Wang A, Wang C, Wang D, Wang F, Wang F, Wang G, Wang H, Wang H-G, Wang H-D, Wang J, Wang K, Wang M, Wang RC, Wang X, Wang XJ, Wang Y-J, Wang Y, Wang Z-B, Wang ZC, Wang Z, Wansink DG, Ward DM, Watada H, Waters SL, Webster P, Wei L, Weihl CC, Weiss WA, Welford SM, Wen L-P, Whitehouse CA, Whitton JL, Whitworth AJ, Wileman T, Wiley JW, Wilkinson S, Willbold D, Williams RL, Williamson PR, Wouters BG, Wu C, Wu D-C, Wu WKK, Wyttenbach A, Xavier RJ, Xi Z, Xia P, Xiao G, Xie Z, Xie Z, Xu D-z, Xu J, Xu L, Xu X, Yamamoto A, Yamamoto A, Yamashina S, Yamashita M, Yan X, Yanagida M, Yang D-S, Yang E, Yang J-M, Yang SY, Yang W, Yang WY, Yang Z, Yao M-C, Yao T-P, Yeganeh B, Yen W-L, Yin J-J, Yin X-M, Yoo O-J, Yoon G, Yoon S-Y, Yorimitsu T, Yoshikawa Y, Yoshimori T, Yoshimoto K, You HJ, Youle RJ, Younes A, Yu L, Yu L, Yu S-W, Yu WH, Yuan Z-M, Yue Z, Yun C-H, Yuzaki M, Zabirnyk O, Silva-Zacarin E, Zacks D, Zacksenhaus E, Zaffaroni N, Zakeri Z, Zeh IIIHJ, Zeitlin SO, Zhang H, Zhang H-L, Zhang J, Zhang J-P, Zhang L, Zhang L, Zhang M-Y, Zhang XD, Zhao M, Zhao Y-F, Zhao Y, Zhao ZJ, Zheng X, Zhivotovsky B, Zhong Q, Zhou C-Z, Zhu C, Zhu W-G, Zhu X-F, Zhu X, Zhu Y, Zoladek T, Zong W-X, Zorzano A, Zschocke J, Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the Regulation of Autophagy: Cross Talk, Shortcuts, and Feedbacks. Molecular and Cellular Biology. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS letters. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JW, Park S, Takahashi Y, Wang HG. The Association of AMPK with ULK1 Regulates Autophagy. PLoS ONE. 2010;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tripathi DN, Chowdhury R, Trudel LJ, Tee AR, Slack RS, Walker CL, Wogan GN. Reactive nitrogen species regulate autophagy through ATM-AMPK-TSC2–mediated suppression of mTORC1. Proceedings of the National Academy of Sciences. 2013;110:E2950–E2957. doi: 10.1073/pnas.1307736110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 65.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The Evolutionarily Conserved Domain of Beclin 1 is Required for Vps34 Binding, Autophagy, and Tumor Suppressor Function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 66.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-Mediated Regulation of Autophagy and Tumorigenesis Through Beclin 1 Phosphorylation. Science (New York, NY) 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Letters. 2010;584:1302–1312. doi: 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 69.Zhang J. Teaching the basics of autophagy and mitophagy to redox biologists Mechanisms and experimental approaches. Redox Biology. 2015;4:242–259. doi: 10.1016/j.redox.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C, Zhao S, Liu JO, Yu L. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem. 2003;278:29278–29287. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- 71.Tanida I, Ueno T, Kominami E. LC3 and Autophagy. In: Deretic V, editor. Autophagosome and Phagosome. Vol. 445. Humana Press; 2008. pp. 77–88. [DOI] [PubMed] [Google Scholar]
- 72.Martinez J, Malireddi RKS, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan JL, Tan H, Peng J, Kanneganti TD, Virgin HW, Green DR. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015;17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Nath S, Dancourt J, Shteyn V, Puente G, Fong WM, Nag S, Bewersdorf J, Yamamoto A, Antonny B, Melia TJ. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat Cell Biol. 2014;16:415–424. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alleman RJ, Katunga LA, Nelson MA, Brown DA, Anderson EJ. The “Goldilocks Zone” from a redox perspective-Adaptive vs. deleterious responses to oxidative stress in striated muscle. Front Physiol. 2014;5:358. doi: 10.3389/fphys.2014.00358. eCollection@2014., 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barbieri E, Sestili P. Reactive oxygen species in skeletal muscle signaling. J Signal Transduct. 2012;2012:982794. doi: 10.1155/2012/982794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jackson MJ. Redox regulation of muscle adaptations to contractile activity and aging. 2015 doi: 10.1152/japplphysiol.00760.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mason S, Wadley GD. Skeletal muscle reactive oxygen species: A target of good cop/bad cop for exercise and disease. Redox Report. 2014 doi: 10.1179/1351000213Y.0000000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Powers SK, Duarte J, Kavazis AN, Talbert EE. Reactive oxygen species are signaling molecules for skeletal muscle adaptation. Experimental Physiology. 2009 doi: 10.1113/expphysiol.2009.050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Powers SK, Jackson MJ. Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol Rev. 2008;88:1243–1276. doi: 10.1152/physrev.00031.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Powers SK, Talbert EE, Adhihetty PJ. Reactive oxygen and nitrogen species as intracellular signals in skeletal muscle. J Physiol. 2011;589:2129–2138. doi: 10.1113/jphysiol.2010.201327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reid MB. Free radicals and muscle fatigue: Of ROS, canaries, and the IOC. Free Radic Biol Med. 2008;44:169–179. doi: 10.1016/j.freeradbiomed.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 82.Guo Y, Gosker HR, Schols AM, Kapchinsky S, Bourbeau J, Sandri M, Jagoe RT, Debigare R, Maltais F, Taivassalo T, Hussain SN. Autophagy in locomotor muscles of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188:1313–1320. doi: 10.1164/rccm.201304-0732OC. [DOI] [PubMed] [Google Scholar]
- 83.Hussain SN, Sandri M. Role of autophagy in COPD skeletal muscle dysfunction. J Appl Physiol (1985) 2013;114:1273–1281. doi: 10.1152/japplphysiol.00893.2012. [DOI] [PubMed] [Google Scholar]
- 84.Lokireddy S, Wijesoma IW, Bonala S, Wei M, Sze SK, McFarlane C, Kambadur R, Sharma M. Myostatin is a novel tumoral factor that induces cancer cachexia. Biochem J. 2012;446:23–36. doi: 10.1042/BJ20112024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.McClung JM, Judge AR, Powers SK, Yan Z. p38 MAPK links oxidative stress to autophagy-related gene expression in cachectic muscle wasting. Am J Physiol Cell Physiol. 2010;298:C542–549. doi: 10.1152/ajpcell.00192.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mofarrahi M, Sigala I, Guo Y, Godin R, Davis EC, Petrof B, Sandri M, Burelle Y, HUSSAIN SNA. Autophagy and Skeletal Muscles in Sepsis. PLoS ONE. 2012;7:e47265. doi: 10.1371/journal.pone.0047265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.O’Leary MF, Hood DA. Denervation-induced oxidative stress and autophagy signaling in muscle. Autophagy. 2009;5:230–231. doi: 10.4161/auto.5.2.7391. [DOI] [PubMed] [Google Scholar]
- 88.Sandri M. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int J Biochem Cell Biol. 2013;45:2121–2129. doi: 10.1016/j.biocel.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smuder AJ, Kavazis AN, Min K, Powers SK. Exercise protects against doxorubicin-induced markers of autophagy signaling in skeletal muscle. J Appl Physiol (1985) 2011;111:1190–1198. doi: 10.1152/japplphysiol.00429.2011. [DOI] [PubMed] [Google Scholar]
- 90.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007:6. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 91.Mofarrahi M, Guo Y, Haspel JA, Choi AM, Davis EC, Gouspillou G, Hepple RT, Godin R, Burelle Y, Hussain SN. Autophagic flux and oxidative capacity of skeletal muscles during acute starvation. Autophagy. 2013;9:1604–1620. doi: 10.4161/auto.25955. [DOI] [PubMed] [Google Scholar]
- 92.Qi Z, He Q, Ji L, Ding S. Antioxidant supplement inhibits skeletal muscle constitutive autophagy rather than fasting-induced autophagy in mice. Oxid Med Cell Longev. 2014;2014:315896. doi: 10.1155/2014/315896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu Y, Palanivel R, Rai E, Park M, Gabor TV, Scheid MP, Xu A, Sweeney G. Adiponectin stimulates autophagy and reduces oxidative stress to enhance insulin sensitivity during high-fat diet feeding in mice. Diabetes. 2015;64:36–48. doi: 10.2337/db14-0267. [DOI] [PubMed] [Google Scholar]
- 94.Xie Q, Deng Y, Huang C, Liu P, Yang Y, Shen W, Gao P. Chemerin-induced mitochondrial dysfunction in skeletal muscle. J Cell Mol Med. 2015;19:986–995. doi: 10.1111/jcmm.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giordano C, Lemaire C, Li T, Kimoff RJ, Petrof BJ. Autophagy-Associated Atrophy and Metabolic Remodeling of the Mouse Diaphragm after Short-Term Intermittent Hypoxia. PLoS ONE. 2015;10:e0131068. doi: 10.1371/journal.pone.0131068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ferraro E, Giammarioli AM, Chiandotto S, Spoletini I, Rosano G. Exercise-induced skeletal muscle remodeling and metabolic adaptation: redox signaling and role of autophagy. Antioxidants & redox signaling. 2014;21:154–176. doi: 10.1089/ars.2013.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lo Verso F, Carnio S, Vainshtein A, Sandri M. Autophagy is not required to sustain exercise and PRKAA1/AMPK activity but is important to prevent mitochondrial damage during physical activity. Autophagy. 2014;10:1883–1894. doi: 10.4161/auto.32154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vainshtein A, Tryon LD, Pauly M, Hood DA. Role of PGC-1α during acute exercise-induced autophagy and mitophagy in skeletal muscle. 2015;308 doi: 10.1152/ajpcell.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481 doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim do H, Hur KY, Kim HK, Ko T, Han J, Kim HL, Kim J, Back SH, Komatsu M, Chen H, Chan DC, Konishi M, Itoh N, Choi CS, Lee MS. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013;19:83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- 101.Lira VA, Okutsu M, Zhang M, Greene NP, Laker RC, Breen DS, Hoehn KL, Yan Z. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J. 2013;27:4184–4193. doi: 10.1096/fj.13-228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carnio S, LoVerso F, Baraibar MA, Longa E, Khan MM, Maffei M, Reischl M, Canepari M, Loefler S, Kern H, Blaauw B, Friguet B, Bottinelli R, Rudolf R, Sandri M. Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep. 2014;8:1509–1521. doi: 10.1016/j.celrep.2014.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krause S. Insights into muscle degeneration from heritable inclusion body myopathies. Frontiers in Aging Neuroscience. 2015;7 doi: 10.3389/fnagi.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xiao Y, Ma C, Yi J, Wu S, Luo G, Xu X, Lin PH, Sun J, Zhou J. Suppressed autophagy flux in skeletal muscle of an amyotrophic lateral sclerosis mouse model during disease progression. Physiological reports. 2015;3 doi: 10.14814/phy2.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16 doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 106.Nogalska A, Wojcik S, Engel WK, McFerrin J, Askanas V. Endoplasmic reticulum stress induces myostatin precursor protein and NF-kappaB in cultured human muscle fibers: relevance to inclusion body myositis. Experimental neurology. 2007;204:610–618. doi: 10.1016/j.expneurol.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kwon I, Lee Y, Cosio-Lima LM, Cho JY, Yeom DC. Effects of long-term resistance exercise training on autophagy in rat skeletal muscle of chloroquine-induced sporadic inclusion body myositis. Journal of exercise nutrition & biochemistry. 2015;19:225–234. doi: 10.5717/jenb.2015.15090710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.De Palma C, Morisi F, Cheli S, Pambianco S, Cappello V, Vezzoli M, Rovere-Querini P, Moggio M, Ripolone M, Francolini M, Sandri M, Clementi E. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell death & disease. 2012;3:e418. doi: 10.1038/cddis.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dobrowolny G, Aucello M, Rizzuto E, Beccafico S, Mammucari C, Boncompagni S, Belia S, Wannenes F, Nicoletti C, Del Prete Z, Rosenthal N, Molinaro M, Protasi F, Fano G, Sandri M, Musaro A. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 110.Rahman M, Mofarrahi M, Kristof AS, Nkengfac B, Harel S, Hussain SN. Reactive oxygen species regulation of autophagy in skeletal muscles. Antioxidants & redox signaling. 2014;20:443–459. doi: 10.1089/ars.2013.5410. [DOI] [PubMed] [Google Scholar]
- 111.Irrcher I, Ljubicic V, Hood DA. Interactions between ROS and AMP kinase activity in the regulation of PGC-1alpha transcription in skeletal muscle cells. Am J Physiol Cell Physiol. 2009;296:C116–123. doi: 10.1152/ajpcell.00267.2007. [DOI] [PubMed] [Google Scholar]
- 112.Yan J, Feng Z, Liu J, Shen W, Wang Y, Wertz K, Weber P, Long J, Liu J. Enhanced autophagy plays a cardinal role in mitochondrial dysfunction in type 2 diabetic Goto-Kakizaki (GK) rats: ameliorating effects of (−)-epigallocatechin-3-gallate. J Nutr Biochem. 2012;23:716–724. doi: 10.1016/j.jnutbio.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 113.Masiero E, Sandri M. Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy. 2010;6:307–309. doi: 10.4161/auto.6.2.11137. [DOI] [PubMed] [Google Scholar]
- 114.Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, Rovira II, Gutkind S, Daniels MP, Komatsu M, Finkel T. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging. 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 116.Azad MB, Chen Y, Gibson SB. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxidants & redox signaling. 2009;11:777–790. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- 117.Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 118.Murphy Michael P. How mitochondria produce reactive oxygen species. Biochemical Journal. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Scherz-Shouval R, Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends in biochemical sciences. 2011;36:30–38. doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 120.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A. 2009;106:6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mitroulis I, Kourtzelis I, Kambas K, Rafail S, Chrysanthopoulou A, Speletas M, Ritis K. Regulation of the autophagic machinery in human neutrophils. Eur J Immunol. 2010;40:1461–1472. doi: 10.1002/eji.200940025. [DOI] [PubMed] [Google Scholar]
- 123.Zhang H, Kong X, Kang J, Su J, Li Y, Zhong J, Sun L. Oxidative stress induces parallel autophagy and mitochondria dysfunction in human glioma U251 cells. Toxicological sciences: an official journal of the Society of Toxicology. 2009;110:376–388. doi: 10.1093/toxsci/kfp101. [DOI] [PubMed] [Google Scholar]
- 124.Byun YJ, Kim SK, Kim YM, Chae GT, Jeong SW, Lee SB. Hydrogen peroxide induces autophagic cell death in C6 glioma cells via BNIP3-mediated suppression of the mTOR pathway. Neurosci Lett. 2009;461:131–135. doi: 10.1016/j.neulet.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 125.Talbert EE, Smuder AJ, Min K, Kwon OS, Szeto HH, Powers SK. Immobilization-induced activation of key proteolytic systems in skeletal muscles is prevented by a mitochondria-targeted antioxidant. J Appl Physiol (1985) 2013;115:529–538. doi: 10.1152/japplphysiol.00471.2013. [DOI] [PubMed] [Google Scholar]
- 126.Liu B, Cheng Y, Zhang B, Bian HJ, Bao JK. Polygonatum cyrtonema lectin induces apoptosis and autophagy in human melanoma A375 cells through a mitochondria-mediated ROS-p38-p53 pathway. Cancer Lett. 2009;275:54–60. doi: 10.1016/j.canlet.2008.09.042. [DOI] [PubMed] [Google Scholar]
- 127.Yuan H, Perry CN, Huang C, Iwai-Kanai E, Carreira RS, Glembotski CC, Gottlieb RA. LPS-induced autophagy is mediated by oxidative signaling in cardiomyocytes and is associated with cytoprotection. Am J Physiol Heart Circ Physiol. 2009;296:H470–479. doi: 10.1152/ajpheart.01051.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hoshino A, Matoba S, Iwai-Kanai E, Nakamura H, Kimata M, Nakaoka M, Katamura M, Okawa Y, Ariyoshi M, Mita Y, Ikeda K, Ueyama T, Okigaki M, Matsubara H. p53-TIGAR axis attenuates mitophagy to exacerbate cardiac damage after ischemia. J Mol Cell Cardiol. 2012;52:175–184. doi: 10.1016/j.yjmcc.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 129.Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jørgensen SB, Richter EA, Wojtaszewski JFP. Role of AMPK in skeletal muscle metabolic regulation and adaptation in relation to exercise. The Journal of Physiology. 2006;574:17–31. doi: 10.1113/jphysiol.2006.109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nakanishi A, Wada Y, Kitagishi Y, Matsuda S. Link between PI3K/AKT/PTEN Pathway and NOX Proteinin Diseases. Aging and disease. 2014;5:203–211. doi: 10.14336/AD.2014.0500203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Leslie NR, Bennett D, Lindsay YE, Stewart H, Gray A, Downes CP. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 2003;22:5501–5510. doi: 10.1093/emboj/cdg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Connor KM, Subbaram S, Regan KJ, Nelson KK, Mazurkiewicz JE, Bartholomew PJ, Aplin AE, Tai YT, Aguirre-Ghiso J, Flores SC, Melendez JA. Mitochondrial H2O2 regulates the angiogenic phenotype via PTEN oxidation. J Biol Chem. 2005;280:16916–16924. doi: 10.1074/jbc.M410690200. [DOI] [PubMed] [Google Scholar]
- 134.Lee SR, Yang KS, Kwon J, Lee C, Jeong W, Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 135.Tan PL, Shavlakadze T, Grounds MD, Arthur PG. Differential thiol oxidation of the signaling proteins Akt, PTEN or PP2A determines whether Akt phosphorylation is enhanced or inhibited by oxidative stress in C2C12 myotubes derived from skeletal muscle. Int J Biochem Cell Biol. 2015;62:72–79. doi: 10.1016/j.biocel.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 136.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Molecular cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 137.Qiao S, Dennis M, Song X, Vadysirisack DD, Salunke D, Nash Z, Yang Z, Liesa M, Yoshioka J, Matsuzawa S-I, Shirihai OS, Lee RT, Reed JC, Ellisen LW. A REDD1/TXNIP pro-oxidant complex regulates ATG4B activity to control stress-induced autophagy and sustain exercise capacity. Nat Commun. 2015;6 doi: 10.1038/ncomms8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sanchez AMJ, Csibi A, Raibon A, Cornille K, Gay S, Bernardi H, Candau R. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. Journal of Cellular Biochemistry. 2012;113:695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 139.Ushio-Fukai M, Alexander RW, Akers M, Yin Q, Fujio Y, Walsh K, Griendling KK. Reactive Oxygen Species Mediate the Activation of Akt/Protein Kinase B by Angiotensin II in Vascular Smooth Muscle Cells. Journal of Biological Chemistry. 1999;274:22699–22704. doi: 10.1074/jbc.274.32.22699. [DOI] [PubMed] [Google Scholar]
- 140.Ahn J, Won M, Choi JH, Kim Y, Jung CR, Im DS, Kyun ML, Lee K, Song KB, Chung KS. Reactive oxygen species-mediated activation of the Akt/ASK1/p38 signaling cascade and p21Cip1 downregulation are required for shikonin-induced apoptosis. Apoptosis. 2013;18:870–881. doi: 10.1007/s10495-013-0835-5. [DOI] [PubMed] [Google Scholar]
- 141.Okoh VO, Felty Q, Parkash J, Poppiti R, Roy D. Reactive Oxygen Species via Redox Signaling to PI3K/AKT Pathway Contribute to the Malignant Growth of 4-Hydroxy Estradiol-Transformed Mammary Epithelial Cells. PLoS ONE. 2013;8:e54206. doi: 10.1371/journal.pone.0054206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reactive Oxygen Species in Skeletal Muscle Signaling. Journal of Signal Transduction. 2012;2012 doi: 10.1155/2012/982794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: crosstalk and redox signalling. Biochemical Journal. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Huang CC, Lee CC, Lin HH, Chen MC, Lin CC, Chang JY. Autophagy-Regulated ROS from Xanthine Oxidase Acts as an Early Effector for Triggering Late Mitochondria-Dependent Apoptosis in Cathepsin S-Targeted Tumor Cells. PLoS ONE. 2015;10:e0128045. doi: 10.1371/journal.pone.0128045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stepkowski TM, Kruszewski MK. Molecular cross-talk between the NRF2/KEAP1 signaling pathway, autophagy, and apoptosis. Free Radic Biol Med. 2011;50:1186–1195. doi: 10.1016/j.freeradbiomed.2011.01.033. [DOI] [PubMed] [Google Scholar]
- 146.Spitali P, Grumati P, Hiller M, Chrisam M, Aartsma-Rus A, Bonaldo P. Autophagy is Impaired in the Tibialis Anterior of Dystrophin Null Mice. PLoS Curr. 2013;5 doi: 10.1371/currents.md.e1226cefa851a2f079bbc406c0a21e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tam BT, Pei XM, Yu AP, Sin TK, Leung KK, Au KK, Chong JT, Yung BY, Yip SP, Chan LW, Wong CS, Siu PM. Autophagic adaptation is associated with exercise-induced fibre-type shifting in skeletal muscle. Acta Physiol (Oxf) 2015;214:221–236. doi: 10.1111/apha.12503. [DOI] [PubMed] [Google Scholar]
- 148.Bentzinger C, Lin S, Romanino K, Castets P, Guridi M, Summermatter S, Handschin C, Tintignac L, Hall M, Ruegg M. Differential response of skeletal muscles to mTORC1 signaling during atrophy and hypertrophy. Skeletal Muscle. 2013;3:6. doi: 10.1186/2044-5040-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Risson V, Mazelin L, Roceri M, Sanchez H, Moncollin V, Corneloup C, Richard-Bulteau H, Vignaud A, Baas D, Defour A. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J Cell Biol. 2009;187 doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yamada E, Bastie CC, Koga H, Wang Y, Cuervo AM, Pessin JE. Mouse skeletal muscle fiber-type-specific macroautophagy and muscle wasting are regulated by a Fyn/STAT3/Vps34 signaling pathway. Cell Rep. 2012;1:557–569. doi: 10.1016/j.celrep.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jannig PR, Moreira JB, Bechara LR, Bozi LH, Bacurau AV, Monteiro AW, Dourado PM, Wisloff U, Brum PC. Autophagy signaling in skeletal muscle of infarcted rats. PLoS One. 2014;9:e85820. doi: 10.1371/journal.pone.0085820. [DOI] [PMC free article] [PubMed] [Google Scholar]