Abstract
We determined the dynamic range, reproducibility, accuracy, genotype bias, and sensitivity of the TaqMan hepatitis C virus (HCV) analyte-specific reagent (ASR). Serum samples were processed using the MagNA Pure LC instrument and run on the COBAS TaqMan 48 analyzer. The performance characteristics of the ASR were also compared with those of the qualitative AMPLICOR and quantitative AMPLICOR MONITOR HCV tests. The ASR exhibited a ≥6-log10 linear dynamic range and excellent reproducibility, with a mean coefficient of variation of 14%. HCV RNA concentration measured with the ASR agreed within an average of 0.42 log10 (2.6-fold) of the labeled concentration with members of a standard reference panel. HCV genotypes 1 to 4 were amplified with similar efficiencies with the ASR. The ASR and AMPLICOR MONITOR viral load results were significantly correlated (r = 0.8898; P < 0.01), but the agreement was poor (mean difference, 0.45 ± 0.35 log10) for 72 HCV RNA-positive clinical samples. However, 98.9% agreement between the ASR and qualitative AMPLICOR test results was found with 60 positive and 29 negative samples. Limiting-dilution experiments demonstrated that the limits of detection for ASR and AMPLICOR tests were 84 and 26 IU/ml, respectively. The performance characteristics of the TaqMan HCV ASR are appropriate for all clinical applications of HCV RNA testing.
Both qualitative and quantitative hepatitis C virus (HCV) RNA tests are used in diagnosis and management of patients with hepatitis C, because no single commercially available test combines high analytical sensitivity with a broad dynamic range. Qualitative nucleic acid amplification tests for detection of HCV RNA in serum are used to confirm the diagnosis of hepatitis C, distinguish active from resolved infection, assess virological response to therapy, and screen blood donors (3, 18, 23). Quantitative tests are used in evaluation of patients being considered for therapy and to assess early response to therapy. A pretreatment viral load of less than 800,000 IU/ml is one of several predictors of a sustained virological response (20, 24). Viral load testing has also been used in early assessment of treatment response. Patients who fail to achieve at least a 2-log10 decline in viral load after 12 weeks of treatment have little chance of a sustained response and can be spared the cost and toxicity of a complete treatment course (9, 17). However, viral load does not predict the progression of hepatitis C and is not associated with the severity of liver disease.
A variety of tests for detection and quantitation of HCV RNA based on different nucleic acid amplification technologies are commercially available. The qualitative AMPLICOR HCV and quantitative AMPLICOR HCV MONITOR version 2.0 tests (Roche Diagnostics Corporation, Indianapolis, Ind.) are based on conventional reverse transcription-PCR in a heterogeneous format (17). The VERSANT HCV RNA qualitative and VERSANT HCV RNA 3.0 quantitative assays (Bayer Healthcare, Tarrytown, N.Y.) are based on transcription-mediated amplification and branched DNA signal amplification, respectively (10, 15).
These tests also differ in their lower limits of detection and dynamic ranges. The qualitative AMPLICOR and VERSANT tests have lower limits of detection of 50 and 5 IU/ml, respectively. Although the lower limits of detection for the quantitative AMPLICOR and VERSANT tests are both approximately 600 IU/ml, the dynamic ranges differ by approximately 1 log10 and are 3.1 and 4.1 log10, respectively. Because of the differences in sensitivity between the qualitative and quantitative assays, many clinical laboratories use a quantitative test to determine viral load and a sensitive qualitative test for diagnosis and test-of-cure. A single test with sensitivity similar to the qualitative tests that accurately quantitates high viral loads would be beneficial for clinical laboratories.
A number of homogeneous TaqMan reverse transcription-PCR assays for detection and quantitation of HCV RNA have been described (12, 13, 19, 21, 29). These tests are very sensitive, have broad dynamic ranges, and provide precise quantitation of viral load. These tests also generate results more rapidly than the earlier heterogeneous tests and are not prone to amplicon carry-over contamination, since the amplification and detection steps are combined in a single closed tube.
Roche Diagnostics Corp. recently developed a TaqMan HCV analyte-specific reagent (ASR). It was designed for the newly released COBAS TaqMan analyzer, a real-time PCR instrument developed for the clinical laboratory. An ASR may be sold to clinical laboratories regulated under the Clinical Laboratory Improvement Amendments (CLIA) of 1998 as qualified to do high-complexity testing. The laboratory is responsible for verifying and validating the test, and the reports should be appended with a standard disclaimer stating that the test was developed by and its performance characteristics determined by the laboratory and that the test has not been cleared or approved by the U.S. Food and Drug Administration (FDA).
This study is the first to report the dynamic range, sensitivity, reproducibility, accuracy, and genotype bias of the ASR. The performance characteristics of the ASR were also compared with those of the qualitative AMPLICOR and quantitative AMPLICOR MONITOR HCV version 2.0 tests.
MATERIALS AND METHODS
Samples.
Serum samples were selected from stored samples that had been submitted to Emory Medical Laboratories for HCV RNA testing. The sera were removed from clots within 4 h of collection and stored at −70°C until needed. A standard reference panel calibrated against the World Health Organization 1st International Standard for HCV RNA (25) was purchased from AcroMetrix, Benicia, Calif. The panel members were assigned concentrations of 50, 500, 5,000, 200,000, 500,000, and 2,000,000 IU/ml by the manufacturer as previously described (11). All dilutions of clinical samples were made in human plasma obtained from a single outdated unit from our blood bank.
Sample preparation.
The starting sample volume for all of the tests was 200 μl. All samples tested in the AMPLICOR MONITOR HCV test were diluted 1:10 in normal human plasma prior to processing. Samples for the other tests were not diluted. HCV RNA was extracted from samples for the AMPLICOR MONITOR HCV test using the manual method according to the manufacturer's instructions. HCV RNA was extracted from serum samples for the AMPLICOR HCV test using the MagNA Pure LC instrument and the total nucleic acid reagent set (Roche Applied Science, Indianapolis, Ind.) as previously described (7).
The MagNA Pure LC instrument and total nucleic acid reagent set were also used to extract HCV RNA for the ASR. The instrument and reagents were used as recommended by the manufacturer with the following exceptions. The ASR quantitation standard (QS) was added to the lysis buffer to achieve a final concentration of 5 copies/μl (0.125 ml of HCV QS to 10 ml of lysis buffer). The purified RNA was eluted in 65 μl of elution buffer. The nominal concentration of the HCV QS in the eluant was 22 copies/μl, as recommended for the TaqMan HCV ASR by the manufacturer.
HCV RNA tests.
The microwell plate AMPLICOR and AMPLICOR MONITOR HCV version 2.0 tests were performed according to the manufacturer's instructions. The ASR consists of an HCV master mix, a QS, and a 50 mM solution of manganese acetate. The master mix contains upstream and downstream primers to the 5′ untranslated region of the HCV genome, fluorescently labeled HCV- and QS-specific oligonucleotide probes, Z05 DNA polymerase, deoxynucleotide triphosphates (dATP, dCTP, dGTP, and dUTP), AmpErase uracil-N-glycosylase, potassium acetate, dimethyl sulfoxide, glycerol, and sodium azide in tricine buffer. The HCV QS contains a noninfectious, protein-encapsulated RNA with the HCV primer binding sequences and a unique probe binding region at a concentration of 400 copies/μl, poly(rA) RNA, EDTA, amaranth dye, and ProClin 300 in sodium phosphate buffer. Single lots of master mix and QS were used throughout the study.
The master mix was activated by the addition of 170 μl of 50 mM manganese acetate, and the activated master mix was used within 60 min of preparation. Fifty microliters of processed sample or calibrator was added to 50 μl of activated master mix. The amplification and detection reactions were started within 30 min of the addition of the sample. Thermal cycle parameters were as follows: two precycles of 5 min at 50°C and 30 min at 59°C, two cycles of 15 s at 95°C and 25 s at 58°C, and 60 cycles of 15 s at 91°C and 25 s at 58°C, followed by a postcycle hold at 40°C.
The ASR was run on the COBAS TaqMan 48 analyzer. It is a real-time PCR instrument designed for the clinical laboratory that consists of two independently programmable, 24-sample thermal cyclers, a halogen light source, two 24-channel fluorescence photometers with four different filter combinations, and Amplilink software. It has a run size of 6 to 48 samples.
The ASR was calibrated by using serial 10-fold dilutions of a well-characterized clinical specimen containing HCV genotype 1 that were tested at seven different levels in quadruplicate in a single run. The starting concentration was assigned using the AMPLICOR HCV MONITOR test. The threshold or elbow values (EVs) for both HCV and QS were stored and used by the instrument software to determine the lot-specific calibration coefficients that were used in calculation of the sample HCV RNA concentrations.
HCV genotyping.
HCV genotypes were determined using a commercially available reverse hybridization, line probe assay (VERSANT HCV Genotype Assay; Bayer Healthcare) according to the manufacturer's instructions. The amplicon from the 5′ untranslated region used in the genotyping assay was generated with the AMPLICOR HCV test.
Data analysis.
Descriptive statistics, correlation coefficients, and regression line equations were calculated with the data analysis tool pack of Microsoft Excel 2000 (Microsoft Corp., Redmond, Wash.). Agreement between viral load values was assessed by the method of Bland and Altman (2). The limits of detection were determined using probit analysis (8).
RESULTS
The sample that was used to calibrate the ASR was assigned a starting concentration of 4 × 106 (6.58 log10) IU/ml with the AMPLICOR HCV MONITOR test. The undiluted sample and six serial 10-fold dilutions were tested in 11 replicates in three separate runs. The mean, standard deviation, and coefficient of variation (CV) of the EVs for each calibrator are shown in Table 1. The EVs were very reproducible over the entire concentration range, with CVs ranging from 0.48 to 2.47%. The ASR detected 100% of the replicates containing greater than or equal to 40 IU/ml and 73% of the replicates containing the lowest concentration tested, 4 IU/ml. The mean EVs and HCV RNA concentrations were highly correlated (r = 0.9961), and the assay showed a linear response over the 6-log10 range of concentrations tested. The calibration coefficients used to calculate the sample HCV RNA concentrations in subsequent experiments were derived from these data.
TABLE 1.
Reproducibility of EVs for the TaqMan HCV ASR calibratorsa
| HCV RNA (IU/ml) | No. detected/no. tested | Mean EV | SD | % CV |
|---|---|---|---|---|
| 4,000,000 | 11/11 | 22.97 | 0.272 | 1.19 |
| 400,000 | 11/11 | 26.38 | 0.199 | 0.75 |
| 40,000 | 11/11 | 29.72 | 0.252 | 0.85 |
| 4,000 | 11/11 | 32.62 | 0.204 | 0.63 |
| 400 | 11/11 | 34.60 | 0.167 | 0.48 |
| 40 | 11/11 | 37.21 | 0.599 | 1.61 |
| 4 | 8/11 | 40.8 | 0.976 | 2.47 |
Eleven replicates at each concentration were tested in three separate runs.
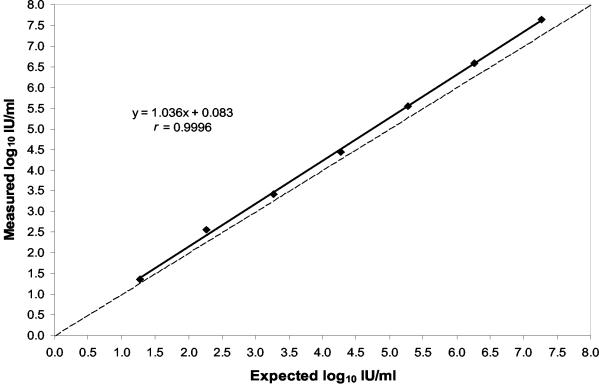
A second set of six serial 10-fold dilutions was prepared from another clinical sample with a higher starting concentration of 18,400,000 (7.27 log10) IU/ml as determined with the AMPLICOR HCV MONITOR test. The undiluted sample and the dilutions were tested in quadruplicate in a single ASR run. The plot of the measured values against the expected values is shown in Fig. 1. The measured and expected values were highly correlated (r = 0.9996) over the 6-log10 concentration range, and the equation for the linear regression line was y = 1.036x + 0.83. The slope of the line closely approximated the ideal slope of 1. The measured values were consistently larger than the expected values, with an average bias of 0.24 log10 (range, 0.1 to 0.37) or 1.7-fold. The average CV for the replicate viral load values in this dilution series was 13.9% (range, 6.6 to 19.9).
FIG. 1.
Measured versus expected concentrations of serial 10-fold dilutions of a sample with a starting concentration of 7.27 log10 IU/ml. The starting concentration was determined with the AMPLICOR HCV MONITOR test. Each point represents the mean of four replicates tested in a single run. The dashed line represents unity.
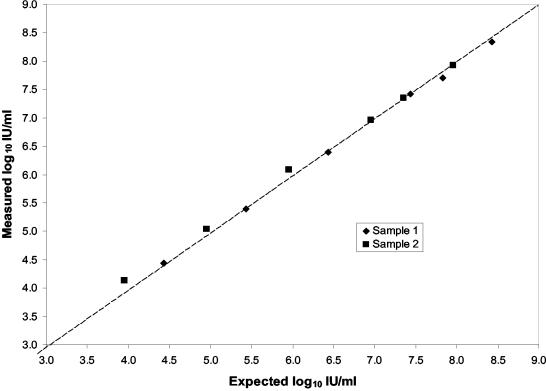
We prepared serial dilutions of two other high-titer clinical samples containing 90,000,000 (7.95 log10) and 270,000,000 (8.43 log10) IU/ml to test the upper limit of the dynamic range of the ASR. The initial concentrations were assigned with the ASR. The equations for the regression lines of the two dilution series were very similar, and the ASR demonstrated no deviation from the ideal response up to the highest concentration tested (Fig. 2). The upper limit of the linear range was at least 7.95 log10 IU/ml.
FIG. 2.
Measured versus expected concentrations of serial dilutions of two high-titer samples. ♦, sample 1; ▪, sample 2. The starting concentrations were determined with the TaqMan HCV ASR (sample 1, 8.43 log10 IU/ml; sample 2, 7.95 log10 IU/ml). Each point represents the average of duplicates tested in a single run. The dashed line represents unity.
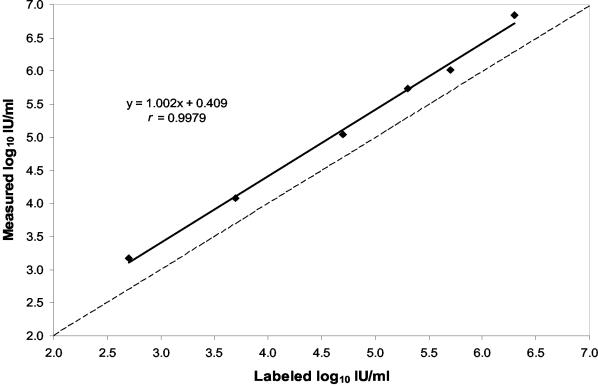
A standard reference panel was used to assess the accuracy of the ASR calibration. The panel consisted of seven members with concentrations ranging from 50 to 2,000,000 IU/ml. The panel was tested in quadruplicate in a single run. HCV RNA was detected in all replicates at each concentration. The viral load values determined for all panel members containing >50 IU/ml were very reproducible, with an average CV of 14%, and were consistently greater than the labeled concentration by an average of 0.42 log10 (2.6-fold) (Table 2). Although HCV RNA was detected in all the replicates of the 50-IU/ml panel member, the viral load value determined by the ASR, 6 IU/ml, was almost 1 log10 less than the labeled concentration and the assay at this concentration was poorly reproducible, with a CV of 53%. The measured versus labeled concentrations for all of the panel members except the 50-IU/ml member are plotted in Fig. 3. The values were highly correlated (r = 0.9979), and the slope of the linear regression line was 1.002. The line shows a consistent positive bias for the values determined with the ASR.
TABLE 2.
Reproducibility and accuracy of the TaqMan HCV ASR determined with a standard reference panel of HCV RNA
| Labeled concn (IU/ml) | Measured concna (IU/ml) | SD | CV (%) | Bias (log10) |
|---|---|---|---|---|
| 2,000,000 | 6,950,000 | 862,300 | 12 | 0.54 |
| 500,000 | 1,050,000 | 159,400 | 15 | 0.32 |
| 200,000 | 538,000 | 70,220 | 13 | 0.43 |
| 50,000 | 109,000 | 3610 | 3 | 0.34 |
| 5,000 | 12,200 | 1940 | 16 | 0.39 |
| 500 | 1,500 | 390 | 26 | 0.48 |
| 50 | 6 | 9 | 53 | −0.92 |
Each concentration was tested in quadruplicate. HCV RNA was detected in all of the replicates.
FIG. 3.
Measured versus labeled concentrations for a standard reference panel of HCV RNA tested with the TaqMan HCV ASR. Each point represents the mean of four replicates tested in a single run. The dashed line represents unity.
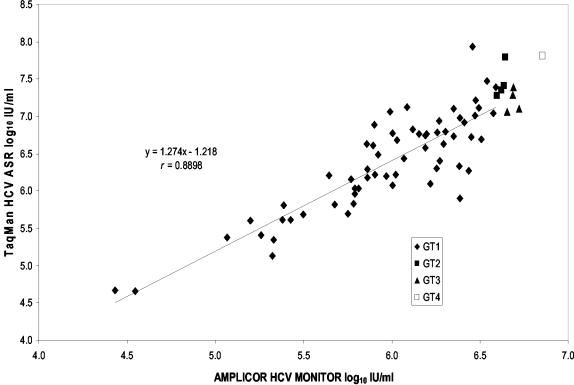
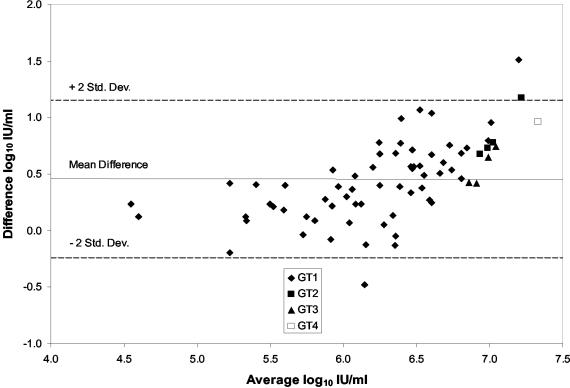
We next examined the correlation and agreement of viral load values determined with the ASR and the AMPLICOR MONITOR test for a total of 71 clinical samples: 61 with genotype 1, 4 with genotype 2, 4 with genotype 3, and 1 with genotype 4 HCV RNA. The population means (ranges) of the viral load values determined with the ASR and the MONITOR test were 6.5 (3.28) and 6.06 (2.43) log10, respectively. The values were significantly correlated (r = 0.8898; P < 0.01), and the linear regression analyses indicated that the slope and intercept were not significantly different from 1 and 0, respectively (Fig. 4). However, the agreement was poor, with a mean difference between values (ASR and MONITOR) of 0.45 log10± 0.35. The limits of agreement (mean difference ± 2 standard deviations) were −0.25 and 1.15 log10.
FIG. 4.
Correlation and linear regression analysis of viral load values obtained with the TaqMan HCV ASR and the AMPLICOR HCV MONITOR test. HCV genotypes are indicated as follows: ♦, genotype 1; ▪, genotype 2; ▴, genotype 3; □, genotype 4.
A plot of the difference versus the average viral load values for the 71 samples is shown in Fig. 5. We estimated the true viral load value for each sample by averaging the results of the two tests, since the true values were not determined with an independent reference test. The ASR values were consistently greater than the MONITOR values; however, the difference between the values measured by the two tests varied with the concentration (r = 0.6156; P < 0.01). The differences between values were consistently less than the mean difference for samples, with average viral load values of <6 log10 IU/ml, equally distributed around the mean difference for samples with average viral load values between 6 and 6.5 log10 IU/ml, and consistently greater than the mean difference for samples, with average viral load values of >6.5 log10 IU/ml.
FIG. 5.
Agreement of viral load values obtained with the TaqMan HCV ASR and the AMPLICOR HCV MONITOR test. Difference log10 IU/ml = ASR value − MONITOR value. Average log10/IU/ml = (ASR value + MONITOR value)/2. HCV genotypes are indicated as follows: ♦, genotype 1; ▪, genotype 2; ▴, genotype 3; □, genotype 4.
Serial 10-fold dilutions of clinical samples containing HCV genotypes 2, 3, and 4 were prepared and tested with the ASR. The EVs were plotted against the log10 of the HCV RNA concentration, and data were analyzed by linear regression (results not shown). The slope of the regression line is a measure of reaction efficiency for real-time PCR. The slopes for genotypes 2, 3, and 4 were −2.95, −2.65, and −3.13, respectively. The regression line slope for the genotype 1 calibrator was −2.73 (Fig. 1). The slopes of the regression lines for the different genotypes were not significantly different. The ASR amplified all of the different genotypes with similar efficiencies.
We tested the 89 clinical samples in parallel with the qualitative AMPLICOR HCV test and the ASR. The results of the two tests agreed for 98.9% of the samples (Table 3). HCV RNA was detected by both tests in 59 samples and not detected by both tests in 29 samples. HCV RNA was detected only with the AMPLICOR test in one sample. The sensitivity and specificity of the ASR with respect to the AMPLICOR test were 98.3 and 100%, respectively.
TABLE 3.
Qualitative results obtained with the AMPLICOR HCV test and the TaqMan HCV ASR for 89 clinical samples
| AMPLICOR result | ASR result
|
Total | |
|---|---|---|---|
| Detected | Not detected | ||
| Positive | 59 | 1 | 60 |
| Negative | 0 | 29 | 29 |
| Total | 59 | 30 | |
The limits of detection of the qualitative AMPLICOR HCV test and the ASR were determined and compared using serial dilutions of a reference standard with a labeled concentration of 5,000 IU/ml (Table 4). HCV RNA was detected in 100% of the replicates at a concentration of 500 IU/ml with the ASR. At concentrations of <500 IU/ml, the detection failure rate with the ASR increased as the concentration of HCV RNA decreased. The AMPLICOR test detected HCV RNA in 100% of replicates at concentrations of ≥50 IU/ml by the AMPLICOR test, and proportionally fewer replicates were detected at lower concentrations. Probit analysis indicated that the concentration at which 95% of the replicates should be positive (limit of detection) was 84 IU/ml for the ASR and 26 IU/ml for the AMPLICOR test.
TABLE 4.
Limits of detection for the AMPLICOR HCV test and the TaqMan HCV ASR
| HCV concn (IU/ml)a | AMPLICOR
|
ASR
|
||
|---|---|---|---|---|
| No. tested | No. (%) positive | No. tested | No. (%) positive | |
| 500 | 8 | 8 (100) | 8 | 8 (100) |
| 100 | 16 | 16 (100) | 16 | 15 (93.5) |
| 50 | 16 | 16 (100) | 16 | 12 (75) |
| 25 | 16 | 12 (75) | 16 | 11 (68.8) |
| 12.5 | 16 | 9 (56.3) | 16 | 9 (56.3) |
Serial dilutions were made from a reference standard with a labeled concentration of 5,000 IU/ml (AcroMetrix) to the indicated concentration.
DISCUSSION
The TaqMan HCV ASR is an example of a reagent that can be marketed to clinical laboratories under the ASR rule (5). Simply stated, an ASR is the active ingredient of an in-laboratory-developed test. The ASR rule was intended to facilitate the transfer of new technology to clinical laboratories, particularly to those high-complexity laboratories capable of using these reagents in the development of new diagnostic tests.
Under the ASR rule manufacturers are not required to seek FDA premarket approval for low-risk ASRs, which include all but blood banking tests, and those used for the diagnosis of potentially deadly infectious diseases (e.g., tuberculosis and AIDS) and genetic disorders. To qualify for the regulatory exemptions, the manufacturer cannot make analytical or clinical performance claims. It also cannot provide clinical laboratories with instructions on how to use the ASR or with appropriate calibrators and controls. In addition, the manufacturer is required to register and list the ASR with the FDA, to meet good manufacturing process standards, to report adverse events, and to restrict the distribution of the ASR to CLIA high-complexity laboratories. The clinical laboratories are required to develop and maintain the analytical performance characteristics of the test in which the ASR is used and to report the test results with a standard disclaimer.
Roche also markets a research-use-only (RUO) version of the TaqMan HCV RNA test. The reagents in an RUO test are calibrated by the manufacturer and come with additional controls and instructions for use. However, clinical laboratories that use the RUO test are still required to determine its local performance characteristics under CLIA 1998. Clinical laboratories may use either the ASR or the RUO kit for all the clinical applications of HCV RNA testing except testing of blood donors.
The TaqMan HCV ASR demonstrated a very broad dynamic range of at least 6 log10 IU/ml. The dynamic range for the ASR may actually be even broader, since with at least one sample we were able to document linearity of the assay up to 270,000,000 IU/ml. Even with the more conservative estimate of 6 log10, the ASR has a much broader dynamic range than either of the other commercially available quantitative HCV assays, the AMPLICOR HCV MONITOR version 2.0 (3.1 log10) and the VERSANT HCV RNA 3.0 (4.1 log10). The dynamic range is comparable to that described for in-laboratory-developed, real-time PCR assays for HCV RNA. We were unable to adequately characterize the performance of the ASR at the upper end of the dynamic range due to the scarcity of clinical samples and reference material with viral loads of in excess of 108 IU/ml. Based on our data, we estimate that the ASR can precisely determine viral loads from 500 to 200,000,000 IU/ml. The broad dynamic range of the ASR makes it well suited for assessing viral loads throughout a course of treatment.
The user defines the calibrators for the ASR. We chose to calibrate it using a clinical sample that was assigned a viral load value with the AMPLICOR HCV MONITOR test. A clinical sample was used rather than standard reference material because of the lack of available high-titer reference material. We used the AMPLICOR HCV MONITOR test to assign the initial concentration because it is commonly used in clinical laboratories and it is calibrated against the World Health Organization HCV international standard. We found that the standard curve was stable when using the same lot of reagents, with an average CV in the EVs of only 1.1% over three runs. A single standard curve can be run once in quadruplicate to generate lot-specific calibration coefficients.
The accuracy of the calibration was checked with a commercially available standard reference panel. The values determined with the ASR were consistently 2.6-fold greater than the labeled concentrations from 100 to 2,000,000 IU/ml. Agreement within threefold is generally considered acceptable when comparing different methods for HCV viral load measurement (28).
The within-run precision of the ASR viral load measurements was assessed with a standard reference panel and serial dilutions of a clinical sample. In each case, the average CV over the dynamic range was approximately 14%. Given this level of variation, a greater-than-twofold (0.3 log10) difference between samples will be statistically significant (P < 0.05) over most of the dynamic range of the assay. At either end of the dynamic range, threefold (0.5 log10) differences will be statistically significant (P < 0.05). The precision of the ASR compares favorably with the precision reported for the AMPLICOR HCV MONITOR version 2.0 test (16) and is similar to that reported for the VERSANT HCV RNA 3.0 assay (1, 30).
We found significant correlation but poor agreement between the results obtained with the ASR and the AMPLICOR MONITOR test for the same clinical samples. The average difference between the results was 0.45 log10, and the difference increased as the viral load increased. This is best explained by differences in the true dynamic ranges of the two tests. Although the upper limit of the linear range claimed by the manufacturer for the AMPLICOR MONITOR test is 850,000 IU/ml, other studies indicate that the test plateaus at concentrations above 500,000 IU/ml (14, 16, 22). Significant variation between values from diluted and undiluted samples was observed in those studies with samples in the range of 500,000 to 850,000 IU/ml. We diluted all samples 1:10 prior to testing with the AMPLICOR MONITOR to permit better quantitation of samples with high viral loads, and we used 8,500,000 IU/ml as the upper limit of the linear range. The sharp increase in the magnitude of the differences between test results with samples with mean viral loads greater than 3,200,000 (6.5 log10) observed here suggests that the dynamic range of the AMPLICOR MONITOR test is more limited than previously reported.
Genotype bias was a significant problem with the first version of the AMPLICOR HCV MONITOR test, due to secondary structures that could form in the target cDNA at the relatively low annealing and extension temperatures used. This was addressed by reformulation of the PCR mixture and modification of the thermal cycle parameters in the version 2.0 test. The version 2.0 test amplifies all HCV genotypes with similar efficiency (6, 16). A prototype of the TaqMan HCV ASR was also shown to amplify RNA transcripts from the different HCV genotypes with comparable efficiencies (13). We demonstrated that the ASR amplification efficiencies were similar for clinical samples with HCV genotypes 1 to 4. Like the AMPLICOR MONITOR version 2.0 and prototype TaqMan tests, the ASR was free of significant genotype bias.
The clinical and analytical sensitivities of the ASR were compared with those for the qualitative AMPLICOR HCV test. We found that the ASR detected all but 1 of the 60 positive clinical samples tested in parallel (sensitivity, 98.3%) and that the limits of detection were similar: 84 IU/ml for the ASR and 26 IU/ml for the AMPLICOR test. Since the same sample volume, nucleic acid extraction protocol, and reaction input volume were used for both tests, the difference in the limits of detection may have resulted from small differences in PCR efficiencies between the tests. The small difference in analytical sensitivity is unlikely to have a major impact on the performance of the ASR when it is used to diagnose active infections, since it is rare for pretreatment viral load values to be less than 100,000 IU/ml.
The analytical sensitivity required to adequately assess end-of-treatment virological responses in HCV infection is not well established. The VERSANT HCV RNA assay, with a 5-IU/ml limit of detection, was able to detect residual serum viral RNA in some patients who had no detectable viral RNA in the AMPLICOR HCV test at the end of treatment with interferon and subsequently experienced a virological relapse (4, 27). However, no difference between the tests was observed with end-of-treatment samples from patients treated with the newer, faster-acting, polyethylene glycol-modified interferon (26). It is unlikely that small differences in analytical sensitivity will be important in the assessment of end-of-treatment virological responses in patients treated with the most effective regimens.
The manual extraction protocol given as an example by the manufacturer in the ASR package insert uses a sample input volume of 500 μl. We used automated processing and a 200-μl input. The larger sample volume could improve the limit of detection by as much as 2.5-fold.
The COBAS TaqMan 48 analyzer is the first real-time PCR instrument designed for the clinical laboratory. The instrument was simple to use and was reliable over the 6-month evaluation period. The software allowed storage of patient demographic and order data, which is helpful for clinical laboratories. However, unlike research instruments, the software did not generate amplification plots or multicomponent views, which can be very helpful in troubleshooting real-time PCR.
In conclusion, the TaqMan HCV ASR and the COBAS TaqMan 48 analyzer are among the first real-time PCR assays and platforms designed specifically for the clinical laboratory. Together they represent a powerful new tool for quantitation of HCV RNA and can provide a single assay platform that has the required combination of analytical sensitivity, dynamic range, and precision for all the current clinical applications of HCV RNA testing.
Acknowledgments
This study was supported in part by a grant from Roche Diagnostics Corp.
REFERENCES
- 1.Beld, M., R. Sentjens, S. Rebers, C. Weegink, J. Weel, C. Sol, and R. Boom. 2002. Performance of the new Bayer VERSANT HCV RNA 3.0 assay for quantitation of hepatitis C virus RNA in plasma and serum: conversion to international units and comparison with the Roche COBAS Amplicor HCV monitor, version 2.0, assay. J. Clin. Microbiol. 40:788-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1998. Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. Morb. Mortal. Wkly. Rep. 47:1-39. [Google Scholar]
- 4.Comanor, L., F. Anderson, M. Ghany, R. Perrillo, E. J. Heathcote, C. Sherlock, I. Zitron, D. Hendricks, and S. C. Gordon. 2001. Transcription-mediated amplification is more sensitive than conventional PCR-based assays for detecting residual serum HCV RNA at end of treatment. Am. J. Gastroenterol. 96:2968-2972. [DOI] [PubMed] [Google Scholar]
- 5.Department of Health and Human Services, Food and Drug Administration. 1997. Medical devices; classification/reclassification; restricted devices; analyte specific reagents. Final rule. Fed. Regist. 62:62243-62259. [PubMed] [Google Scholar]
- 6.Doglio, A., C. Laffont, F. X. Caroli-Bosc, P. Rochet, and J.-C. Lefebvre. 1999. Second generation of the automated Cobas Amplicor HCV assay improves sensitivity of hepatitis C virus RNA detection and yields results that are more clinically relevant. J. Clin. Microbiol. 37:1567-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiebelkorn, K. R., B. G. Lee, C. E. Hill, A. M. Caliendo, and F. S. Nolte. 2002. Clinical evaluation of an automated nucleic acid isolation system. Clin. Chem. 48:1613-1615. [PubMed] [Google Scholar]
- 8.Finney, D. J. 1971. Probit analysis, 3rd ed. Cambridge University Press, London, England.
- 9.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 10.Germer, J. J., P. J. Heimgartner, D. M. Ilstrup, W. S. Harmsen, G. D. Jenkins, and R. Patel. 2002. Comparative evaluation of the VERSANT HCV RNA 3.0, QUANTIPLEX HCV RNA 2.0, and COBAS AMPLICOR HCV MONITOR version 2.0 assays for quantification of hepatitis C virus RNA in serum. J. Clin. Microbiol. 40:495-500. [Erratum, 40:1885.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorgensen, P. A., and P. D. Neuwald. 2001. Standardized hepatitis C virus RNA panels for nucleic acid testing assays. J. Clin. Virol. 20:35-40. [DOI] [PubMed] [Google Scholar]
- 12.Kawai, S., O. Yokosuka, T. Kanda, F. Imazeki, Y. Maru, and H. Saisho. 1999. Quantification of hepatitis C virus by TaqMan PCR: comparison with HCV Amplicor Monitor assay. J. Med. Virol. 58:121-126. [DOI] [PubMed] [Google Scholar]
- 13.Kleiber, J., T. Walter, G. Haberhausen, S. Tsang, R. Babiel, and M. Rosenstraus. 2000. Performance characteristics of a quantitative, homogeneous TaqMan RT-PCR test for HCV RNA. J. Mol. Diagn. 2:158-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konnick, E. Q., M. Erali, E. R. Ashwood, and D. R. Hillyard. 2002. Performance characteristics of the COBAS Amplicor hepatitis C virus (HCV) Monitor, version 2.0, international unit assay and the National Genetics Institute HCV Superquant assay. J. Clin. Microbiol. 40:768-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krajden, M., R. Ziermann, A. Khan, A. Mak, K. Leung, D. Hendricks, and L. Comanor. 2002. Qualitative detection of hepatitis C virus RNA: comparison of analytical sensitivity, clinical performance, and workflow of the Cobas Amplicor HCV test version 2.0 and the HCV RNA transcription-mediated amplification qualitative assay. J. Clin. Microbiol. 40:2903-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Gutekunst, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, S. S., E. J. Heathcote, K. R. Reddy, S. Zeuzem, M. W. Fried, T. L. Wright, P. J. Pockros, D. Haussinger, C. I. Smith, A. Lin, and S. C. Pappas. 2002. Prognostic factors and early predictability of sustained viral response with peginterferon alfa-2a (40KD). J. Hepatol. 37:500-506. [DOI] [PubMed] [Google Scholar]
- 18.Legler, T. J., J. Riggert, G. Simson, C. Wolf, A. Humpe, U. Munzel, A. Uy, M. Kohler, and K. H. Heermann. 2000. Testing of individual blood donations for HCV RNA reduces the residual risk of transfusion-transmitted HCV infection. Transfusion 40:1192-1197. [DOI] [PubMed] [Google Scholar]
- 19.Martell, M., J. Gomez, J. I. Esteban, S. Sauleda, J. Quer, B. Cabot, R. Esteban, and J. Guardia. 1999. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J. Clin. Microbiol. 37:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHutchinson, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M.-H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 21.Mercier, B., L. Burlot, and C. Ferec. 1999. Simultaneous screening for HBV DNA and HCV RNA genomes in blood donations using a novel TaqMan PCR assay. J. Virol. Methods 77:1-9. [DOI] [PubMed] [Google Scholar]
- 22.Morishima, C., M. Chung, K. W. Ng, D. J. Brambilla, and D. R. Gretch. 2004. Strengths and limitations of commercial tests for hepatitis C virus RNA quantification. J. Clin. Microbiol. 42:421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Institutes of Health. 2002. Management of hepatitis C: 2002. NIH Consensus and State-of-the-Science Statements 19(3):1-46. [PubMed] [Google Scholar]
- 24.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, and J. Albrecht. 1998. Randomised trial of interferon α2b plus ribavirin for 48 weeks or for 24 weeks verus interon α2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 25.Saldanha, J., N. Lelie, A. Heath, et al. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 26.Sarrazin, C., D. A. Hendricks, F. Sedarati, and S. Zeuzem. 2001. Assessment, by transcription-mediated amplification, of virologic response in patients with chronic hepatitis C virus treated with peginterferon α2a. J. Clin. Microbiol. 39:2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarrazin, C., G. Teuber, R. Kokka, H. Rabenau, and S. Zeuzem. 2000. Detection of residual hepatitis C virus RNA by transcription-mediated amplification in patients with complete virologic response according to polymerase chain reaction-based assays. Hepatology 32:818-823. [DOI] [PubMed] [Google Scholar]
- 28.Schirm, J., A. M. van Loon, E. Valentine-Thon, P. E. Klapper, J. Reid, and G. M. Cleator. 2002. External quality assessment program for qualitative and quantitative detection of hepatitis C virus RNA in diagnostic virology. J. Clin. Microbiol. 40:2973-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi, T., A. Katsume, T. Tanaka, A. Abe, K. Inoue, K. Tsukiyama-Kohara, R. Kawaguchi, S. Tanaka, and M. Kohara. 1999. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 116:636-642. [DOI] [PubMed] [Google Scholar]
- 30.Trimoulet, P., P. Halfon, E. Pohier, H. Khiri, G. Chene, and H. Fleury. 2002. Evaluation of the VERSANT HCV RNA 3.0 assay for quantification of hepatitis C virus RNA in serum. J. Clin. Microbiol. 40:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]