Abstract
The cytokine IFN-γ coordinates macrophage activation and is essential for control of pathogens including Mycobacterium tuberculosis. However, the mechanisms by which IFN-γ controls M. tuberculosis infection are only partially understood. Here, we show that the transcription factor HIF-1α is an essential mediator of IFN-γ dependent control of M. tuberculosis infection both in vitro and in vivo. M. tuberculosis infection of IFN-γ activated macrophages results in a synergistic increase in HIF-1α protein levels. This increase in HIF-1α levels is functionally important, as macrophages lacking HIF-1α are defective for IFN-γ dependent control of infection. RNA-seq profiling demonstrates that HIF-1α regulates nearly half of all IFN-γ inducible genes during infection of macrophages. In particular, HIF-1α regulates production of important immune effectors including inflammatory cytokines and chemokines, eicosanoids, and nitric oxide (NO). In addition, we find that during infection HIF-1α coordinates a metabolic shift to aerobic glycolysis in IFN-γ activated macrophages. We find that this enhanced glycolytic flux is crucial for IFN-γ dependent control of infection in macrophages. Furthermore, we identify a positive feedback loop between HIF-1α and aerobic glycolysis that amplifies macrophage activation. Finally, we demonstrate that HIF-1α is crucial for control of infection in vivo as mice lacking HIF-1α in the myeloid lineage are strikingly susceptible to infection, and exhibit defective production of inflammatory cytokines and microbicidal effectors. In conclusion, we have identified HIF-1α as a novel regulator of IFN-γ dependent immunity that coordinates an immunometabolic program essential for control of M. tuberculosis infection in vitro and in vivo.
Introduction
Mycobacterium tuberculosis, the causative agent of tuberculosis, infects 2 billion people worldwide and is responsible for more deaths annually than any other single bacterial pathogen (1). Interferon-γ (IFN-γ) activation of macrophages leads to restriction of M. tuberculosis growth and is crucial for successful immunity. Patients lacking components of the IFN-γ signaling pathway are highly susceptible to mycobacterial infection (2). Similarly, mice lacking IFN-γ rapidly succumb to infection with M. tuberculosis (3, 4). Some of the proposed anti-bacterial responses induced by IFN-γ include nutrient restriction (5), enhanced production of antimicrobial peptides (6, 7), autophagy (8), expression of cell intrinsic restriction factors including interferon inducible GTPases (9, 10), and production of nitric oxide (NO) by inducible nitric oxide synthase (iNOS). NO has bactericidal activity against M. tuberculosis (11) and is essential for host defense against M. tuberculosis infection in mice (12), accounting for a substantial portion of the susceptibility of IFN-γ deficient mice.
The transcription factor hypoxia inducible factor-1α (HIF-1α) canonically functions to induce glycolytic gene expression under conditions of hypoxia. More recently, HIF-1α has been implicated in macrophage function. HIF-1α contributes to the transition to aerobic glycolysis and expression of genes associated with M1 polarization in response to LPS stimulation (13). In the context of sepsis, HIF-1α was demonstrated to mediate a transition from a pro-inflammatory to an immunosuppressive phenotype while maintaining antimicrobial and protective functions (14). Further, HIF-1α has been identified as important for control of Group A Streptococcus (GAS), Pseudomonas aeruginosa, and uropathogenic E. coli (15–17). The susceptibility of HIF-1α deficient mice to infection has been attributed to an inability to produce the ATP required for migration to sites of inflammation (15, 18) and to decreased production of iNOS and antimicrobial peptides (15–17). Intriguingly, recent studies suggest that HIF-1α may play a role in host defense against mycobacteria. Exogenously increasing the levels of active HIF-1α using pharmacological or genetic tools in zebrafish embryos enhances bactericidal activity against Mycobacterium marinum (19). Mice lacking HIF-1α in the myeloid lineage exhibit more rapid progression of hypoxic granulomatous lesions in the liver following intravenous infection with Mycobacterium avium (20). These data suggest the intriguing possibility that HIF-1α may be an important mediator of resistance to M. tuberculosis infection. Importantly, HIF-1α is thought to be important in the context of hypoxia or during innate immune responses to infection, and has not previously been shown to be involved in IFN-γ dependent immunity.
In this study, we demonstrate that HIF-1α is required for host defense against infection with virulent M. tuberculosis. Mice lacking HIF-1α in the myeloid lineage are strikingly susceptible to infection. Surprisingly, we do not find evidence that HIF-1α is important for innate defense of macrophages against M. tuberculosis. In contrast, we find that HIF-1α is critical for IFN-γ dependent control of M. tuberculosis infection. RNA-seq reveals that approximately half of the transcriptional response to IFN-γ during M. tuberculosis infection requires HIF-1α. Further, HIF-1α deficient macrophages are impaired for important effector functions including production of NO, PGE2, as well as inflammatory cytokines and chemokines. In addition to regulating these key immune effector functions, we find that HIF-1α regulates a metabolic transition to aerobic glycolysis in IFN-γ activated macrophages during M. tuberculosis infection. We show that this transition to aerobic glycolysis is required for IFN-γ dependent control of M. tuberculosis infection of macrophages. In addition, we identify a positive feedback loop between HIF-1α and glycolytic flux that reinforces IFN-γ mediated activation of macrophages and control of M. tuberculosis infection.
Materials and Methods
Ethics Statement
All procedures involving the use of mice were approved by the University of California, Berkeley IACUC, the Animal Care and Use Committee (protocol number R353-1113B). All protocols conform to federal regulations, the National Research Council’s Guide for the Care and Use of Laboratory Animals and the Public Health Service’s (PHS’s) Policy on Humane Care and Use of Laboratory Animals.
Reagents
Recombinant mouse IFN-γ (485 MI/CF) was obtained from R&D systems, Minneapolis, MN, and was used at indicated concentrations. 2-deoxyglucose (2-DG) and D-glucose were obtained from Sigma Aldrich, St Louis, MO and were used at indicated concentrations. U-13C-glucose was obtained from Cambridge Isotope Laboratories, Andover, MA. Dimethyloxalylglycine (DMOG) was obtained from Cayman Chemical, Ann Arbor, MI and was used at indicated concentrations.
Mice and cell culture
WT mice were C57BL/6 and were obtained from the Jackson Laboratory, Bar Harbor, ME. All knockout mice were backcrossed to C57BL/6. B6.129-Hif1atm3Rsjo/J mice were obtained from the Jackson Laboratory and were crossed with B6.129P2-Lyz2tm1(cre)Ifo/J to generate mice that had Hif1a deletion targeted to the myeloid lineage. B6.129P2-Nos2tm1Lau/J mice were obtained from the Jackson Laboratory and were bred in house. Bone marrow derived macrophages (BMDM) were obtained by flushing cells from the femurs and tibias of mice and culturing in DMEM with 10% FBS and 10% supernatant from 3T3-M-CSF cells (BMDM media) for 6 days with feeding on day 3. After differentiation, BMDM continued to be cultured in BMDM media containing M-CSF.
Bacterial culture
The M. tuberculosis strain Erdman was used for all experiments. M. tuberculosis was grown in Middlebrook 7H9 liquid media supplemented with 10% ADS (albumin-dextrose-saline), 0.4% glycerol, and 0.05% Tween-80 or on solid 7H10 agar plates supplemented with 10% Middlebrook OADC (BD) and 0.4% glycerol. The TB-lux strain used for measuring bacterial growth was derived from an Erdman strain, and was cultured as described above.
In vitro infections
BMDM were plated into 96-well or 24-well plates with 5×104 and 3×105 macrophages per well respectively, and were allowed to adhere and rest for 24 hours. BMDM were then treated with vehicle or IFN-γ (1.25 ng/mL) overnight and then infected in DMEM supplemented with 5% horse serum and 5% FBS at an MOI of 5 unless otherwise noted. After a 4 hour phagocytosis period, infected BMDM were washed with PBS before replacing with BMDM media. For experiments with DMOG, 2-DG or galactose, these reagents were added to the BMDM media after the 4 hour phagocytosis. For 2-DG and galactose treatment, this was done to minimize the amount of time that the BMDM experienced glycolytic inhibition. For IFN-γ pretreated wells, IFN-γ was also added after infection at the same concentration. To measure intracellular growth of M. tuberculosis, cells were infected with TB-lux (Erdman) and luminescence was measured at 32°C immediately following the 4 hour phagocytosis, PBS wash, and media replacement. Luminescence was then read again at the noted time points. All growth was normalized to day 0 luminescence readings for each infected well, and is presented as fold change in luminescence compared to day 0. For enumeration of CFU, M. tuberculosis Erdman strain was used, and infected BMDM were washed with PBS, lysed in water with 0.1% Triton-X 100 for ten minutes, and serial dilutions were prepared in PBS with 0.05% Tween-80 and were plated onto 7H10 plates.
In vivo infections
Cohorts of age and sex matched wild-type, Nos2−/−, and Hif1afl/flLysMcre+/+ mice were infected by aerosol route with M. tuberculosis strain Erdman. All mice were on the C57BL/6 background, and were 7–10 weeks of age when infected, with cohorts of 10–12 mice for each genotype for time to death experiments. Aerosol infection was done using a Nebulizer and Full Body Inhalation Exposure System (Glas-Col, Terre Haute, IN). 9mL of an OD600=.01 culture was loaded into the nebulizer. This resulted in ~400 CFU per mouse 1 day after infection. Mice were weighed the day of infection, and weights were followed until a humane 15% weight loss cutoff was reached at which point the mice were euthanized. For CFU, one lung lobe (the largest) was homogenized in PBS+.05%Tween-80, and serial dilutions were plated on 7H10 plates. For CFU experiments, cohorts of 4–5 mice were used for each genotype at each timepoint. For qRT-PCR experiments, lung lobes from 3–5 mice per genotype were pooled, and a cell suspension was obtained by pressing the lungs through a 40um filter. The cells were then washed and CD11b+ cells were purified by MACS magnetic bead separation using CD11b MicroBeads (130-049-601, Miltenyi Biotec, San Diego, CA) as described in the manufacturer’s protocol. Cells were purified to ~95% CD11b+. For qPCR, purified CD11b+ cells were lysed in TRIzol and qRT-PCR was performed as described below. For NO measurements from CD11b+ cells ex vivo, the same procedure described above was performed, and CD11b+ cells were re-plated in 96-well plates with 3×105 cells per well, and griess assays were performed on the supernatants at the indicated timepoints.
Griess Assays
The griess reaction was used to detect nitrite in the supernatants of BMDM or ex vivo CD11b+ cells as a proxy for NO production. Briefly, a solution of .2% napthylethylenediamine dihydrochloride was mixed 1:1 with a 2% sulfanilamide, 4% phosphoric acid solution. 50uL of supernatant from cultured cells was added to 50uL of this mixture, and absorbance was measured at 546nm. Concentrations were determined by comparing to a standard curve of nitrite in BMDM media.
Western blots
Infected BMDM were washed with PBS, lysed in 1x SDS-PAGE buffer on ice, and heat sterilized for 30 min at 100°C. Total protein lysates were analyzed by SDS-PAGE using pre-cast Tris-HCl criterion gels (Bio-Rad, Hercules, CA). The following primary antibodies were used: rabbit antibody against HIF-1α (NB100-479, Novus Biologicals, Littleton, CO and also D2U3T, Cell Signaling Technology, Danvers, MA), goat antibody against mouse IL-1b (AF-401-NA, R&D systems, Minneapolis, MN). HRP conjugated secondary antibodies were used. Western Lightning Plus-ECL chemiluminescence substrate (PerkinElmer, Waltham, MA) was used and blots were developed on film or using a ChemiDoc MP System (Bio-Rad, Hercules, CA). Blots were stripped using 0.2 M NaOH then washed in ddH2O and TBST before blocking and reprobing for actin as a loading control, using an HRP conjugated rabbit antibody against β-actin (13E5, Cell Signaling Technology, Danvers, MA).
qRT-PCR and RNA-seq
For q-RT-PCR, 3×105 BMDM were seeded in 24-well dishes and infected as described. At 24h post-infection, cells were washed with room temperature PBS and lysed in 500uL TRIzol (Invitrogen Life Technologies, Carlsbad, CA). Total RNA was extracted using chloroform (100 μL) and the aqueous layer was further purified using RNeasy spin columns (Qiagen, Limburg, Germany). For qPCR, cDNA was generated from 1 μg of RNA using Superscript III (Invitrogen Life Technologies, Carlsbad, CA) and Oligo-dT primers. Select genes were analyzed using Maxima SYBR green qPCR master mix (Thermo Scientific, Waltham, MA). Each sample was analyzed in triplicate on a CFX96 Real-time PCR detection system (Bio-Rad, Hercules, CA). CQ values were normalized to values obtained for actin and relative changes in gene expression were calculated using the ΔΔCQ method.
For RNA-seq, three independent experiments were performed. For each experiment, BMDM were seeded in 24-well dishes and infection and RNA preparation were performed as described. For each sample in each experiment two duplicate wells were pooled. RNA-seq was performed at the Genome Center and Bioinformatics Core Facility at the University of California, Davis. SR50 reads were run on an Illumina HiSeq, with ~30 million reads per sample. Data analysis was performed by the UC Davis bioinformatics group using FastQC for read quality assessment, Sythe and Sickle for Illumina adapter and quality trimming, and Tophat2 for read alignment. Raw counts were derived from alignments using a STSeq-count python script (21). Tests of differential expression were conducted using a multifactorial model in edgeR/limma(voom). Data was uploaded to NCBI: http://www.ncbi.nlm.nih.gov/sra/SRP075696.
Glucose assays and Lactate Assays
Lactate accumulation in supernatants of BMDM was measured using the Lactate Assay Kit (MAK064, Sima-Aldritch, St Louis, MO) as described in the manufacturer’s protocol. Glucose depletion from the media was measured using the Glucose (HK) assay kit (GAHK20, Sigma Aldrich, St Louis, MO). The protocol was modified to perform the assays in 96 well plates with 100uL reactions instead of 1mL reactions in cuvettes as described in the manufacturer’s protocol. Glucose consumption was calculated by measuring glucose levels in the media after infection and subtracting from glucose measured from cell-free media.
ELISAs
For IL-1b ELISAs, supernatants from BMDM in 24 well plates were used. A mouse IL-1b ELISA kit (DY401, R&D systems, Minneapolis, MN) was used as described in the manufacturer’s protocol. For PGE2 ELISAs, the PGE2 EIA Kit (EA02, Oxford Biomedical Research, Rochester Hills, MI) was used as described in the manufacturer’s protocol.
Metabolomic profiling by LC-MS/MS
Preparation of lysates for metabolomics profiling followed published protocols (22). In brief, 4×106 BMDM were plated in 6 cm dishes in DMEM containing 10% FBS, 10 mM glucose, 4 mM L-glutamine, and 20 ng/mL recombinant GM-CSF (Cell Signaling Technologies, Danvers, MA). BMDM were infected at an MOI of 1 as described above. At 24h post infection, cells were washed with ice cold PBS and immediately lysed with ice cold 40:40:20 MeCN/MeOH/H2O and immediately placed on ice. D3-Serine (1 nmol) was added to each sample as an external standard. Samples were vigorously vortexed and sonicated and insoluble material was removed by centrifugation. An aliquot of the supernatant (20 μl) was analyzed by SRM-based LC/MS. Polar metabolite separation was achieved with a Luna normal-phase NH2 column (50 × 4.6 mm, with 5-μm-diameter particles; Phenomenex, Torrance, CA). Mobile phase A was composed of 100% acetonitrile, and mobile phase B consisted of water and acetonitrile in a 95:5 ratio. Solvent modifier 0.2% ammonium hydroxide with 50 mM ammonium acetate was used to assist ion formation and to improve the LC resolution in negative ionization mode. The gradient started at 0% B and increased linearly to 100% B over the course of 30 min with a flow rate of 0.7 mL/min. MS analysis was performed with an electrospray ionization (ESI) source on an Agilent 6430 QQQ LC-MS/MS (Agilent Technologies, Santa Clara, CA). The capillary voltage was set to 3.0 kV, and the fragmentor voltage was set to 100 V. The drying gas temperature was 350°C, the drying gas flow rate was 10 L/min, and the nebulizer pressure was 35 psi. Representative metabolites were quantified by SRM of the transition from precursor to product ions at associated optimized collision energies.
Results
HIF-1α is required for IFN-γ mediated control of M. tuberculosis infection
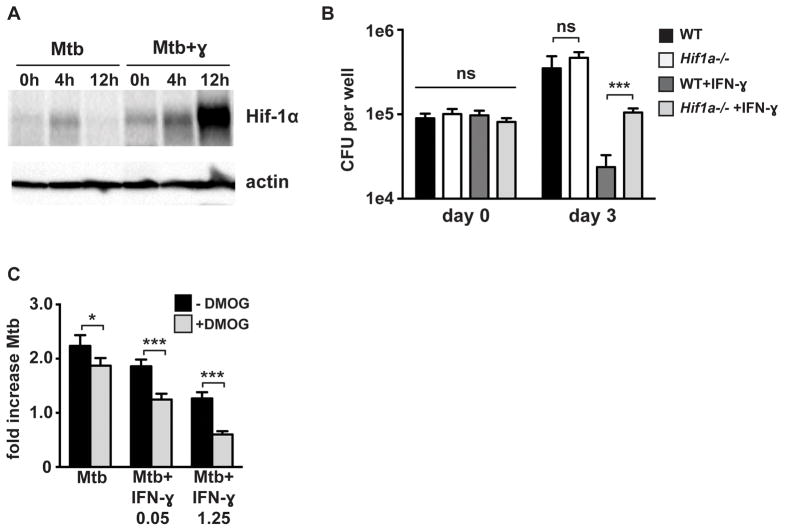
To characterize the role of HIF-1α in macrophages during M. tuberculosis infection, HIF-1α protein levels were first assayed by western blot following infection with M. tuberculosis. Infection of bone marrow derived macrophages (BMDM) with M. tuberculosis resulted in accumulation of HIF-1α. However, the induction was weak and transient, with accumulation of HIF-1α that peaked at 4 hours after infection and was undetectable by 12 hours after infection (Figure 1A). This was surprising given that HIF-1α protein levels increase substantially in macrophages treated with LPS and during infection with several bacterial species (13, 15). However, in the context of IFN-γ stimulation, M. tuberculosis infection resulted in a substantially more robust and prolonged increase in HIF-1α protein levels (Figure 1A). The increase in HIF-1α protein levels observed with infection of IFN-γ activated macrophages is synergistic, as neither infection nor IFN-γ treatment alone induced substantial accumulation of HIF-1α (Figure S1A).
Figure 1. HIF-1α is required for IFN-γ based control of M. tuberculosis replication in macrophages.
(A) Timecourse of HIF-1α protein stabilization by western blotting after infection of resting and IFN-γ activated WT BMDM with M. tuberculosis at MOI=5 with the 0h timepoint reflecting the end of the 4h phagocytosis period. (B) WT and Hif1a−/− BMDM were infected with M. tuberculosis at MOI=5 and bacterial replication was monitored with and without IFN-γ treatment by plating for CFU. CFU on day 0 and day 3 are shown. (C) Resting and IFN-γ activated WT BMDM were infected with the TB-lux strain of M. tuberculosis and treated with 200 μM DMOG after phagocytosis of bacteria. Bacterial growth was assessed by reading RLU immediately after phagocytosis and at 72h after infection and fold change is shown. For all experiments error bars represent the standard deviation of a minimum of quadruplicate wells and a representative experiment of a minimum of 3 is shown. p-values were determined using an unpaired t-test. ***p≤0.001, *p≤0.05.
Next, growth of M. tuberculosis in wild-type and HIF-1α deficient macrophages BMDM was compared both in resting and IFN-γ activated macrophages. Bacterial numbers were enumerated by counting CFU at multiple time points after infection. After the initial phagocytosis period, CFU bacterial numbers were equivalent across all genotypes and conditions (Figure 1B). Over a 3 day timecourse, BMDM are a relatively restrictive environment for M. tuberculosis replication, with only a 2.5-fold increase in CFU observed, with wild-type and HIF-1α deficient BMDM able to restrict M. tuberculosis replication to the same degree in the absence of IFN-γ stimulation (Figure 1B). However, following IFN-γ stimulation, wild-type BMDM are able to kill M. tuberculosis, while bacterial numbers remain constant in the HIF-1α deficient BMDM (Figure 1B). HIF-1α therefore regulates processes in IFN-γ activated macrophages that enable bacterial killing.
HIF-1α is constitutively transcribed and translated and protein levels are governed by the activity of prolyl hydroxylases that mark HIF-1α for ubiquitination and degradation. Inhibition of prolyl hydroxlases by low oxygen, metabolic intermediates, or small molecule inhibitors causes HIF-1α stabilization and accumulation (23). To test whether pharmacological stabilization of HIF-1α in the absence of IFN-γ activation would impact M. tuberculosis replication in macrophages, BMDM were infected with M. tuberculosis and treated with the prolyl hydroxylase inhibitor DMOG. DMOG was tested in the presence of IFN-γ at both a standard activating concentration (1.25ng/mL), and at a subactivating concentration (0.05ng/mL). The addition of 200uM DMOG enhanced HIF-1α levels under both conditions, as well as in resting macrophages infected with M. tuberculosis (Figure S1B). To assess the impact on bacterial replication a reporter strain that constitutively expresses the bacterial luciferase encoding luxCDABE operon (TB-lux, gift from the Cox lab) was utilized. Luminescence from this strain was linear with bacterial number in axenic culture and during infection of macrophages and expression of the lux genes did not attenuate growth (Figure S1C,D). Exogenously stabilizing HIF-1α during M. tuberculosis infection of resting macrophages slightly reduced M. tuberculosis growth (Figure 1C). However, the addition of DMOG to IFN-γ activated macrophages resulted in a larger decrease in bacterial numbers (Figure 1C) indicating that pharmacological stabilization of HIF-1α can activate microbicidal mechanisms effective against M. tuberculosis more robustly in the context of IFN-γ. The fact that the enhancement of bacterial restriction occurs both at effective IFN-γ concentrations as well as at lower, sub-activating IFN-γ concentrations suggests that artificial HIF-1α stabilization might be of therapeutic utility in the context of an insufficient IFN-γ response to M. tuberculosis infection.
HIF-1α is a key mediator of IFN-γ dependent gene expression
HIF-1α has a large number of potential target genes. A comprehensive analysis of HIF-1α target genes in the context of infection has not been performed. Therefore, RNA-seq was used to identify HIF-1α dependent changes in the macrophage transcriptome during infection with M. tuberculosis (data available at http://www.ncbi.nlm.nih.gov/sra/SRP075696). Infection of macrophages with M. tuberculosis resulted in changes in expression in 3330 genes, and the addition of IFN-γ to M. tuberculosis infected macrophages results in differential expression of 2595 genes relative to M. tuberculosis infection alone. In resting M. tuberculosis infected macrophages HIF-1α contributed to altered expression in only 118 genes, or 3.5% of all regulated genes. In IFN-γ activated macrophages, however, HIF-1α contributed to regulation of 1191 genes during infection, demonstrating that HIF-1α is responsible for approximately ~45% of IFN-γ induced alterations in the macrophage transcriptome 24 hours after M. tuberculosis infection.
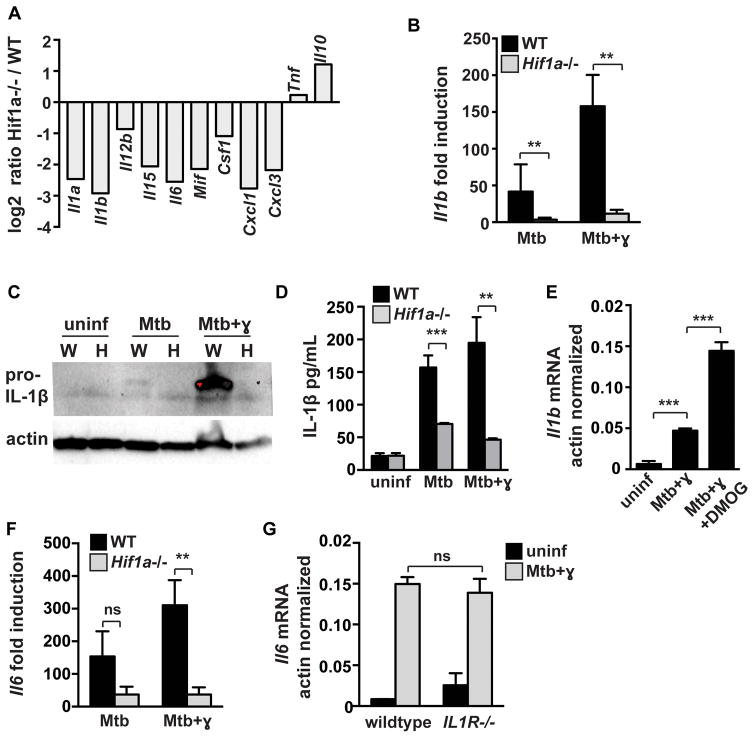
Prominent among genes that were downregulated in HIF-1α deficient macrophages during infection with IFN-γ activation were inflammatory cytokines and chemokines including Il1a, Il1b, Il6 and the neutrophil chemoattractant Cxcl1 (Figure 2A). Loss of HIF-1α did not result in a global defect in transcription of cytokine and chemokine genes. Tnf levels were unaffected by HIF-1α deficiency, and IL10 levels were increased (Figure 2A). The cytokine IL-1 is essential for control of M. tuberculosis infection in mice. HIF-1α contributes to IL-1 transcription downstream of LPS activation (13), however a role for HIF-1α in expression of IL-1 during M. tuberculosis infection has not been demonstrated. Interestingly, levels of Il1b mRNA were dependent on HIF-1α in both resting and IFN-γ activated macrophages. This defect translated to a significant defect in pro-IL-1β protein production as assayed by western blotting from cell lysates (Figure 2C) and by ELISA from cell supernatants (Figure 2D). Furthermore, enhancing HIF-1α stabilization with DMOG further increased Il1b mRNA levels in M. tuberculosis infected and IFN-γ activated macrophages (Figure 2E). Of note, we observed that IFN-γ increased levels of pro-IL-1β in cell lysates and had no impact on IL-1β levels in cell supernatants. This contrasts with several reports that have demonstrated that IFN-γ suppresses IL-1β production in the context of TLR stimulation or M. tuberculosis infection (24, 25). Nevertheless, our data clearly indicate that HIF-1α is required for production of IL-1 during M. tuberculosis infection in both resting and IFN-γ activated macrophages. Thus, while HIF-1α only contributes to control of M. tuberculosis in infected macrophages in the context of IFN-γ activation, the low levels of HIF-1α observed in the absence of IFN-γ do promote transcription of a small number of immunologically important genes.
Figure 2. HIF-1α regulates cytokine and chemokine production.
Resting and IFN-γ activated WT and Hif1a−/− BMDM were infected with M. tuberculosis at MOI=5. RNA was prepared for RNA-seq at 24h post infection. (A) RNA-seq data showing transcript levels of cytokines and chemokines in Hif1a−/− BMDM relative to WT BMDM on a log2 scale. Data shown is from macrophages treated with IFN-γ and infected with M. tuberculosis. (B) RNA-seq data showing fold induction of Il1b transcript over untreated macrophages in WT and Hif1a−/− BMDM (C) Western blotting for pro-IL-1b from cell lysates at 12h after infection in wild-type [W] and Hif1a−/− [H] macrophages. (D) ELISA for IL1b from cell supernatants at 36h after infection. (E) qPCR data showing actin normalized Il1b transcript in WT BMDM with DMOG treatment. (F) RNAseq data showing fold induction of Il6 transcript over untreated macrophages in WT and Hif1a−/− BMDM (G) qPCR data showing actin normalized Il6 transcript levels in WT and IL1R deficient BMDM. For RNA-seq, RNA was prepared from 3 independent infections. All other experiments were repeated 2–3 times and representative experiments are shown. Error bars represent standard deviation. p values were determined using an unpaired t-test. ***p≤0.001, **p≤0.01, *p≤0.05.
IL-6 expression was also found to be dependent on HIF-1α during M. tuberculosis infection (Figure 2F). To test whether the deficient expression of other cytokines and chemokines in the HIF-1α deficient macrophages was downstream of an autocrine effect of IL-1 on macrophage activation, we tested whether IL-6 expression was altered in macrophages lacking IL1R. However, IL-6 levels were unaffected by the absence of IL1R (Figure 2G) supporting the hypothesis that HIF-1α plays a direct role in regulating the expression of cytokine and chemokine genes.
HIF-1α regulates production of prostaglandins and nitric oxide in M. tuberculosis infected macrophages
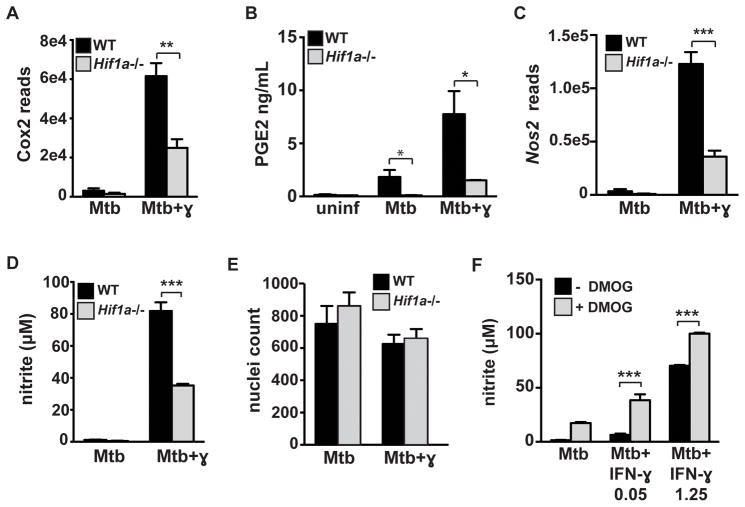
A recent study demonstrated that one important function of IL-1 during M. tuberculosis infection is to promote the production of prostaglandin E2 (PGE2) (26), a critical component of host immunity to M. tuberculosis (27–29). PGE2 is an eicosanoid derived from arachadonic acid via the enzymes COX2 and PGES. The defective IL-1 production in HIF-1α deficient macrophages suggests that there might also be a defect in PGE2 production in HIF-1α deficient macrophages. Eicosanoids have been shown to be important in macrophages for cell intrinsic control of M. tuberculosis replication and for productive and balanced inflammatory responses (28, 30, 31). However, eicosanoid production during M. tuberculosis infection of macrophages has only previously been characterized in the absence of IFN-γ. Interestingly, Cox2 expression levels and PGE2 production in M. tuberculosis infected BMDM was dramatically enhanced by IFN-γ stimulation (Figure 3A, 3B). In addition, Cox2 expression was partially dependent on HIF-1α in IFN-γ activated M. tuberculosis infected macrophages (Figure 3A). This decrease in Cox2 expression led to a significant defect in PGE2 production in HIF-1α deficient macrophages (Figure 3B). These data identify the production of enhanced levels of PGE2 as a potential mechanism of IFN-γ dependent control of M. tuberculosis, and demonstrate that HIF-1α is essential for PGE2 production.
Figure 3. HIF-1α is required for full activation of IFN-γ dependent cell intrinsic immune responses.
Resting and IFN-γ activated WT and Hif1a−/− BMDM were infected with M. tuberculosis at MOI=5. RNA was prepared for RNA-seq at 24h post infection. (A) RNA-seq data showing expression levels of Cox2 (official name Ptgs2) in WT and Hif1a−/− BMDM. (B) PGE2 ELISA from macrophage supernatants at 36h after infection. (C) RNA-seq data showing expression levels of Nos2 in WT and Hif1a−/− BMDM (D) Griess assay for nitric oxide production in WT and Hif1a−/− BMDM. (E) Nuclei counts after DAPI staining in WT and Hif1a−/− BMDM 24h after infection. (F) Resting and IFN-γ activated BMDM were infected with M. tuberculosis and treated with 200 μM DMOG after phagocytosis of bacteria and a Griess assay was performed 72h after infection. For RNA-seq, RNA was prepared from 3 independent infections. All other experiments are representative of 3 or more. Error bars represent standard deviation. p-values were determined using an unpaired t-test. ***p≤0.001, **p≤0.01, *p≤0.05.
Following IFN-γ activation and M. tuberculosis infection, HIF-1α deficient BMDM had lower levels of iNOS transcript (Nos2) than wild-type macrophages (Figure 3C). This observation is consistent with the observation that HIF-1α can bind at the Nos2 promoter (32). This defect at the transcript level corresponded to a ~50% defect in NO production (Figure 3D) and was not a result of decreased cell viability of the HIF-1α deficient macrophages (Figure 3E). Finally, the addition of DMOG to resting and IFN-γ activated macrophages enhanced NO production, further confirming the importance of HIF-1α for functional responses of macrophages (Figure 3F). Taken together our results implicate HIF-1α as a crucial regulator of IFN-γ dependent inflammatory responses as well as cell intrinsic immune responses to M. tuberculosis.
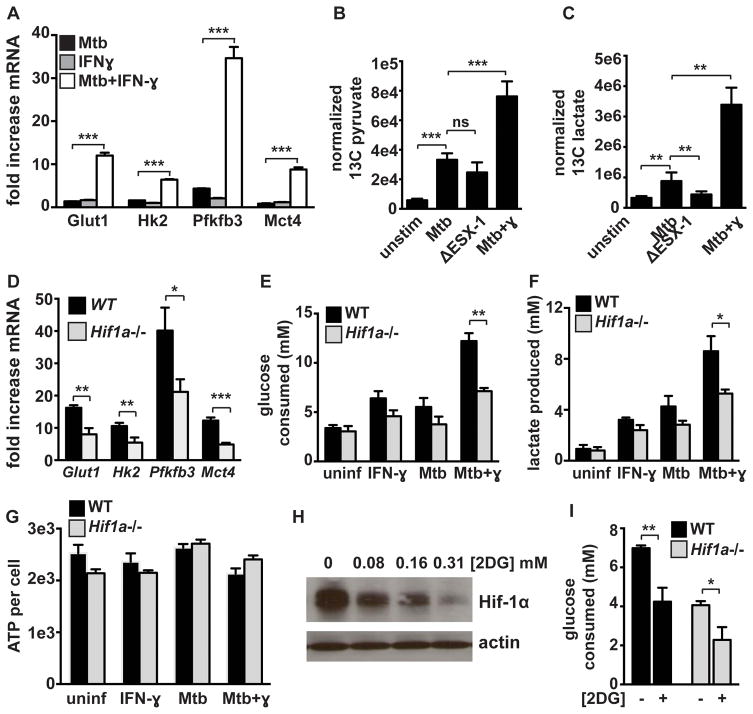
Metabolic profiling during M. tuberculosis infection reveals increased levels of aerobic glycolysis in IFN-γ activated macrophages
RNA-seq revealed that infection of IFN-γ activated macrophages with M. tuberculosis causes a dramatic increase in expression of numerous glycolytic genes relative to the increase in gene expression observed with infection alone (Figure S2). We confirmed the increased expression of four of these genes, Glut1, Hk2, Pfkfb3, and Mct4 by qPCR and found that the combination of IFN-γ and M. tuberculosis infection dramatically and synergistically increased expression of these glycolytic genes (Figure 4A). Aerobic glycolysis in resting macrophages infected with M. tuberculosis is thought to promote bacterial replication. Thus, the observation that glycolysis was further increased in IFN-γ activated macrophages, a bactericidal environment for M. tuberculosis, is unexpected. To confirm these results, high-resolution tandem mass spectrometry was used to examine steady state levels of glycolytic intermediates in infected cells (33). M. tuberculosis infection of BMDM that had been activated with IFN-γ prior to infection resulted in a substantial increase in steady state levels of glycolytic intermediates compared to M. tuberculosis infection alone (Figure S3). Metabolic flux analysis using 13C labeled glucose [U-13C-glucose] demonstrated a modest increase in levels of 13C pyruvate and lactate upon infection of resting macrophages with M. tuberculosis (Figure 4B, 4C). In agreement with previous work, this increase was partially dependent on the ESX-1 alternative secretion system (Figure 4B, 4C). However, analysis of 13C labeled pyruvate and 13C lactate levels confirmed that IFN-γ pre-activation resulted in greatly enhanced glycolytic flux relative to infection of resting macrophages (Figure 4B, 4C). Taken together these results demonstrate that increased glycolytic flux is associated with macrophages that are able to control infection with M. tuberculosis.
Figure 4. HIF-1α mediates the transition to aerobic glycolysis in M. tuberculosis infected macrophages but is not required for maintenance of ATP production.
(A) Fold increase in transcript levels over resting BMDM for the glycolytic genes Glut1, Hk2, Pfkfb3, and Mct4 were measured by qPCR in WT BMDM. (B and C) BMDM infected with M. tuberculosis were cultured in media containing 13C labeled glucose for 24h at which time lysates were prepared and 13C labeled pyruvate (B) and lactate (C) were detected by LC-MS/MS. Values represent the average of quintuplicate wells and error is standard deviation. (D) RNAseq data showing fold increase over resting BMDM of glycolytic genes in WT and Hif1a−/− BMDM treated with IFN-γ and infected with M. tuberculosis. (E) Glucose consumption after 36h by WT and Hif1a−/− BMDM. (F) Lactate production after 24h by WT and Hif1a−/− BMDM. (G) ATP levels were measured using a luciferase based assay (CellTiter-Glo) and were normalized to cell number measured by DAPI staining and counting of nuclei. (H) HIF-1α western blot 12h after M. tuberculosis infection of IFN-γ treated BMDM, treated with increasing dose of 2-DG following 4h phagocytosis period. (I) Glucose consumption 24h post infection of WT and Hif1a−/− BMDM with and without 2DG treatment. Data in (A),(D),(E),(F) and (H) are representative of 3 or more experiments, data in (G) and (I) are representative of 2 experiments. Error bars represent standard deviation. p-values were determined using an unpaired t-test. ***p≤0.001, **p≤0.01, *p≤0.05
HIF-1α regulates metabolic transitions in IFN-γ activated and M. tuberculosis infected macrophages
In keeping with the known role of HIF-1α in regulating aerobic glycolysis, the majority of glycolytic genes had decreased expression in HIF-1α deficient BMDM relative to wild-type BMDM after M. tuberculosis infection of IFN-γ activated BMDM (Figure 4D). HIF-1α deficient BMDM also had significantly decreased glucose utilization during M. tuberculosis infection of IFN-γ activated macrophages (Figure 4E). Furthermore, HIF-1α deficient macrophages produced significantly less lactate upon infection (Figure 4F). The observation that HIF-1α deficient macrophages are not able to engage in aerobic glycolysis upon activation has previously been linked to a defect in ATP production in HIF-1α deficient peritoneal macrophages (18), which results in a defect in macrophage trafficking to sites of inflammation. A similar inability to produce ATP during infection could explain the defect in IFN-γ dependent control of M. tuberculosis infection observed in HIF-1α deficient BMDM. To test whether ATP production is compromised in HIF-1α deficient BMDM, ATP levels during M. tuberculosis infection in wild-type and HIF-1α deficient BMDM were measured. No difference in ATP levels between wild-type and HIF-1α deficient BMDM was observed in M. tuberculosis infected BMDM either in the presence or absence of IFN-γ (Figure 4G). This demonstrates that the observed defects in M. tuberculosis control in these macrophages does not result from major perturbations in cellular energetics.
HIF-1α is sensitive to changes in oxygen levels as well as changes in metabolite levels. HIF-1α stabilization is promoted by metabolites associated with aerobic glycolysis, including succinate and lactate (13, 34). 2-deoxyglucose (2-DG) is a glucose analog that is commonly used to inhibit or block flux through glycolysis. To test whether HIF-1α stabilization in M. tuberculosis infected and IFN-γ activated macrophages is promoted by flux through glycolysis, we treated macrophages with 2-DG and measured HIF-1α levels by western blotting. Limiting glycolytic flux dramatically reduced HIF-1α protein levels (Figure 4H). Thus, there is a positive feedback loop between HIF-1α and aerobic glycolysis which links aerobic glycolysis to IFN-γ dependent activation of macrophages. As expected, treatment of IFN-γ activated and M. tuberculosis infected macrophages with 2DG resulted in a significant decrease in glucose uptake from the culture media, confirming that 2DG limits glycolytic flux in this system (Figure 4I).
Flux through glycolysis supports IFN-γ dependent control of M. tuberculosis infection in macrophages
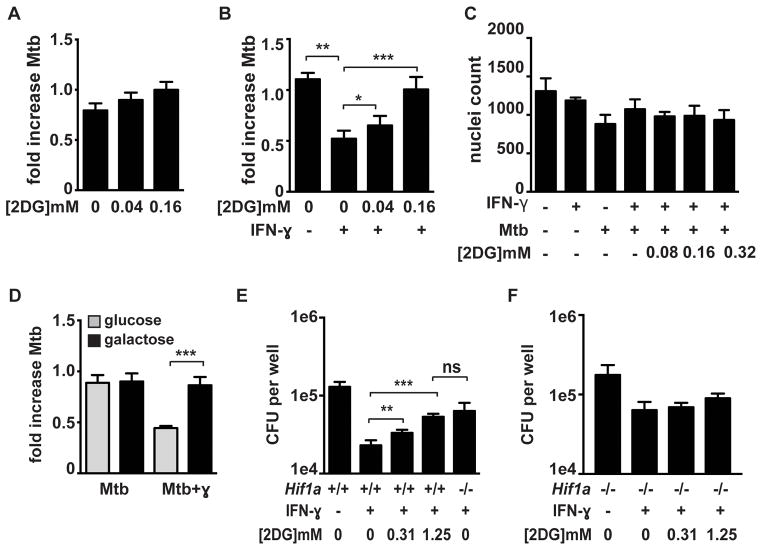
Previous reports have suggested that M. tuberculosis induces aerobic glycolysis in resting macrophages as a virulence strategy to promote bacterial growth (35, 36). Our data indicate that IFN-γ activation of macrophages, which restricts M. tuberculosis growth, also greatly enhances glycolytic flux. We therefore sought to determine whether the induction of aerobic glycolysis altered the ability of M. tuberculosis to replicate and/or survive in macrophages. To evaluate the role of aerobic glycolysis, resting and IFN-γ activated macrophages were infected with M. tuberculosis and treated with 2-DG and bacterial survival was assessed at 24 hours after infection. 2-DG treatment did not substantially alter M. tuberculosis growth or host control in resting BMDM (Figure 5A). In contrast, we found that the addition of 2-DG to IFN-γ activated BMDM resulted in enhanced bacterial survival, reversing IFN-γ dependent killing (Figure 5B). To confirm that the doses of 2-DG used had no impact on macrophage viability, microscopy was used to enumerate the number of cells surviving under each condition, a measurement of macrophage viability that is not confounded by possible alterations in metabolism (Figure 5C). To confirm that aerobic glycolysis is necessary for IFN-γ dependent killing, the capacity for cells to enhance glycolytic flux was also limited by culturing the macrophages on galactose rather than glucose as the carbon source in the media (37, 38). Macrophages cultured in galactose were unable to restrict the growth of M. tuberculosis in an IFN-γ dependent manner (Figure 5D). However culturing macrophages on galactose did not impact infection in resting macrophages (Figure 5D). Although macrophage viability was not impacted by 24h of culture with 2DG or galactose, by 48h we observed differences in macrophage survival that precluded analysis of bacterial loads at later timepoints. To circumvent this issue, and validate the importance of flux through glycolysis, we treated macrophages for 24h with 2DG followed by restoration of standard glucose containing media. We then measured bacterial loads at three days after infection, a time point at which we see a large difference in bacterial numbers between resting and IFN-γ activated macrophages, compared with the only two fold difference at 24h post infection. In addition, three days post infection is the timepoint at which the defect in HIF-1α deficient macrophages is most apparent. We found that 2DG treatment in the first 24h after infection resulted in impaired IFN-γ dependent control at 3d after infection. The effect of 2DG was dose dependent, with the maximal concentration of 2DG used resulting in a defect of IFN-γ dependent control that was equivalent to that found in HIF-1α deficient macrophages (Figure 5E). Interestingly, treatment with 2DG did not further impair the microbicidal activity of HIF-1α deficient macrophages (Figure 5F). Taken together, these findings demonstrate that macrophages infected with M. tuberculosis increase rates of glycolysis, that IFN-γ addition enhances this effect, and that aerobic glycolysis is necessary for IFN-γ dependent restriction of M. tuberculosis.
Figure 5. Enhanced flux through glycolysis is required for IFN-γ dependent control of M. tuberculosis infection.
Resting (A) and IFN-γ activated (B) BMDM were infected with the TB-lux strain of M. tuberculosis and treated with 2DG immediately after the 4h phagocytosis period. Bacterial growth was assessed by reading RLU immediately after phagocytosis and at 24h after infection and fold change is shown. (C) Macrophage viability was assessed using DAPI staining of nuclei and microscopy 24h after infection. (D) Resting and IFN-γ activated macrophages were infected with the TB-lux strain of M. tuberculosis and were switched to glucose free media containing galactose immediately after the 4h phagocytosis period and fold growth was determined at 24h after infection. (E and F) WT and Hif1a−/− BMDM were infected with M. tuberculosis at MOI=5 and bacterial replication was monitored by plating for CFU. 2DG treatment at the indicated concentrations began after the 4h phagocytosis and 2DG was washed out 24h later. CFU 72h post infection is shown. Representative experiments of 3 or more are shown for (A–D) and representative of 2 experiments for (E) and (F). Error bars are standard deviation from 3–6 replicate wells for TB-lux data, 3 replicate wells for nuclei counts, and 5 replicate wells for CFU. p-values were determined using an unpaired t-test, ***p≤0.001, **p≤0.01, *p≤0.05
HIF-1α is crucial for control of M. tuberculosis infection in vivo
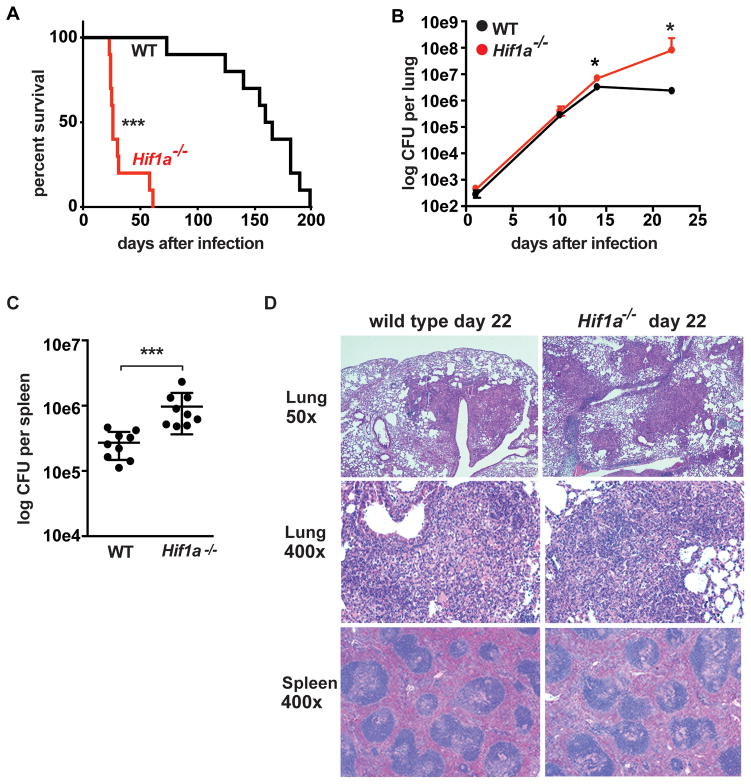
To assess the role of HIF-1α during M. tuberculosis infection in vivo, mice deficient for HIF-1α in the myeloid lineage (Hif1afl/flLysMcre+/+ [hereafter Hif1a−/−]) and wild-type mice were infected with M. tuberculosis via the aerosol route. Hif1a−/− mice exhibited rapid weight loss, with ~75% of the Hif1a−/− mice succumbing to infection within 30 days (Figure 6A). The increased susceptibility of Hif1a−/− mice was also reflected in bacterial burden in the lungs. The earliest timepoint with a difference in bacterial burden was 14 days after infection, with a small but statistically significantly higher bacterial burden in the Hif1a−/− mice compared to wild-type. By 22 days after infection there was more than 10-fold higher bacterial burden in the Hif1a−/− mice (Figure 6B). Additionally, Hif1a−/− mice had higher burdens in spleens at 22 days after infection (Figure 6C). Histological analysis was also performed on lungs and spleens from wild-type and Hif1a−/− mice 22 days after infection. Both wild-type and HIF-1α deficient mice exhibited acute to subacute neutrophilic and histiocytic inflammation, peribronchiolar to perivascular pneumonia, and lymphocytic perivascular cuffing (Figure 6D). The HIF-1α deficient lungs had a more necrotizing character and slightly more area affected (42.6% vs 32.3%), however the differences were relatively modest despite the ~1 log increased bacterial burden in the lungs at this timepoint. Interestingly, the HIF-1α deficient mice do not have large necrotic lesions, as has been reported for IFN-γ deficient mice (39). No differences were observed in HIF-1α deficient spleens compared with wild type. Taken together, these data support the idea that there is not a major defect in immune recruitment to the lungs in Hif1a−/− mice, but rather there is a defect in control of bacterial replication in infected macrophages.
Figure 6. HIF-1α is required for control of M. tuberculosis infection in vivo.
WT and Hif1a−/− mice on the C57BL/6 background were infected with ~400 CFU of the virulent M. tuberculosis strain Erdman via the aerosol route. (A) Survival following aerosol infection is shown for WT and Hif1a−/− mice. Experiment shown is representative of three experiments using 10–12 mice per genotype. (B) Bacterial loads in the lungs of WT and Hif1a−/− mice were enumerated by plating for CFU. Timecourse is representative of 3 experiments with 4–5 mice used per group at each timepoint. (C) Bacterial loads in the spleens at 21d after infection. Data from two pooled experiments are shown. (D) Lung tissues were fixed, embedded in paraffin, sectioned, and stained with haematoxylin and eosin. Images were obtained at 50x and 400x magnification. Error bars represent standard deviation. For survival curves p values were determined using the Mantel-Cox log. For CFU, p-values were determined using Mann-Whitney U. ***p<0.001, *p≤0.05.
HIF-1α regulates expression of immunologically important genes in vivo
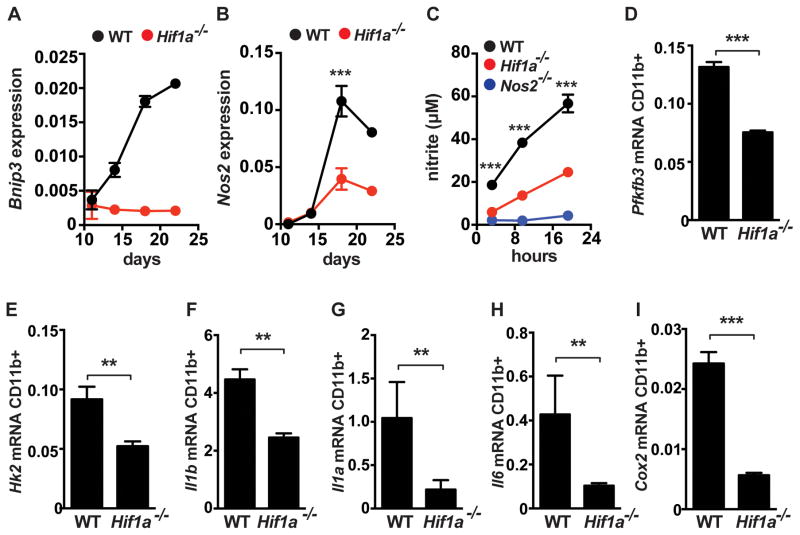
Our in vitro data suggests that HIF-1α activity is enhanced in the context of IFN-γ stimulation of macrophages. To test whether this is also true in vivo we isolated CD11b+ macrophages from lungs of infected mice and examined the expression of HIF-1α target genes over time. Bnip3 is a canonical HIF-1α target gene. We found that Bnip3 expression increased between day 11 after infection and day 22 after infection, a timing that mirrors the development of the IFN-γ dependent T cell response (Figure 7A). As expected, macrophages from Hif1a−/− mice had much lower levels of Bnip3 that did not increase with the onset of IFN-γ signaling (Figure 7A). Expression of iNOS increased over time in a partially HIF-1α dependent manner (Figure 7B). Furthermore, HIF-1α deficient CD11b+ cells plated ex vivo were deficient for NO production (Figure 7C). In addition, we confirmed that inflammatory cytokines and regulators of aerobic glycolysis were dependent on HIF-1α for expression in vivo. At 21d after infection we found that expression of glycolytic enzymes (Pfkfb3, Hk2), inflammatory cytokines (Il1a, Il1b, Il6) and Cox2 were all significantly lower in HIF-1α deficient CD11b+ cells than wild type (Figure 7D–7I). Furthermore, we found that expression of these genes was largely independent of HIF-1α until ~18d after infection, coinciding with the onset of IFN-γ dependent immunity (Figure S4). These results confirm that RNA-seq profiling of HIF-1α deficient macrophages in vitro is predictive of HIF-1α activity in vivo, and that IFN-γ activation enhances HIF-1α activity both in vitro and in vivo.
Figure 7. HIF-1α regulates immune effectors in vivo.
WT and Hif1a−/− mice on the C57BL/6 background were infected with ~400 CFU of the virulent M. tuberculosis strain Erdman via the aerosol route. CD11b+ cells were isolated from lungs of infected mice at the indicated timepoints and analyzed by qPCR for expression of Bnip3 (A) and Nos2 (B). (C) isolated CD11b+ cells were plated and the supernatants were assayed for nitric oxide production by Griess assay at the indicated timepoints. (D–I) qPCR from isolated from CD11b+ cells at 21 days post-infection in WT and Hif1a−/− mice. Data shown in (C–I) is represtentative of 2–3 experiments. p-values were determined using an unpaired t-test, ***p≤0.001, **p≤0.01.
Discussion
In this report we identify HIF-1α as an essential mediator of IFN-γ dependent immunity to M. tuberculosis. We find that Hif1a−/− mice are strikingly susceptible to M. tuberculosis infection in vivo. This places HIF-1α among a surprisingly short list of genes essential for control of M. tuberculosis infection in vivo. We find that in macrophages HIF-1α is required for the production of immune effectors including NO, IL-1 and PGE2. In addition, we demonstrate that HIF-1α is required for a transition to aerobic glycolysis in IFN-γ activated and M. tuberculosis infected macrophages and that aerobic glycolysis is crucial for IFN-γ dependent control of M. tuberculosis replication. Finally, we find that the immune effectors regulated by HIF-1α in vitro are also regulated by HIF-1α in vivo during infection with M. tuberculosis.
HIF-1α is emerging as an important regulator of immune responses to and defense against bacterial infection. In particular, two recent studies using different mycobacterial pathogens raised the possibility that HIF-1α might be required for defense against M. tuberculosis infection. First, it was demonstrated that pharmacological stabilization of HIF-1α during infection of zebrafish embryos with M. marinum leads to a reduced bacterial burden at early timepoints (19). Second, HIF-1α was found to play a role in a mouse model of granuloma caseation in livers of mice infected with M. avium (20). This granuloma formation depends upon hypoxia but is independent of effectors of IFN-γ based immunity, including NO. In this model, the lack of HIF-1α leads to more rapid necrosis of granulomatous lesions and a modest increase in bacterial numbers in livers and spleens that emerged by around 60 days after infection. Interestingly, our data indicate that HIF-1α is much more important for defense against M. tuberculosis than might have been predicted from these studies as we observed a dramatic susceptibility to aerosol infection with most HIF-1α deficient mice succumbing to infection within 30 days. As mouse lungs are not hypoxic during infection with M. tuberculosis (40–42) our data demonstrate that HIF-1α is not simply required for defense in areas of hypoxic inflammation, but is a mediator of IFN-γ dependent immunity regardless of oxygen availability. In addition, our data support the hypothesis that pharmacological stabilization of HIF-1α might be beneficial in clinical settings where IFN-γ production is impaired.
HIF-1α has been identified as important for control of Group A Streptococcus (GAS), Pseudomonas aeruginosa, and uropathogenic E. coli (15–17). For these infections HIF-1α has been described to regulate innate responses of macrophages, and is crucial for the production of NO and antimicrobial peptides by infected macrophages. Although several antimicrobial peptides have been proposed to contribute to host defense against M. tuberculosis, we did not observe that expression of these antimicrobial peptides was dependent on HIF-1α (not shown). Furthermore, our data indicate that HIF-1α is primarily important for control of M. tuberculosis in IFN-γ activated macrophages, thus extending the role of HIF-1α to a mediator of adaptive immunity. In this context, HIF-1α regulates both cell intrinsic and inflammatory responses of IFN-γ activated macrophages. The production of NO is a critical mediator of IFN-γ dependent control, and we find that maximal production of NO requires HIF-1α. Interestingly, we also find that the production of the prostaglandin PGE2 is dramatically increased by IFN-γ stimulation of M. tuberculosis infected macrophages. PGE2 is known to mediate cell intrinsic control of M. tuberculosis infection in the absence of IFN-γ stimulation, however a possible role for PGE2 in cell intrinsic control of infection in IFN-γ activated macrophages has not been established. We find that HIF-1α is required for PGE2 production, potentially due to a defect in Cox2 expression in HIF-1α deficient macrophages. We also find that HIF-1α is important for expression of numerous cytokines and chemokines in IFN-γ activated macrophages including Il1a, Il1b and Il6. Interestingly, IL-1 is one of the few cytokines that appear to be regulated by HIF-1α in both resting and IFN-γ activated macrophages infected with M. tuberculosis. In summary, our data suggests that HIF-1α plays a far more important role in the expression of important immune defenses against bacterial pathogens than has been previously appreciated. Determining the relative importance of each of these factors to the dramatic susceptibility of HIF-1α deficient mice will be the subject of future studies.
We also identify aerobic glycolysis as a novel component of IFN-γ dependent immunity. It was recently published that infection of unstimulated macrophages with M. tuberculosis induces aerobic glycolysis, and that this is a pathogenesis strategy employed by M. tuberculosis (35, 36). We also find that infection with M. tuberculosis induces a modest increase in glycolytic flux and lactate production in macrophages in the absence of IFN-γ, and that this is indeed ESX-1 dependent. However, we observe a much more dramatic shift to aerobic glycolysis in the presence of IFN-γ. In previous studies, treatment of macrophages with glycolytic inhibitors caused a decrease in M. tuberculosis replication in resting macrophages (35, 36). In contrast, we observed that glycolytic inhibition with 2-DG reversed control of M. tuberculosis replication in the context of IFN-γ activation, but had no impact in the absence of IFN-γ. One possible explanation for the different observations include the use of primary murine cells in our study as opposed to transformed human cell lines that have metabolic perturbations at baseline. An additional possibility is our use of significantly lower 2-DG concentrations that limit flux through glycolysis without impacting macrophage viability. It is also possible that in the context of a permissive growth environment M. tuberculosis actively induces and perhaps slightly benefits from heightened flux through glycolysis and concomitant nutrient availability, but when an infected macrophage is also activated with IFN-γ as part of an adaptive immune response, the immunometabolic program it adopts includes and is dependent upon aerobic glycolysis to differentiate into an effective microbicidal cell.
Our data supports a model in which HIF-1α contributes to a shift to aerobic glycolysis in M. tuberculosis infected and IFN-γ activated macrophages. However, as yet unidentified mechanisms also promote glycolysis, as the defect in HIF-1α deficient macrophages is only partial. Furthermore the fact that HIF-1α stabilization requires aerobic glycolysis suggests that a HIF-1α independent mechanism initially induces enhanced glycolytic flux, leading to stabilization of HIF-1α, which further promotes aerobic glycolysis by enhancing expression of glycolytic genes. The importance of aerobic glycolysis for IFN-γ dependent control of M. tuberculosis is likely at least partially explained by a positive feedback loop between glycolytic flux and HIF-1α that we identify in which aerobic glycolysis supports the immune effector functions mediated by HIF-1α. However, it is possible that aerobic glycolysis also mediates HIF-1α independent functions in IFN-γ dependent control of M. tuberculosis infection and gene expression in activated macrophages, for example via regulation of translation (38, 43).
In conclusion, we identify HIF-1α as an essential component of immunity to M. tuberculosis in vitro and in vivo. We further find that HIF-1α coordinates an immunometabolic program in IFN-γ activated and M. tuberculosis infected macrophages required for the transition to aerobic glycolysis and the production of numerous immune effectors including NO, IL-1 and PGE2. Finally, we demonstrate that aerobic glycolysis also contributes to HIF-1α stabilization, suggesting that positive feedback amplifies IFN-γ dependent activation of macrophages during M. tuberculosis infection.
Supplementary Material
Acknowledgments
SAS gratefully acknowledges the Searle Scholars Program and NIH 1R01AI113270 for support.
The authors gratefully acknowledge Tim Eubank for the kind gift of Hif1a−/− macrophages; Russell Vance for LysMcre and Nos2−/− mice; Jeffrey Cox and Paolo Manzanillo for the TB-lux strain; Russell Vance, Daniel Portnoy, and Jordan Price for helpful discussions; Katherine Chen for technical assistance. We thank Lutz Froenicke (DNA Technologies and Expression Analysis Core, UC Davis) and Monica Britton, Joseph Fass and Blythe Durbin (Genome Center and Bioinformatics Core Facility, UC Davis) for RNA-seq and data analysis. We thank Denise M. Imai-Leonard, DVM, PhD, DACVP (UC Davis Comparative Pathology Laboratory) for histological analysis.
References
- 1.Floyd K. World Health Organization, editor. Global Tuberculosis Report 2012. WHO Press; 2012. pp. 1–100. [Google Scholar]
- 2.Abel L, Casanova JL. Genetic predisposition to clinical tuberculosis: bridging the gap between simple and complex inheritance. American journal of human genetics. 2000;67:274. doi: 10.1086/303033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell. 2013;155:1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci USA. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabri M, Stenger S, Shin DM, Yuk JM, Liu PT, Realegeno S, Lee HM, Krutzik SR, Schenk M, Sieling PA, Teles R, Montoya D, Iyer SS, Bruns H, Lewinsohn DM, Hollis BW, Hewison M, Adams JS, Steinmeyer A, Zugel U, Cheng G, Jo EK, Bloom BR, Modlin RL. Vitamin D Is Required for IFN-γ-Mediated Antimicrobial Activity of Human Macrophages. Sci Transl Med. 2011;3:104ra102–104ra102. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 9.MacMicking JD. Immune Control of Tuberculosis by IFN-γ-Inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 10.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A Family of IFN-γ-Inducible 65-kD GTPases Protects Against Bacterial Infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 11.Chan J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci USA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O’Neill LAJ. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalova IN, Lim JY, Chittezhath M, Zinkernagel AS, Beasley F, Hernández-Jiménez E, Toledano V, Cubillos-Zapata C, Rapisarda A, Chen J, Duan K, Yang H, Poidinger M, Melillo G, Nizet V, Arnalich F, López-Collazo E, Biswas SK. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α. Immunity. 2015;42:484–498. doi: 10.1016/j.immuni.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J Clin Invest. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin AE, Beasley FC, Olson J, Keller N, Shalwitz RA, Hannan TJ, hultgren SJ, nizet V. Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Escherichia coli Infection. PLOS Pathogens. 2015;11:e1004818. doi: 10.1371/journal.ppat.1004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger EA, McClellan SA, Vistisen KS, Hazlett LD. HIF-1α Is Essential for Effective PMN Bacterial Killing, Antimicrobial Peptide Production and Apoptosis in Pseudomonas aeruginosa Keratitis. PLOS Pathogens. 2013;9:e1003457. doi: 10.1371/journal.ppat.1003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cramer T, Yamanishi Y, Clausen BE, Förster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V. HIF-1α is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elks PM, Brizee S, van der Vaart M, Walmsley SR, van Eeden FJ, Renshaw SA, Meijer AH. Hypoxia Inducible Factor Signaling Modulates Susceptibility to Mycobacterial Infection via a Nitric Oxide Dependent Mechanism. PLOS Pathogens. 2013;9:e1003789. doi: 10.1371/journal.ppat.1003789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cardoso MS, Silva TM, Resende M, Appelberg R, Borges M. Lack of the transcription factor hypoxia-inducible factor (HIF)-1α in macrophages accelerates the necrosis of Mycobacterium avium-induced granulomas. Infect Immun. 2015 doi: 10.1128/IAI.00144-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamin DI, Cozzo A, Ji X, Roberts LS, Louie SM, Mulvihill MM, Luo K, Nomura DK. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc Natl Acad Sci USA. 2013;110:14912–14917. doi: 10.1073/pnas.1310894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J-W, Bae S-H, Jeong J-W, Kim S-H, Kim K-W. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp Mol Med. 2004;36:1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 24.Mishra BB, Rathinam VAK, Martens GW, Martinot AJ, Kornfeld H, Fitzgerald KA, Sassetti CM. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1β. Nat Immunol. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eigenbrodm T, Bode KA, Dalpke AH. Early inhibition of IL-1β expression by IFN-γ is mediated by impaired binding of NF-κB to the IL-1β promoter but is independent of nitric oxide. J Immunol. 2013;190:6533–6541. doi: 10.4049/jimmunol.1300324. [DOI] [PubMed] [Google Scholar]
- 26.Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL, Gonzales J, Derrick SC, Shi R, Kumar NP, Wei W, Yuan X, Zhang G, Cai Y, Babu S, Catalfamo M, Salazar AM, Via LE, Barry CE, Sher A. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 2014;511:99–103. doi: 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Divangahi M, Desjardins D, Nunes-Alves C, Remold HG, Behar SM. Eicosanoid pathways regulate adaptive immunity to Mycobacterium tuberculosis. Nat Immunol. 2010;11:751–758. doi: 10.1038/ni.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen M, Divangahi M, Gan H, Shin DSJ, Hong S, Lee DM, Serhan CN, Behar SM, Remold HG. Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J Exp Med. 2008;205:2791–2801. doi: 10.1084/jem.20080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Divangahi M, Chen M, Gan H, Desjardins D, Hickman TT, Lee DM, Fortune S, Behar SM, Remold HG. Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat Immunol. 2009;10:899–906. doi: 10.1038/ni.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tobin DM, Vary JC, Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King M-C, Hawn TR, Moens CB, Ramakrishnan L. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 2010;140:717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobin DM, Roca FJ, Oh SF, McFarland R, Vickery TW, Ray JP, Ko DC, Zou Y, Bang ND, Chau TTH, Vary JC, Hawn TR, Dunstan SJ, Farrar JJ, Thwaites GE, King M-C, Serhan CN, Ramakrishnan L. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell. 2012;148:434–446. doi: 10.1016/j.cell.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung F, Palmer LA, Zhou N, Johns RA. Hypoxic regulation of inducible nitric oxide synthase via hypoxia inducible factor-1 in cardiac myocytes. Circ Res. 2000;86:319–325. doi: 10.1161/01.res.86.3.319. [DOI] [PubMed] [Google Scholar]
- 33.Kopp F, Komatsu T, Nomura DK, Trauger SA, Thomas JR, Siuzdak G, Simon GM, Cravatt BF. The Glycerophospho Metabolome and Its Influence on Amino Acid Homeostasis Revealed by Brain Metabolomics of GDE1(−/−) Mice. Chemistry & Biology. 2010;17:831–840. doi: 10.1016/j.chembiol.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colegio OR, Chu N-Q, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, Cline GW, Phillips AJ, Medzhitov R. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh V, Jamwal S, Jain R, Verma P, Gokhale R, Rao KVS. Mycobacterium tuberculosis-Driven Targeted Recalibration of Macrophage Lipid Homeostasis Promotes the Foamy Phenotype. Cell Host Microbe. 2012;12:669–681. doi: 10.1016/j.chom.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 36.Mehrotra P, Jamwal SV, Saquib N, Sinha N, Siddiqui Z, Manivel V, Chatterjee S, Rao KVS. Pathogenicity of Mycobacterium tuberculosis is expressed by regulating metabolic thresholds of the host macrophage. PLOS Pathogens. 2014;10:e1004265. doi: 10.1371/journal.ppat.1004265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Research. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 38.Chang C-H, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O’Sullivan D, Huang SC-C, van der Windt GJ, Blagih J, Qiu J. Posttranscriptional Control of T Cell Effector Function by Aerobic Glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nandi B, Behar SM. Regulation of neutrophils by interferon-γ limits lung inflammation during tuberculosis infection. Journal of Experimental Medicine. 2011;208:2251–2262. doi: 10.1084/jem.20110919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, Lee EG, Park S-K, Raleigh JA, Cho S-N, McMurray DN, Flynn JL, Barry CE. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aly S, Wagner K, Keller C, Malm S, Malzan A, Brandau S, Bange F-C, Ehlers S. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J Pathol. 2006;210:298–305. doi: 10.1002/path.2055. [DOI] [PubMed] [Google Scholar]
- 42.Klinkenberg LG, Sutherland LA, Bishai WR, Karakousis PC. Metronidazole lacks activity against Mycobacterium tuberculosis in an in vivo hypoxic granuloma model of latency. The Journal of Infectious Diseases. 2008;198:275–283. doi: 10.1086/589515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su X, Yu Y, Zhong Y, Giannopoulou EG, Hu X, Liu H, Cross JR, Rätsch G, Rice CM, Ivashkiv LB. Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat Immunol. 2015;16:838–849. doi: 10.1038/ni.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.