Abstract
In central and northern Wisconsin methicillin-resistant Staphylococcus aureus (MRSA) was first detected in 1989. Over the next 10-year period, 581 MRSA isolates were collected, 17.2% of which came from patients who were treated at five Native American clinics. These isolates were typed by SmaI-macrorestricted pulsed-field gel electrophoresis (PFGE). The PFGE patterns clustered the isolates into six major clonal groups (MCGs), i.e., MCGs 1 to 6, and 19 minor clonal groups (mCGs). The 25 clonal groups were represented by 109 unique PFGE types. Sixty-five percent of the MCG-2 isolates were recovered from patients who were treated at Native American clinics. Ninety-four percent of the MCG-2 isolates harbored the staphylococcal cassette chromosome mec (SCCmec) IVa. These isolates also had PFGE profiles that were clonally related to the midwestern community-associated MRSA (CA-MRSA) strain, MW2. The representative isolates from MCG-2 had the multilocus sequence type allelic profile 1-1-1-1-1-1-1 and contained pvl genes. They were also susceptible to various antibiotics, a finding consistent with the CA-MRSA phenotype. SCCmec IV was also present in other mCGs. Unlike MCG-2, isolates from the remaining five MCGs harbored SCCmec II and were resistant to multiple antibiotics, suggesting their nosocomial origin. The 19 mCGs were represented by diverse SCCmec types and three putative new variants referred to as SCCmec Ib, IIa, and IIb.
Since the first reported case of methicillin-resistant Staphylococcus aureus (MRSA) from the United Kingdom in 1961, MRSA has continued to spread in different parts of the world (9, 22, 26, 37, 44). In U.S. hospitals, MRSA infections have been steadily increasing. Surveillance data have suggested that the percentage of staphylococcal hospital infections due to MRSA has increased from 2.4% in 1975 to 54.5% in 1999 (32, 35). Genotyping of MRSA is important in order to determine outbreaks, the dissemination of virulent strains, and the understanding of its changing epidemiology. In the past, MRSA was confined to nosocomial settings, but in recent years, community-associated MRSA (CA-MRSA) has become a significant concern in the United States and other parts of the world (8, 11, 26, 44). The problem is further compounded by the fact that the ecologic distinction between hospital-acquired MRSA (HA-MRSA) and CA-MRSA seems to be decreasing (8, 39).
A number of molecular tools, such as pulsed-field gel electrophoresis (PFGE), ribotyping, and multilocus sequence typing (MLST) have been utilized to type MRSA strains (4, 12, 25, 36, 42). A great deal of epidemiologic studies on MRSA has been reported from different parts of the world. These studies have focused largely on characterizing different circulating MRSA clones and discussing their evolutionary relationships (5, 9, 13, 19-21, 23, 28). Molecular and epidemiologic distinctions have been made between MRSA strains that are typically acquired in a nosocomial or community-associated setting (30). Nosocomial MRSA isolates are represented by numerous genomic backgrounds; harbor primarily staphylococcal cassette chromosome mec (SCCmec) types I, II, III, and occasionally IV; have resistance to multiple antibiotics; and produce multiple staphylococcal toxins (38). CA-MRSA isolates, on the other hand, are represented by diverse PFGE-based genetic profiles, harbor SCCmec IV, and are susceptible to most antibiotics, except for β-lactams (10, 38). The majority of CA-MRSA strains that cause necrotizing pneumonia produce Panton-Valentine-leukocidin (PVL) toxins (24). The pvl genes also have been reported to be present in 77% of the CA-MRSA isolates (30).
Cases of CA-MRSA in the United States have been reported from Alaska (3), Chicago (15, 18, 41), Dallas (1, 17), Los Angeles (7), Minnesota and North Dakota (6, 29), and Nebraska (14). In central and northern Wisconsin, a relatively rural area in the United States, MRSA was not seen until 1989. Many clinical specimens containing MRSA that were sent to Marshfield Laboratories for identification reportedly came from five Native American clinics in central and northern Wisconsin. We undertook a comprehensive molecular epidemiologic study of these MRSA isolates collected between 1989 and 1999 from Wisconsin because specimens dating back to the early 1990s reflect the early phases of the CA-MRSA problem in the United States. We characterized and determined the clonal relationships of CA- and HA-MRSA isolates by PFGE, SCCmec typing, MLST, and the presence or absence of the pvl genes.
(A portion of the data discussed here was presented at the 2002 International Conference on Emerging Infectious Diseases meeting in Atlanta, Ga.)
MATERIALS AND METHODS
S. aureus collection and their identification.
MRSA clinical strains (n = 581) were collected from 77 healthcare facilities that included clinics (64%) and hospitals and nursing homes (36%) located in northern and central Wisconsin. The isolates were collected from 1989 to 1999. The frequency distribution of isolates in each year of the study was as follows: 1989 (n = 1), 1990 (n = 5), 1991 (n = 10), 1992 (n = 44), 1993 (n = 89), 1994 (n = 44), 1995 (n = 62), 1996 (n = 53), 1997 (n = 49), 1998 (n = 95), and 1999 (n = 129). The geographic origin of these isolates and other demographic data are described elsewhere (M. E. Stemper, S. K. Shukla, and K. D. Reed, submitted for publication). S. aureus strains were identified by colony morphology, Gram stain, and positive tests for catalase and coagulase. The SCCmec control strains—COL, PER34, BK2464, ANS46, and HDE288—were kindly provided by Herminia de Lencastre of Rockefeller University. The MW2 strain was kindly provided by Timothy Naimi of the Minnesota Department of Health. This study was reviewed and approved by the Institutional Review Board of Marshfield Clinic Research Foundation.
Antimicrobial susceptibilities.
An antibiogram for seven drugs—erythromycin (ERY), gentamicin (GEN), rifampin (RIF), ciprofloxacin (CIP), clindamycin (CLI), tetracycline (TET), and trimethoprim-sulfamethoxazole (SXT)—was determined by using the Vitek System (bioMérieux, Inc., Durham, N.C.). Isolates were considered methicillin resistant if the MIC for oxacillin was ≥4 μg/ml as determined by the E-test method according to the National Committee on Clinical Laboratory Standards (31).
PFGE.
PFGE was done on 581 MRSA isolates. The PFGE method used in the present study was essentially adapted from previously described methods (4, 27). Briefly, plugs were prepared with 250 μl of cell suspension, 4 μl of lysostaphin enzyme (1 U/μl), and an equal volume of molten 1.8% SeaPlaque agarose. Plugs were placed in lysis buffer for 4 h at 37°C. Incubation proceeded overnight at 55°C in proteinase K buffer, followed by four washes with Tris-EDTA buffer. A portion of the plug was digested with 35 U of SmaI enzyme. Separation was performed in 1% SeaKem agarose in a CHEF-DRIII system (Bio-Rad Laboratories, Hercules, Calif.) for 20 h at 200 V with switching times ramped from 5 to 40 s. Gels were poststained with ethidium bromide solution, imaged by using GelDoc 2000 (Bio-Rad Laboratories), and saved as TIFF files for analysis. The genetic relatedness of strains, shown as a dendrogram, was based on analysis of PFGE patterns of isolates by Multianalyst Fingerprinting Plus software version 1.1 (Bio-Rad Laboratories). The digested DNA from a global standard control strain, S. aureus NCTC 8325, was run in every fifth lane to normalize the PFGE band pattern of clinical strains. The dendrogram was created by using the Dice coefficient and the unweighted pair group method with a 1.5% position tolerance. The tolerance, expressed as a percentage, is the numerical value for the positional difference between two bands that are considered matching for defining relatedness.
SCCmec typing.
A total of 169 isolates were characterized for their SCCmec genotype. This included all unique PFGE types (n = 109), 32.9% (n = 155) of the strains in major clonal group 2 (MCG-2), and nine random isolates from other clonal groups. The determination of SCCmec types was done by modifying the multiplex PCR assay described by Oliveira and de Lencastre (34). The modified SCCmec multiplex protocol was as follows. The assay was carried out in a 50-μl reaction that contained 1.4× Tris buffer, 2 mM Mg2+, 200 mM concentrations of deoxynucleoside triphosphates, 0.5 U of Taq polymerase, a 50 nM concentration (each) of primer set KDPF1 and KDPR1, 100 nM concentrations of the primer sets mecAP4-mecAP7, RIF4F3-RIF4F9, DCSF2-DCSR1, and IS431P4-pUB110R1, 150 nM concentrations of the primers mecIP2-mecIP3, IS431P4-pT181R1, 200 nM concentrations of RIF5F10-RIF5R13 and CIF2F2-CIF2R2, and 3 μl of the template DNA. The primer sequences are described by Oliveira and de Lencastre (34). The reaction mix was set up in a PE 9700 thermocycler for initial denaturation at 94°C for 4 min; followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s, and elongation at 68°C for 90 s; with a final extension at 72°C for 4 min. PCR products were resolved in 2.5% SeaKem agarose gels and run at 80 V for 4 h. The following MRSA strains were used as a positive control for SCCmec types: COL (type I), PER34 (type Ia), BK2464 (type II), ANS46 (type III), and HDE288 (type IV). The definitions of the SCCmec-specific DNA loci A to G that were used to define the SCCmec variants are described by Oliveira and de Lencastre (34). The SCCmec subtyping was done by the method of Okuma et al. (33).
pvl PCR and sequencing.
A total of 156 isolates that included all isolates from the six MCGs and representative isolates from the minor clonal groups (mCGs) were screened for the presence of the pvl genes. The pvl PCR assay was performed as described by Lina et al. (24). Two independent pvl amplicons were randomly selected for sequencing to confirm their identity.
MLST.
Two representative strains from each of the six MCGs were selected for MLST to see how well they correlated with the strain clonality determined by PFGE. MLST was done by the procedure described by Enright et al. (12). The sequencing was done on an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.).
Case definitions of CA-MRSA and HA-MRSA.
Since we had no access to information about the patients' prior medical records, we were not able to determine the timeline of the exposure to MRSA and/or whether the source patients were significantly exposed to nosocomial settings prior to acquiring MRSA. Therefore, we were limited to define the terms, CA-MRSA and HA-MRSA, based on the currently described molecular characteristics associated with these phenotypes. Thus, an isolate was referred to as a CA-MRSA if it met the following criteria: (i) its SmaI-digested PFGE pattern was identical to or within one to six bands difference (one or two mutation events) with that of the well-described midwestern CA-MRSA strain, MW2, (ii) the isolate harbored both the type IV SCCmec and the pvl genes, and (iii) the isolate was sensitive to multiple antibiotics. An isolate was considered to be a HA-MRSA if (i) it was resistant to multiple antibiotics; (ii) the isolate harbored SCCmec type I, II, III, or sometimes IV; and (iii) the isolate lacked the pvl genes. As expected, we did find a small number of isolates (3.4%, n = 581) that did not clearly meet either one of the criteria. It is possible that some true CA-MRSA cases may have been missed by our strict molecular definition.
RESULTS
Clonality by PFGE.
Eighty-two percent of the Wisconsin MRSA strains were clustered into six MCGs, referred to as MCG-1, MCG-2, MCG-4, MCG-7, MCG-10, and MCG-18, based on six DNA band difference criteria (42). Isolates within each MCG had a genetic similarity index of ≥80%. An MCG was defined as a clonal group that was represented by at least 5% (n = 581) of the total isolates. In addition to MCGs, 19 mCGs were identified. An mCG was defined as a group with sporadic isolates that were represented by <5% of the total isolates. Of 581 MRSA isolates, 109 unique PFGE types were identified based on a single band difference (42).
Clonality by MLST.
The MLST allelic profile was determined on two representative isolates from all six MCGs. Five sequence types (STs)—ST1, ST5, ST45, ST225, and ST373—were identified, of which the first four have been previously reported (Table 1). Individual isolates from the same or closely related MCGs had similar allelic profiles. For example, isolates from MCGs 1, 7, and 18, which had a similarity index value of ∼75% by PFGE analysis (Fig. 1), had an MLST allelic profile of 1-4-1-4-12-1-10 (ST 5). Interestingly, the MCG-10 was represented by two STs, ST225 and the newly identified ST373 (Table 1). The allelic profiles of ST225 and ST373 were identical except for the different alleles in the gmk (4 versus 49) and the tpi (25 versus 1) locus (Table 1). The two MCG-4 isolates had the ST45. The MCG-2 isolates had an allelic profile of 1-1-1-1-1-1-1, the same as the profile for a midwestern hypervirulent strain, MW2.
TABLE 1.
Representative multilocus STs from each major clonal group and their relationships to SCCmec types
| Isolate | MCG | Allelic profile | ST | SCCmec type(s)
|
|
|---|---|---|---|---|---|
| na | mec type(s) (%) | ||||
| WI-MRSA 33 | 2 | 1-1-1-1-1-1-1 | 1 | 51 | IVa (94.1), IV (2), Varb (3.9) |
| WI-MRSA 34 | 2 | 1-1-1-1-1-1-1 | 1 | ||
| WI-MRSA 24 | 1 | 1-4-1-4-12-1-10 | 5 | 9 | II (33.3), IIa (66.7) |
| WI-MRSA 363 | 1 | 1-4-1-4-12-1-10 | 5 | ||
| WI-MRSA 214 | 7 | 1-4-1-4-12-1-10 | 5 | 19 | II (78.9); IIa (21) |
| WI-MRSA 507 | 7 | 1-4-1-4-12-1-10 | 5 | ||
| WI-MRSA 207 | 18 | 1-4-1-4-12-1-10 | 5 | 12 | II (91.7); IIa (8.3) |
| WI-MRSA 440 | 18 | 1-4-1-4-12-1-10 | 5 | ||
| WI-MRSA 75 | 4 | 10-14-8-6-10-3-2 | 45 | 10 | II (80); IV (20) |
| WI-MRSA 290 | 4 | 10-14-8-6-10-3-2 | 45 | ||
| WI-MRSA 302 | 10 | 1-4-1-4-12-25-10 | 225 | 11 | II (100) |
| WI-MRSA 280 | 10 | 1-4-1-49-12-1-10 | 373 | ||
n = number of isolates from six major clonal groups that were typed for the presence of SCCmec.
Var, SCCmec variants.
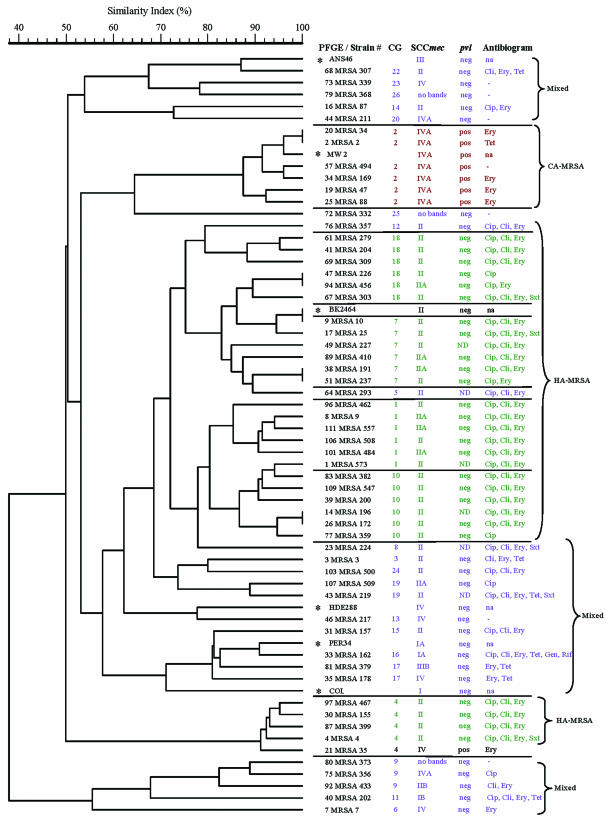
FIG. 1.
Dendrogram showing the genetic relatedness of representative isolates from 6 MCGs and 19 mCGs based on the SmaI macrorestriction pattern of genomic DNA. The corresponding SCCmec genotypes, antibiograms, and the presence or absence of pvl features are presented next to the strain numbers. PFGE types and strain identification for each isolate are shown in the first column. The scale above the dendrogram shows the percentage of the similarity index among different strains. Strain numbers with asterisks represent SCCmec MRSA positive control or reference strains. The MRSA isolates belonging to MCGs and mCGs are demarcated by horizontal lines. Properties associated with CA-MRSA, HA-MRSA, and mCGs are in red, green, and blue, respectively. No bands indicates that no amplicons in the SCCmec multiplex assay were detected with the primers used in the present study. Abbreviations: CG, clonal group; ND, not determined; neg, negative; pos, positive; na, not available.
Analysis of SCCmec types.
We determined the SCCmec types of 169 isolates that included all 109 unique PFGE types, 32.9% of MCG-2 isolates (n = 155), and 9 random isolates from other clonal groups. The aim was to determine (i) whether there was a preponderance of a specific SCCmec type in a majority of the unique PFGE types, including the major clonal types, and (ii) whether the majority of the MCG-2 isolates were of SCCmec type IV. SCCmec types II, III, and IV, subtypes Ia, IIIb, and IVa, and putative new variants of type I (named Ib) and type II (named IIa and IIb) were identified from this collection of isolates based on the presence or absence of expected SCCmec locus-specific PCR products in a multiplex assay to determine SCCmec types (34). Two isolates (WI-MRSA 291 and WI-MRSA 434) harbored SCCmec variants that could not be assigned to any known SCCmec due to the lack of enough specificity with any one type. Among isolates with unique PFGE types (n = 109), the type II SCCmec was the most common form (51.38%), followed by putative new variants (14.68%), IV (13.8%), IVa (12.8%), Ia (2.75%), type III (0.9%), and IIIb (0.92%). Of these, 2.75% did not give any bands when tested for SCCmec type. As shown above, SCCmec II and IIa together were found in 62.2% of the unique PFGE types, more than twice the number of the PFGE types that harbored SCCmec IV. Two isolates showed SCCmec profiles that did not match with any known I to IV SCCmec types (data not shown).
Identification of new SCCmec variants.
Twenty-one MRSA isolates clustered into four new SCCmec variants (Table 2). Fifteen isolates were identified as SCCmec IIa since they lacked the DNA locus G present in the left junction between IS431 and pUB110 (35). One isolate was positive for loci B (internal to the kdp operon) and D, a region in the dcs gene only. We referred to this isolate as SCCmec IIb clone. Four genetic loci—B, C (a mecI region), D, and G—are distinctive features of the SCCmec II. Three isolates were positive for loci D and G only, the specific markers for SCCmec I and Ia. Since these isolates lacked the locus A, a region downstream of the pls gene, an additional specific marker for SCCmec I, we have tentatively named the clone as SCCmec Ib. Two isolates belonging to the MCG-2 were positive for loci D, a region in the dcs gene, and E, a region between the integrated plasmid pI258 and transposon Tn554. The locus D is one marker for SCCmec types I, II, and IV, whereas the locus E is the marker for SCCmec II. We consider this clone a new mec type. All of these SCCmec variants were positive for the mecA internal control.
TABLE 2.
Characteristics of SCCmec variants
| New SCCmec variant | Loci presenta | No. of isolates | Antibiogram | mec type |
|---|---|---|---|---|
| Ib | D, G | 1 | CIP, CLI, ERY, TET | Class B1 mec |
| Ib | D, G | 1 | CIP, CLI, ERY, TET, SXT | WTc |
| Ib | D, G | 1 | CIP, CLI, ERY, TET, SXT, GEN, RIF | WT |
| IIa | B, C, D | 15 | CIP, CLI, ERY | WT |
| IIb | B, D | 1 | CLI, ERY | Class B1 mec |
| New | D, E | 2 | -b | Class B1 mec |
Definitions of loci A to G are based on the criteria of Oliveira and de Lencastre (34).
Sensitive to all seven antibiotics tested (see Materials and Methods).
WT, wild type.
Determination of CA- and HA-MRSA based on the genotypic properties.
A representative group of Wisconsin MRSA isolates from the 6 MCGs (n = 30) and 19 mCGs (n = 23) was compared to the available SCCmec types, pvl genes, and antibiogram data (Fig. 1). Available data from six reference strains—ANS46, BK2464, COL, HDE288, MW2, and PER34—were also included for comparison. The dendrogram was created on the basis of the PFGE profile only. This comparison created a clustering of isolates in three distinct groups referred to as CA-MRSA, HA-MRSA, and mixed types. The figure shows that the isolates in four PFGE-based related MCGs—MCG-18, MCG-7, MCG-1, and MCG-10—harbored either SCCmec II or IIa and were resistant to CIP, CLI, and ERY. They were also negative for the pvl genes by PCR. The MCG-4, which was somewhat distantly related to the four previous MCGs, had the same genetic markers and antibiogram with the exception of the WI-35 strain that harbored SCCmec IV and pvl genes. The genotypic trait, such as the presence of SCCmec II, multidrug resistance, and the absence of pvl genes suggest that the MRSA isolates in these MCGs were HA-MRSA. The strain WI-35 probably represents an example of a HA-MRSA strain that had acquired the phenotypic and some genotypic features typically associated with CA-MRSA.
The molecular characteristics of MCG-2 isolates shown in Fig. 1 were different from other MCGs. Of the 51 MCG-2 isolates that were SCCmec genotyped, 94% were type IVa, 4% were SCCmec variants, and 2% were not subtyped beyond type IVa. These isolates also had pvl genes and were sensitive to multiple antibiotics, including CLI and fluoroquinolones. Most strains from the MCG-2 had the SmaI-restricted PFGE pattern that were indistinguishable from, or closely related to, the MW2 strain, the prototype of a CA-MRSA strain. The remaining isolates of MCG-2 had a PFGE banding pattern within the six bands difference of MW2. Like MW2, representative isolates belonging to the MCG-2 had the allelic profile 1-1-1-1-1-1-1 (Fig. 1 and Table 1). The majority of these isolates came from five outpatient clinics in Wisconsin, which primarily served Native American patients (Stemper et al., submitted). The presence of the SCCmec IV and the pvl genes and the lack of resistance to multiple antibiotics are features that are strongly suggestive of CA-MRSA (30). The fact that Wisconsin MCG-2 isolates had these features suggested that they were indeed CA-MRSA.
The representative isolates from the 19 mCGs (Fig. 1, shown in the blue font) were clustered into three unrelated PFGE based groups. All four SCCmec types or their variants such as Ia, Ib, IIIb, and IVa were represented in these three groups. The antibiogram profiles of MRSA isolates in these groups were variable and ranged from being susceptible to all antibiotics tested to resistant to several antibiotics such as CIP, CLI, ERY, TET, GEN, and RIF. Interestingly, isolates in these three clusters lacked the pvl genes. Thus, these three clusters show a heterogeneous mixture of MRSA that have molecular characteristics representative of both the CA-MRSA and the HA-MRSA.
DISCUSSION
Our PFGE data show the presence of several MRSA major and minor clones circulating in the rural communities of northern and central Wisconsin. The representative isolates from MCGs 1, 7, and 18 had the same ST, ST5, suggesting those strains may have originated from a single clone. Additional evidence that supports this conclusion was based on the presence of a similar antibiogram, the same SCCmec, and the lack of pvl genes. The MLST allelic profiles of the representative isolates of MCG-10 were represented by STs 225 and 373 due to differences in alleles of gmk and tpi genes. A single base change (in WI-MRSA 280) at nucleotide position 329 (T→A) in the gmk gene created the new ST373 (Table 1).
MLST data also suggest the presence of at least two international clones, ST5 and ST45, described as the New York/Japan clone and the Berlin clone, respectively, in Wisconsin (13, 43). ST45 has been reported from several European countries and in a CA-MRSA from western Australia (http://saureus.mlst.net/). The ST225, which is related to ST5, has been reported in MRSA isolates from the United States (13). Our ST373 is a new ST represented in the midwestern United States. When we combine the MLST and SCCmec data to define clonality, we could identify five clonal groups: ST1-IV, ST5-II, ST45-II, ST225-II, and ST373-II. Of these five, ST1-IV (CA-MRSA), ST5-II (New York/Japan clone), and ST45-11 have been described before (2, 13). To the best of our knowledge, the remaining two combinations have not been reported yet. Therefore, they may well represent some of the HA-MRSA clones still limited to the midwestern United States.
One of the significant features that emerged from the molecular characterization of MRSAs from rural Wisconsin was the presence of a single clonal group, MCG-2, that had very distinct genotypic features of CA-MRSA. The majority of these isolates came from clinics primarily visited by Native American communities. This fact strongly suggests the presence of predominantly a single CA-MRSA clone circulating in the Native American communities of Wisconsin. Our molecular data show that almost all CA-MRSA present in Wisconsin are identical or clonally related to the hypervirulent strain, MW2, the strain that caused fatal septicemia and septic arthritis in a 16-month-old girl in North Dakota, in 1998 (6). The presence of CA-MRSA in the late 1990s in a rural Native American community in the midwestern United States has been described previously based on the patient record (16). However, our data suggest the presence of the MW2 related clone in a Native American population in the midwestern United States since the early 1990s. Another marker for the Wisconsin CA-MRSA is the presence of the class B1 mec complex in mec DNA. About 25% of these isolates had this deleted version of the mec complex (40).
The fact that SCCmec IV was present in several PFGE types suggests the possibility of multiple and independent acquisition of this mec genotype in different MRSA genetic backgrounds. However, a high percentage of the SCCmec IV had the same mecA promoter mutation and the same type of deletion in the mec complex suggests that source(s) of SCCmec IV in Wisconsin MRSA isolates may be limited (40). It is likely to be from a single or a very small number of source organism(s).
In conclusion, several important observations were made from the present study of a collection of temporal isolates from a predominantly rural environment where the dynamics of nosocomial and community-based spread of infectious diseases is expected to be different from the coastal U.S. cities and hospitals. First, the molecular evidence that CA-MRSA coming out of the Native American clinics in northern and central Wisconsin were clones of the virulent strain MW2. Second, it seems that there are three closely related clones of HA-MRSA circulating in hospitals and long-term care institutions in Wisconsin. Third, almost all CA-MRSA isolates contain SCCmec IV and pvl genes, but not all SCCmec IV harboring MRSA would be CA-MRSA. It appears that the pvl genes and SCCmec IV together offers some selective advantage for CA-MRSA (43). In addition, SCCmec type IV was integrated into some isolates of other mCGs representing multidrug-sensitive nosocomially related isolates. Whether the acquisition of SCCmec IV by nosocomial isolates provides them any advantage in colonization remains to be seen. Genetically, it should be more favorable for an S. aureus strain to acquire an approximately 21- to 24-kb genetic element with two functional recombinase genes than for an approximately 67-kb element to gain mecA based methicillin resistance. Smaller genetic elements, such as SCCmec IV, can be packaged in phage more efficiently than larger genetic elements.
Acknowledgments
The Marshfield Clinic Research Foundation provided financial support for this study.
We thank Alice Stargardt and Linda Weis for help with preparation of the manuscript.
REFERENCES
- 1.Adcock, P. M., P. Pastor, F. Medley, J. E. Patterson, and T. V. Murphy. 1998. Methicillin-resistant Staphylococcus aureus in two child care centers. J. Infect. Dis. 178:577-580. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Baggett, H. C., T. W. Hennessy, R. Leman, C. Hamlin, D. Bruden, A. Reasonover, P. Martinez, and J. C. Butler. 2003. An outbreak of community-onset methicillin-resistant Staphylococcus aureus skin infections in southwestern Alaska. Infect. Control Hosp. Epidemiol. 24:397-402. [DOI] [PubMed] [Google Scholar]
- 4.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branger, C., C. Gardye, J. O. Galdbart, C. Deschamps, and N. Lambert. 2003. Genetic relationship between methicillin-sensitive and methicillin-resistant Staphylococcus aureus strains from France and from international sources: delineation of genomic groups. J. Clin. Microbiol. 47:2946-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus-Minnesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep. 48:707-710.[Online.] http://www.cdc.gov/mmwr/preview/mmwrhtml/mm4832a2.htm. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2003. Methicillin-resistant Staphylococcus aureus infections among competitive sports participants-Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000-2003. Morb. Mortal. Wkly. Rep. 52:793-795. [PubMed] [Google Scholar]
- 8.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daum, R. S., T. Ito, K. Hiramatsu, F. Hussain, K. Mongkolrattanothai, M. Jamklang, and S. Boyle-Vavra. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344-1347. [DOI] [PubMed] [Google Scholar]
- 11.Dufour, P., Y. Gillet, M. Bes, G. Lina, F. Vandenesch, D. Floret, J. Etienne, and H. Richet. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35:819-824. [DOI] [PubMed] [Google Scholar]
- 12.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fey, P. D., B. Said-Salim, M. E. Rupp, S. H. Hinrichs, D. J. Boxrud, C. C. Davis, B. N. Kreiswirth, and P. M. Schlievert. 2003. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank, A. L., J. F. Marcinak, P. D. Mangat, and P. C. Schreckenberger. 1999. Community-acquired and clindamycin-susceptible methicillin-resistant Staphylococcus aureus in children. Pediatr. Infect. Dis. J. 18:993-1000. [DOI] [PubMed] [Google Scholar]
- 16.Groom, A. V., D. H. Wolsey, T. S. Naimi, K. Smith, S. Johnson, D. Boxrud, K. A. Moore, and J. E. Cheek. 2001. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA 286:1201-1205. [DOI] [PubMed] [Google Scholar]
- 17.Haley, R. W., N. B. Cushion, F. C. Tenover, T. L. Bannerman, D. Dryer, J. Ross, P. J. Sanchez, and J. D. Siegel. 1995. Eradication of endemic methicillin-resistant Staphylococcus aureus infections from a neonatal intensive care unit. J. Infect. Dis. 171:614-624. [DOI] [PubMed] [Google Scholar]
- 18.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 19.Hiramatsu, K. 1995. Molecular evolution of MRSA. Microbiol. Immunol. 39:531-543. [DOI] [PubMed] [Google Scholar]
- 20.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistance Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 21.Hiramatsu, K., N. Kondo, and T. Ito. 1996. Genetic basis for molecular epidemiology of MRSA. J. Infect. Chemother. 2:117-129. [DOI] [PubMed] [Google Scholar]
- 22.Jevons, M. P. 1961. “Celbenin”-resistant staphylococci. Br. Med. J. 1:124-125. [Google Scholar]
- 23.Kreiswirth, B., J. Kornblum, R. D. Arbeit, W. Eisner, J. N. Maslow, A. McGeer, D. E. Low, and R. P. Novick. 1993. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 259:227-230. [DOI] [PubMed] [Google Scholar]
- 24.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 25.Lipuma, J. J. 1998. Molecular tools for epidemiologic study of infectious diseases. Pediatr. Infect. Dis. J. 17:667-675. [DOI] [PubMed] [Google Scholar]
- 26.Maguire, G. P., A. D. Arthur, P. J. Boustead, B. Dwyer, and B. J. Currie. 1996. Emerging epidemic of community-acquired methicillin-resistant Staphylococcus aureus infection in the Northern Territory. Med. J. Aust. 164:721-723. [DOI] [PubMed] [Google Scholar]
- 27.Matushek, M. G., M. J. Bonten, and M. K. Hayden. 1996. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J. Clin. Microbiol. 10:2598-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musser, J. M., and V. Kapur. 1992. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J. Clin. Microbiol. 30:2058-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naimi, T. S., K. H. LeDell, D. J. Boxrud, A. V. Groom, C. D. Steward, S. K. Johnson, J. M. Besser, C. O'Boyle, R. N. Danila, J. E. Cheek, M. T. Osterholm, K. A. Moore, and K. E. Smith. 2001. Epidemiology and clonality of community-acquired methicillin-resistance Staphylococcus aureus in Minnesota, 1996-1998. Clin. Infect. Dis. 33:990-996. [DOI] [PubMed] [Google Scholar]
- 30.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976-2984. [DOI] [PubMed] [Google Scholar]
- 31.National Committee on Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S9. Ninth supplement, vol. 19. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 32.National Nosocomial Infections Surveillance System. 1999. National Nosocomial Infections Surveillance (NNIS) System report: data summary from January 1990-May 1999. Am. J. Infect. Control 27:520-532. [DOI] [PubMed] [Google Scholar]
- 33.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panlilio, A. L., D. H. Culver, R. P. Gaynes, S. Banerjee, T. S. Henderson, J. S. Tolson, and W. J. Martone. 1992. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975-1991. Infect. Control Hosp. Epidemiol. 13:582-586. [DOI] [PubMed] [Google Scholar]
- 36.Prevost, G., B. Jaulhac, and Y. Piemont. 1992. DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J. Clin. Microbiol. 30:967-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riley, T. V., and I. L. Rouse. 1995. Methicillin-resistant Staphylococcus aureus in Western Australia, 1983-1992. J. Hosp. Infect. 29:177-188. [DOI] [PubMed] [Google Scholar]
- 38.Said-Salim, B., B. Mathma, and B. N. Kreiswirth. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging pathogen. Infect. Control Hosp. Epidemiol. 24:451-455. [DOI] [PubMed] [Google Scholar]
- 39.Saiman, L., M. O'Keefe, P. L. Graham III, F. Wu, B. Said-Salim, B. Krieswirth, A. LaSala, P. M. Schlievert, and P. Della-Latta. 2003. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin. Infect. Dis. 37:1313-1319. [DOI] [PubMed] [Google Scholar]
- 40.Shukla, S. K., S. V. Ramaswamy, J. M. Conradt, M. E. Stemper, R. A. Reich, K. D. Reed, and E. A. Graviss. 2004. Identification of novel polymorphisms in mec genes and a new mec complex type in methicillin-resistant Staphylococcus aureus from rural Wisconsin. Antimicrob. Agents Chemother. 48:3080-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suggs, A. H., M. C. Maranan, S. Boyle-Vavra, and R. S. Daum. 1999. Methicillin-resistant and borderline methicillin-resistant asymptomatic Staphylococcus aureus colonization in children without identifiable risk factors. Pediatr. Infect. Dis. 18:410-414. [DOI] [PubMed] [Google Scholar]
- 42.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi, T., Y. Yokota, J. Terajima, T. Hayashi, M. Aepfelbacher, M. Ohara, H. Komatsuzawa, H. Watanabe, and M. Sugai. 2002. Clonal association of Staphylococcus aureus causing bullous impetigo and the emergence of new methicillin-resistant clonal groups in Kansai district in Japan. J. Infect. Dis. 185:1511-1516. [DOI] [PubMed] [Google Scholar]