Abstract
The p51 gene that encodes the major antigenic 51-kDa protein in Neorickettsia risticii was identified in strains of Neorickettsia sennetsu and the Stellantchasmus falcatus agent but not in Neorickettsia helminthoeca, suggesting that p51-based diagnosis would be useful to distinguish among them. groESL sequencing results delineated the phylogenic relationships among Neorickettsia spp.
Neorickettsia spp. are obligatory intracellular bacteria and belong to the family Anaplasmataceae, in the order Rickettsiales. Currently, three species are recognized in the genus Neorickettsia, namely, N. risticii, N. sennetsu, and N. helminthoeca (4). The ecology and transmission of Neorickettsia spp. are unique among bacteria, in that this agent parasitizes both trematodes and mammals (17, 18). In mammals, these bacteria reside within cytoplasmic vacuoles, primarily in monocytes in the blood and in macrophages of lymphoid or other tissues, and they can cause systemic diseases. N. helminthoeca causes salmon poisoning disease, an acute and highly fatal disease of domestic and wild canidae (17). N. risticii causes Potomac horse fever, an acute diarrheal disease of horses (18). N. sennetsu (6, 10) causes human sennetsu rickettsiosis. In addition, the SF agent isolated directly from the metacercaria of Stellantchasmus falcatus trematodes that encyst within gray mullet fish (7, 21, 22) belongs to the genus Neorickettsia. The adult stage of S. falcatus can parasitize the human intestine (8). Despite the wide environmental distribution of Neorickettsia spp. and their importance to public health and veterinary medicine, few molecular and antigenic markers have been identified for this group of bacteria.
N. risticii, N. sennetsu, SF agent, and N. helminthoeca are antigenically cross-reactive, and inoculation with N. sennetsu protects horses from Potomac horse fever (15, 21). However, other than approximate molecular sizes, the nature of these cross-reacting antigens is unknown. A 51-kDa protein (P51) is the major antigen recognized in horses with Potomac horse fever (19). P51 is encoded by the p51 gene, which is not found in any other bacteria based on a search of the GenBank database, and has been found in all N. risticii strains identified to date (2, 5, 9, 11). P51 is predicted to be an outer membrane protein by PSORT analysis (http://psort.nibb.ac.jp/). Therefore, p51 may be a diagnostically important gene for N. risticii. However, several critical issues have never been addressed. These include the following: (i) whether the p51 gene exists in other Neorickettsia species, (ii) whether the P51 protein is expressed by the organisms isolated from cultures that can be used as diagnostic antigens, and (iii) whether P51 is antigenically cross-reactive among Neorickettsia spp. Therefore, the present study was designed to address these questions and to delineate the phylogenic relationships among Neorickettsia spp. Neorickettsia strains used in this study are shown in Table 1.
TABLE 1.
Neorickettsia species used in this study
| Species | Strain | Origin | Source or reference |
|---|---|---|---|
| N. risticii | IllinoisT | Horse blood, Md. | ATCC |
| N. sennetsu | MiyayamaT | Human blood, Japan | G. Dascha |
| N. sennetsu | 11098 | Human blood, Malaysia | G. Dasch |
| N. sennetsu | Nakazaki | Human blood, Japan | S. Yamamotob |
| N. sennetsu | Kawano | Human blood, Japan | S. Yamamoto |
| SF agent | Hirose | Metacercaria, Japan | 21 |
| SF agent | Oregon | Trout, Oreg. | This study |
| N. helminthoeca | OregonT | Salmon, Oreg. | 16 |
| N. helminthoeca | California 1 | Dog blood, Calif. | This study |
| N. helminthoeca | California 2 | Dog blood, Calif. | This study |
Centers for Disease Control and Prevention, Atlanta, Ga.
Miyazaki Prefecture Institute for Public Health and Environment.
p51 and murE.
We obtained a total of seven new DNA sequences comprising p51, the intergenic space, and an upstream open reading frame. These include the following: a 2,463-bp DNA fragment (relative positions 75 to 2,534 of N. risticii 90-12) of N. risticii IllinoisT; 2,438-bp fragments (relative positions 22 to 2,443 of N. risticii 90-12) of the N. sennetsu MiyayamaT, Nakazaki, and 11098 strains; a 1,481-bp fragment (relative positions 899 to 2,376 of N. risticii 90-12) of the Kawano strain of N. sennetsu; and two 2,417-bp fragments (relative positions 22 to 2,444 of N. risticii 90-12) from the SF agent Hirose in DH82 cells that were frozen in 1994 (21) and the Oregon strain isolated from the frozen spleen of a dog, which was fed with trout caught in Oregon in 1990.
Unexpectedly, the p51 DNA sequences among four N. sennetsu strains, including three strains from Japan (Miyayama, Nakazaki, and Kawano), and one strain from Malaysia (11098) shared 100% identity. The p51 sequences of an SF strain from Japan and an SF agent strain from the state of Oregon shared 99.3% identity. The 1,461 bp (including gaps) of almost-complete p51s were compared among four N. risticii strains, four N. sennetsu strains, and two SF agent strains, showing clustering within each species (Fig. 1). Although more-divergent p51 sequences from other N. risticii strains are available (2, 9, 11, 14), they are too short to be included in this comparison.
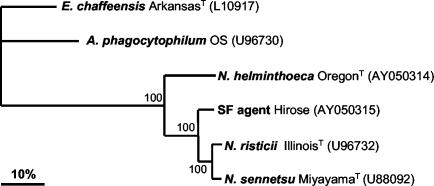
FIG. 1.
Phylogram of Neorickettsia spp. based on p51 sequence comparisons. GenBank accession numbers are shown in parentheses. Numbers beside the internal nodes indicate the percentage of 1,000 bootstrap replicates that supported the branch. Bar, the percentage of divergence.
The partial open reading frame was located 408 bp upstream of p51 in the N. risticii IllinoisT and 90-12 strains, 425 bp upstream of p51 in the N. sennetsu MiyayamaT, 11098, and Nakazaki strains, and 418 bp upstream of p51 in the Hirose and Oregon SF agents. This element was identified as the murE gene (BlastP E values of 9e-07 to murE of Brucella melitensis), which encodes UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diamino-pimelate ligase. Comparison of a 418-bp fragment of the murE sequence (relative positions 10011 to 10428 of Rickettsia rickettsii murE; BlastP E values of 4e-06; GenBank accession number AJ293314) in these seven Neorickettsia spp. showed identity levels ranging from 84.0 to 100.0% (Fig. 2).
FIG. 2.
Phylogram of Neorickettsia spp. based on murE sequence comparison. GenBank accession numbers are shown in parentheses. Numbers beside the internal nodes indicate the percentage of 1,000 bootstrap replicates that supported the branch. Bar, the percentage of divergence.
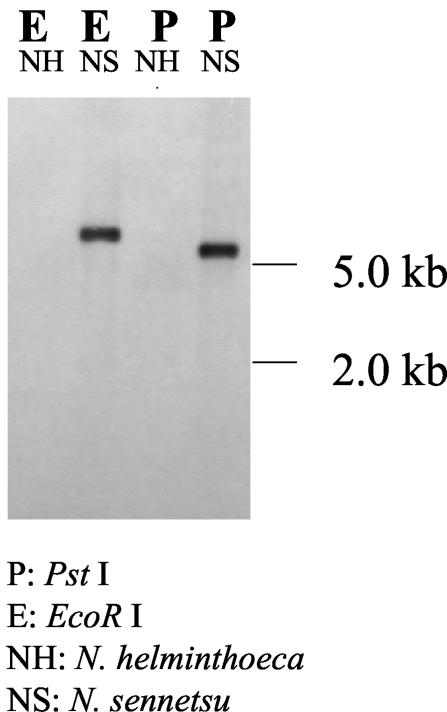
The p51 gene in N. helminthoeca OregonT as well as in the two new strains from blood specimens of two dogs that were naturally infected in California in 2003 was not detectable by PCR using primers specific to the conserved regions of p51. 16S rRNA sequences from these two strains (443 bp; GenBank accession numbers AY510029 and AY510030) were 99% identical to that of N. helminthoeca OregonT. To confirm this result, Southern blot analysis was performed using genomic DNA extracted by the phenol-chloroform method from organisms purified by Percoll gradient centrifugation (12). The blot was hybridized with digoxigenin-labeled N. risticii IllinoisT p51 (1,153 bp, corresponding to nucleotides 894 to 2046 of the N. sennetsu Miyayama p51). As positive control, genomic DNA from N. sennetsu was digested with either PstI or EcoRI, whereupon a single clear band was observed. In contrast, when genomic DNA from N. helminthoeca OregonT was digested with either of these two restriction enzymes, no band was detected (Fig. 3). This result supports the fact that there is no ortholog of p51 in N. helminthoeca or that p51 in this organism is highly divergent from that of N. sennetsu or N. risticii.
FIG. 3.
Southern blot analysis. Samples of genomic DNA from N. sennetsu (NS) and N. helminthoeca (NH) were digested with either EcoRI (E) or PstI (P). The DNA probe used to detect the p51 gene was amplified from N. sennetsu genomic DNA and was labeled by digoxigenin, as described in the text. No p51 band was detected in N. helminthoeca OregonT, whereas the N. sennetsu MiyayamaT positive control showed a clear band upon digestion with either EcoRI or PstI.
Western immunoblot analysis.
To determine whether P51 is expressed by Neorickettsia spp. isolated from cell cultures and whether it is antigenically cross-reactive, we cloned and expressed the entire p51 from N. risticii IllinoisT, which encodes a mature protein with a molecular mass of 52.8 kDa (after cleavage of a 20-amino-acid signal peptide), by using a pET30a expression vector (Novagen, Inc., Madison, Wis.) as described elsewhere (12).
Histidine-tagged recombinant N. risticii IllinoisT (rP51; Mr 57,128.57 fusion protein) was purified, and an antibody specific to rP51 was prepared in rabbits as described elsewhere (12). Western blot analysis revealed that rabbit anti-rP51 serum strongly reacted with both purified rP51 and with 51-kDa native proteins of N. risticii IllinoisT, N. sennetsu MiyayamaT, and N. sennetsu 11908 cultured in P388D1 cells (Fig. 4). Thus, P51 protein is expressed by the N. risticii IllinoisT and N. sennetsu strains in cell culture and is a major cross-reacting antigen between N. risticii and N. sennetsu.
FIG. 4.
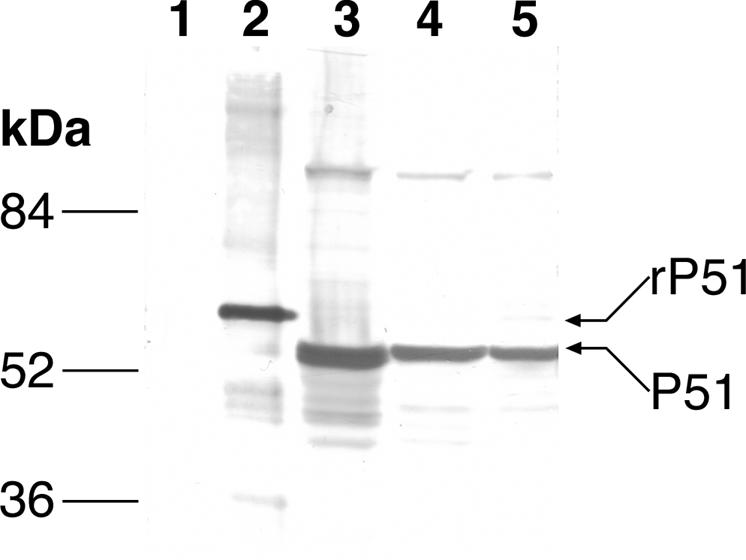
Detection of P51 levels by Western blot analysis using rabbit anti-N. risticii IllinoisT rP51 serum. Lane 1, P388D1 cells (negative control); lane 2, N. risticii IllinoisT rP51; lane 3, native N. risticii IllinoisT; lane 4, N. sennetsu 11908; and lane 5, N. sennetsu MiyayamaT antigens.
groESL sequences.
New sequences totaling 1,914 bp of N. helminthoeca OregonT groESL (relative positions 191 to 2,104 bp of N. sennetsu MiyayamaT groESL) and 2,112 bp of SF agent Hirose groESL (relative positions 30 to 2,142 bp of N. sennetsu MiyayamaT groESL) were obtained. The intergenic spaces between groES and groEL of N. helminthoeca OregonT, SF agent Hirose, N. risticii IllinoisT, and N. sennetsu MiyayamaT were all −1 bp (i.e., they overlapped). Comparison of 1,296 bp of groESL (relative positions 436 to 1,731 bp of N. sennetsu MiyayamaT groESL, including 70 bp of groES and 1,227 bp of groEL with 1-bp overlaps) revealed that a cluster of SF agent Hirose, N. risticii IllinoisT, and N. sennetsu MiyayamaT (identity, 91.2 to 95.4%) is separated from N. helminthoeca OregonT (identity, 77.8 to 78.5%) (Fig. 5). We also compared the citrate synthase (gltA) DNA sequences (relative positions 1 to 1,323 of N. risticii IllinoisT) available from GenBank. gltA DNA sequences from N. sennetsu MiyayamaT (GenBank no. AF304148) and N. risticii IllinoisT (GenBank no. AF304147) shared 94% identity, but there was only 54.3 to 56.2% identity between N. sennetsu MiyayamaT or N. risticii IllinoisT and N. helminthoeca OregonT (GenBank no. AF304149).
FIG. 5.
Phylogram of groEL of Neorickettsia spp. and two other members of the family Anaplasmataceae. GenBank accession numbers are shown in parentheses. Numbers beside the internal nodes indicate the percentage of 1,000 bootstrap replicates that supported the branch. Bar, the percentage of divergence.
The present study revealed that N. helminthoeca is distinct from the other known Neorickettsia species and that p51 can serve as a molecular and antigenic marker to distinguish this species. This is consistent with newly obtained groESL sequences, gltA sequences, and the previous observation showing that 16S rRNA gene sequences among N. risticii, N. sennetsu, and the SF agent share close similarity and are divergent from the 16S rRNA gene sequence of N. helminthoeca (13, 21). Significant differences in levels of genetic and antigenic divergence were found among N. risticii and N. sennetsu strains. It has been previously shown that N. risticii strains isolated from horses are both genetically and antigenically diverse (2, 3, 9, 11). The greatest differences in the nucleotide sequence of 16S rRNA are found between N. risticii IllinoisT and N. risticii Bunn (14 different bases), followed by N. risticii 081 (10 different bases). Even within the same geographic region, there are diverse strains. For example, comparison of the 16S rRNA sequence in the 081 strain isolated in the state of Ohio is significantly different from that of the other known Ohio strains, and the protein compositions also differ, as determined by Western blot analysis with monoclonal and polyclonal antibodies (3, 20). Sequences of p51 segments among N. risticii strains in the United States are up to ∼20% divergent (2, 9, 11, 14). Compared to N. risticii, much less is known about the genetic and antigenic diversity of N. sennetsu strains. The 16S rRNA gene sequences of N. sennetsu MiyayamaT and N. sennetsu 11908 are identical (1), and the present study shows that the p51 sequences of N. sennetsu MiyayamaT (Japanese 1950 isolate), 11908, Nakazaki, and Kawano were identical, suggesting that N. sennetsu human isolates are nearly identical clones. Culturing and sequencing of p51 sequences derived from N. sennetsu MiyayamaT, N. sennetsu 11908, and Nakazaki strains were repeated independently 1 year later from different stock sources with a different laboratory member verifying the results. N. sennetsu 11908 was originally isolated in culture (Malaysian 1970 isolate) in the laboratory of M. Ristic at the University of Illinois and was later transferred to the laboratory of E. Weiss at the Naval Medical Research Center (Bethesda, Md.), and stocks frozen on 28 February 1989 were used in this study. The N. sennetsu, Nakazaki, and Kawano strains had been kept only at the Miyazaki Prefecture Institute for Public Health and Environment, Miyazaki, Japan, where the Miyayama or 11908 strains were never received. This suggests that only a single strain of N. sennetsu identified so far is pathogenic to humans, whereas diverse strains of N. risticii can infect and cause disease in horses. The N. sennetsu Miyayama strain is not pathogenic to horses and protects the horses from N. risticii challenge (15). Thus, the present study contributes to understanding these closely related Neorickettsia species of different mammalian host pathogenicity.
Nucleotide sequence accession numbers.
GenBank accession numbers of sequences obtained in this study are shown in the text and the figures.
Acknowledgments
This work was supported by grant R01AI47885 from the National Institutes of Health and grant 99-35204-8531 from the United States Department of Agriculture.
We appreciate Shogo Yamamoto for providing N. sennetsu Nakazaki and Kawano strains and Gregory Dasch for providing N. sennetsu Miyayama and 11098 strains. We also appreciate Ahmet Unver for his help in immunizing the rabbit.
REFERENCES
- 1.Anderson, B. E., J. E. Dawson, D. C. Jones, and K. H. Wilson. 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J. Clin. Microbiol. 29:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlough, J. E., G. H. Reubel, J. E. Madigan, L. K. Vredevoe, P. E. Miller, and Y. Rikihisa. 1998. Detection of Ehrlichia risticii, the agent of Potomac horse fever, in freshwater stream snails (Pleuroceridae: Juga spp.) of Northern California. J. Appl. Environ. Microbiol. 64:2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaichanasiriwithaya, W., Y. Rikihisa, S. Yamamoto, S. M. Reed, L. E. Perryman, T. B. Crawford, and G. Palmer. 1994. Antigenic, morphologic, and molecular characterization of new Ehrlichia risticii isolates. J. Clin. Microbiol. 32:3026-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumler, J. S., A. F. Barbet, C. P. J. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia, and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 5.Dutta, S. K., R. Vemulapalli, and B. Biswas. 1998. Association of deficiency in antibody response to vaccine and heterogeneity of Ehrlichia risticii strains with Potomac horse fever vaccine failure in horses. J. Clin. Microbiol. 36:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda, T., T. Kitao, and Y. Keida,. 1954. Studies on the causative agent of “Hyuganetsu” disease. I. Isolation of the agent and its inoculation trial in human beings. Med. Biol. 32:200-209. [Google Scholar]
- 7.Fukuda, T., T. Sasahara, and T. Kitao. 1973. Studies on the causative agent of Hyuganetsu disease. XI. Characteristics of rickettsia-like organism isolated from metacercaria of Stellantochasmus falcatus parasitic in grey mullet. J. Jpn. Assoc. Infect. Dis. 47:474-482. [DOI] [PubMed] [Google Scholar]
- 8.Hong, S. J. 2000. A human case of Stellantchasmus falcatus infection in Korea. Korean J. Parasitol. 38:25-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanter, M., J. Mott, N. Ohashi, B. Fried, S. Reed, Y. C. Lin, and Y. Rikihisa. 2000. Analysis of 16S rRNA and 51-kilodalton antigen gene and transmission in mice of Ehrlichia risticii in virgulate trematodes from Elimia livescens snails in Ohio. J. Clin. Microbiol. 38:3349-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misao, T., and Y. Kobayashi. 1954. Studies on infectious mononucleosis. I. Isolation of etiologic agent from blood, bone marrow, and lymph node of a patient with infectious mononucleosis by using mice. Tokyo Iji Shinshi. 71:683-686. [Google Scholar]
- 11.Mott, J., Y. Muramatsu, E. Seaton, C. Martin, S. Reed, and Y. Rikihisa. 2002. Molecular analysis of Ehrlichia risticii in adult aquatic insects in Pennsylvania, in horses infected by ingestion of insects, and isolated in cell culture. J. Clin. Microbiol. 40:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohashi, N., N. Zhi, Y. Zhang, and Y. Rikihisa. 1998. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect. Immun. 66:132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pretzman, C., D. Ralph, D. Stotherd, P. Fuerst, and Y. Rikihisa. 1995. 16SrRNA sequence relatedness of Neorickettsia helminthoeca to Ehrlichia risticii and Ehrlichia sennetsu. Int. J. Syst. Bacteriol. 45:207-211. [DOI] [PubMed] [Google Scholar]
- 14.Reubel, G. H., J. E. Barlough, and J. E. Madigan. 1998. Production and characterization of Ehrlichia risticii, the agent of Potomac horse fever, from snails (Pleuroceridae: Juga spp.) in aquarium culture and genetic comparison to equine strains. J. Clin. Microbiol. 36:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rikihisa, Y., C. I. Pretzman, G. C. Johnson, S. M. Reed, S. Yamamoto, and F. Andrews. 1988. Clinical and immunological responses of ponies to Ehrlichia sennetsu and subsequent Ehrlichia risticii challenge. Infect. Immun. 56:2960-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rikihisa, Y., H. Stills, and G. Zimmerman. 1991. Isolation and continuous culture of Neorickettsia helminthoeca in a macrophage cell line. J. Clin. Microbiol. 29:1928-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rikihisa, Y., and G. Zimmerman. 1995. Salmon poisoning disease, p. 297-300. In R. W. Kirk and J. D. Bonagura (ed.), Current veterinary therapy. XII. Small animal practice. W. B. Saunders, Philadelphia, Pa.
- 18.Rikihisa, Y. 2004. Rickettsial diseases, p. 96-109. In S. M. Reed, W. M. Bayly, and D. C. Sellon (ed.), Equine internal medicine, 2nd ed. W. B. Saunders, Philadelphia, Pa.
- 19.Vermulapalli, R., B. Biswas, and S. K. Dutta. 1998. Cloning and molecular analysis of genes encoding two immunodominant antigens of Ehrlichia risticii. Microb. Pathol. 24:361-372. [DOI] [PubMed] [Google Scholar]
- 20.Wen, B., Y. Rikihisa, P. A. Fuerst, and W. Chaichanasiriwithaya. 1995. Analysis of 16SrRNA genes of ehrlichial organisms isolated from horses with clinical signs of Potomac horse fever. Int. J. Syst. Bacteriol. 45:315-318. [DOI] [PubMed] [Google Scholar]
- 21.Wen, B., Y. Rikihisa, S. Yamamoto, N. Kawabata, and P. A. Fuerst. 1996. Characterization of the SF agent, an Ehrlichia sp. isolated from the fluke Stellantchasmus falcatus, by 16S rRNA base sequence, serological, and morphological analyses. Int. J. Syst. Bacteriol. 46:149-154. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto, S. 1978. Studies on the causative agent of Hyuga fever: cultivation of rickettsia-like organisms isolated from metacercariae of Stellantchasmus falcatus in tissue culture cell and their antigenic relation to Rickettsia sennetsu. J. Jpn. Assoc. Infect. Dis. 52:240-245. (Abstract in English.) [DOI] [PubMed] [Google Scholar]