Abstract
The transcription factor retinoid acid-related orphan receptor gamma t (RORγt) directs the differentiation of T helper 17 (Th17) cells. Th17 cells mediate pathological immune responses responsible for autoimmune diseases, including psoriasis and multiple sclerosis. Previous studies have focused on RORγt target genes and their function in Th17 differentiation. Here, we studied post-transcriptional regulation of RORγt and identified a functional ubiquitination site, lysine 446 (K446). Mutation of K446 to arginine (K446R) to prevent ubiquitination greatly enhanced recruitment of steroid receptor co-activator 1 (SRC1), a co-activator critical for RORγt activity. Correspondingly, the K446R mutation potentiated Th17 differentiation. We also showed that the ubiquitin specific protease 15 (USP15) interacted with RORγt, removed ubiquitin from K446, and stimulated RORγt activity by enhancing co-activator SRC1 recruitment. Knockdown of USP15 or expression of inactive USP15 impaired Th17 differentiation, suggesting a positive role for USP15-mediated deubiquitination of RORγt in Th17 differentiation. Therefore, ubiquitination of K446 limits RORγt-mediated Th17 differentiation by inhibiting the recruitment of co-activator SRC1. Our study will inform the development of treatments that target RORγt ubiqutination pathways to limit Th17-mediated autoimmunity.
Introduction
CD4+-derived T helper 17 (Th17) cells contribute to protective immunity against pathogens (1–3), as well as pathological immune responses responsible for autoimmune diseases, including psoriasis and multiple sclerosis (4, 5). Retinoid acid-related orphan receptor gamma t (RORγt) is the well-known transcription factor that directs Th17 differentiation (6, 7). Activation of naïve T cells in the presence of TGFβ and IL-6 is sufficient for up-regulation of RORγt and subsequent differentiation into Th17 cells (8, 9). Addition of IL-23 and IL-1 further potentiates Th17 differentiation (10–12). Mutation of the RORγt gene causes severe immunodeficiency in both mice (6) and humans (13). Because it is a transcription factor, most previous studies have focused on RORγt target genes critical for the regulation of Th17 differentiation, and have demonstrated that RORγt directly stimulates the expression of genes, including IL-17 (6, 7, 14, 15). However, little is known about the mechanisms responsible for post-transcriptional regulation of RORγt. Because Th17 cells produce the effector cytokines including IL-17A, IL-17F, IL-22, and GM-CSF to mediate pathological inflammation (16–18), targeting Th17 cells is considered a potential treatment for autoimmune disease (19). Pharmaceutical and academic scientists have developed RORγt inhibitors for treatment of Th17-dependent autoimmunity (20–24). Therefore, understanding the post-transcriptional regulation of RORγt will not only reveal the basic mechanisms for regulating Th17 differentiation, but may also facilitate the development of novel therapeutics for the treatment of autoimmunity.
RORγt is a member of the steroid nuclear receptor superfamily and consists of three domains (25, 26): 1) a conserved DNA-binding domain with two zinc finger motifs responsible for DNA-binding; 2) a conserved ligand-binding domain with a carboxy-terminal activation function 2 (AF2) motif responsible for recruiting steroid receptor co-activators (SRC) such as SRC1 (27, 28); and 3) a hinge domain in between the DNA-binding and ligand-binding domains. Upon ligand binding, RORγt binds to target DNA and recruits SRC1 that associate with histone acetyltransferases and methyltransferases to stimulate target gene expression (29). Regulation of co-activator recruitment is an important step for controlling RORγt activity; mutation within the AF2 motif that prevents the recruitment of SRC1 abrogates RORγt gene-stimulating activity (28, 30). Furthermore, pharmacological RORγt inhibitors developed to date inhibit RORγt activity by interfering with the interaction between RORγt and SRC1 (21, 30, 31).
Ubiquitination is a post-transcriptional modification that regulates many aspects of cellular function (32). Ubiquitin ligase enzymes catalyze the addition of ubiquitin to lysines of the protein substrates, whereas deubiqutinases are responsible for removing ubiquitin. There are five families of deubiquitinases, and ubiquitin specific proteases (USPs) are the largest of these families (33). One of the functions of ubiquitination is to regulate protein stability by inducing protein degradation. Evidence indicates that RORγt stability can be regulated by ubiquitination (34–37). Another important role for ubiquitination is to regulate protein-protein interactions. It remains unknown whether ubiquitination plays a role in regulating the interactions between RORγt and its associated proteins in Th17 differentiation. In this study, we identified a ubiquitination site, RORγt K446, that negatively regulates Th17 differentiation by controlling the interaction between RORγt and SRC1. Mutation of K446 to arginine (K446R) stimulates RORγt activity by enhancing RORγt-SRC1 interaction. Correspondingly, K446R is more potent than wild-type (WT) RORγt in rescuing Th17 differentiation when expressed in RORγt−/− T cells. Furthermore, ubiquitin specific protease 15 (USP15) binds RORγt to deubiquitinate K446, leading to stimulation of RORγt activity by promoting the RORγt-SRC1 interaction. Moreover, knockdown of UPS15, or expression of inactive USP15, impaired Th17 differentiation. We thus identified a RORγt ubiquitination site, K446, which is deubiquitinated by USP15 to control RORγt interaction with co-activator SRC1 and ultimately regulates Th17 differentiation.
Materials & Methods
Mice
Wild-type (WT) and RORγt−/− C57BL/6 mice have been described previously (38). Mouse care and experimental procedures were performed under specific pathogen-free conditions following institutional guidance and approved protocols from the Institutional Animal Care and Use Committee at the Beckman Research Institute, City of Hope (no. 07023).
Plasmids and antibodies
A retroviral expression plasmid [murine stem cell virus (MSCV)-IRES-GFP] was used to clone RORγt (Mus musculus, NM_001293734.1) or SRC1 (Mus musculus, NM_010881.2) cDNA. Specific point mutations of RORγt were introduced by using a mutagenesis kit (Agilent Technologies, CA, USA). LMP vector-based retroviral USP15 (Mus musculus, NM_027604.4) and USP45 (Mus musculus, NM_152825.2) short hairpin RNA (shRNA)-expressing vectors (LMP-USP15 shRNA1 and shRNA2; LMP-USP45 shRNA1 and shRNA2) were constructed by using following oligonucleotide sequences: shUSP15-1, TGCTGTTGACAGTGAGCGACCAGACTGTGGAACAAGTATATAGTGAAGCCACAGATGTATA TACTTGTTCCACAGTCTGGCTGCCTACTGCCTCGGA; and shUSP15-2, TGCTGTTGACAGTGAGCGCGGACAGGTGTTAGTGATAGAATAGTGAAGCCACAGATGTATT CTATCACTAACACCTGTCCTTGCCTACTGCCTCGGA. shUSP45-1, TGCTGTTGACAGTGAGCGCCCACTTTCTCCTAAAGTTCTTTAGTGAAGCCACAGATGTAAAG AACTTTAGGAGAAAGTGGTTGCCTACTGCCTCGGA and shUSP45-2, TGCTGTTGACAGTGAGCGAGCATCTTTATTGGCGAATTAATAGTGAAGCCACAGATGTATTAATTCGCCAATAAAGATGCGTGCCTACTGCCTCGGA. USP15 was amplified from pCMV6-Kan/Neo-USP15 (Origene Technology Inc, MD, USA) and inserted into pMSCV-IRES-GFP. USP15 cysteine 298 was mutated to alanine using a mutagenesis kit (Agilent Technologies, CA, USA). Hamster anti-RORγ antibody was purchased from Biolegend (Biolegend, CA, USA). Rabbit anti-USP15 antibody was purchased from Bethyl Laboratories, Inc (Bethyl Laboratories, TX, USA). Mouse anti-SRC1 antibody, mouse anti-Actin antibody, mouse anti-HA antibody and mouse anti-FLAG antibody were purchased from Sigma-Aldrich (Sigma-Aldrich Inc., MO, USA). Rabbit anti-GFP antibody was purchased from Life Technologies (Life Technologies Inc., CA, USA).
Cell culture, transient transfection and reporter assay
EL-4 cells were cultured in RPMI1640 supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. 293T cells were cultured in DMEM supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells (2 × 105 in each well of a 24 well-plate) were transfected with the reporter plasmid (100 ng), pSV40-Renilla luciferase vector (50 ng), and expression vectors (0.5 μg) by using BioT (Bioland Scientific LLC, CA, USA). The total amount of transfected DNA was kept constant by adjusting the amount of the empty vector. Cells were collected after 24 h and lysed in 200 μl lysis buffer (137 mM NaCl, 50 mM Tris-HCl, 0.5% NP-40); luciferase activities were measured by the Dual Luciferase system, according to the manufacturer’s instructions (Promega, WI, USA), and normalized against Renilla luciferase activities. “Folds of stimulation” represents normalized luciferase activity divided by the result of reporter-only groups.
T cell isolation and in vitro Th17 differentiation
CD4+ T cells were isolated from spleens of 6- to 10-wk-old C57BL/6 mice by negative selection using a CD4+ T cell isolation kit (Miltenyi Biotec, Bergisch-Gladbach, Germany) and replicate suspensions of 4 × 105/ml iDMEM with 10% FBS, 100 U/ml penicillin G, and 50 μg/ml streptomycin were cultured in 24-well plates that had been precoated with 0.2 mg/ml goat anti-hamster Ab. The medium was supplemented with 0.25 μg/ml hamster anti-CD3 (145-2C11; eBioscience, CA, USA), 1μg/ml hamster anti-CD28 (37.51; eBioscience, CA, USA), 2 ng/ml TGF-β1 (eBioscience), and 20 ng/ml IL-6 (eBioscience). T cells were cultured for 3 days at 37°C with 5% CO2.
Flow cytometry
For intracellular cytokine staining, cells obtained from in vitro cultures were incubated for 4–5 h with 50 ng/ml PMA, 750 ng/ml Ionomycin (both Sigma-Aldrich Inc., MO, USA) and 10 μg/ml Brefeldin A (BD) in a tissue culture incubator at 37°C. Cells were resuspended in Fixation/Permeabilization solution (BD Cytofix/Cytoperm Kit, BD Pharmingen, USA), and intracellular cytokine staining was performed according to the manufacturer’s protocol. Cells were intracellularly stained with anti-mouse IL17 (TC11-18H10.1) and analyzed using a BD FACSCanto II flow cytometer (BD Biosciences, CA, USA) and FlowJo software (Tree Star, OR, USA).
Retroviral packaging and transduction
Retroviral expression plasmids were transfected into Plat-E packaging cells using Lipofectamine 2000 (Invitrogen Life Technologies, CA, USA). After 48 h, viral supernatants were collected, passed through 0.4-μm filters, and stored at −80°C until use. For transduction, naïve T cells were first activated with 0.25 μg/ml hamster anti-CD3 (145-2C11; eBioscience, CA, USA), 1μg/ml hamster anti-CD28 (37.51; eBioscience, CA, USA) in a 24-well plate precoated with goat anti-hamster Ab for 24 h, then spin-infected with viral supernatants (2500 rpm, 30°C for 2 h) in the presence of 8 μg/ml polybrene (Sigma-Aldrich, MO, USA). After spin infection, indicated cytokines were added to the culture media to induce Th17 differentiation.
RT-PCR and quantitative PCR
Total RNA was prepared using TRIzol (Invitrogen Life Technologies, CA, USA), and cDNA was prepared by using Tetro cDNA synthesis kit (Bioline Inc, London, UK). Primers for RT-PCR (5′ to 3′) were as follows: IL17A Forward: TTTAACTCCCTTGGCGCAAAA, IL17A Reverse: CTTTCCCTCCGCATTGACAC; IL17F Forward: TGCTACTGTTGATGTTGGGAC, IL17F Reverse: AATGCCCTGGTTTTGGTTGAA; CCR6 Forward: CCTGGGCAACATTATGGTGGT, CCR6 Reverse: CAGAACGGTAGGGTGAGGACA; CCL20 Forward: GCCTCTCGTACATACAGACGC, CCL20 Reverse: CCAGTTCTGCTTTGGATCAGC; IRF4 Forward: TCCGACAGTGGTTGATCGAC, IRF4 Reverse: CCTCACGATTGTAGTCCTGCTT; RORα Forward: GTGGAGACAAATCGTCAGGAAT, RORα Reverse: TGGTCCGATCAATCAAACAGTTC; AHR Forward: AGCCGGTGCAGAAAACAGTAA, AHR Reverse: AGGCGGTCTAACTCTGTGTTC; Beta-Actin Forward: GAGTCCTACGACATCATCGCT, Beta-Actin Reverse: CCGACATAGTTTGGGAAACAGT.
Isolation of active deubiquitinases with the HA-VS-Ub Probe
Th17 cells, differentiated in vitro as described above, were lysed in 0.1% NP-40, 150 mM NaCl, 20 mM CaCl2, 50 mM Tris pH 7.4 buffer containing 1 mM phenylmethanesulfonyl fluoride (PMSF). Sample preparation was performed as described (39). Briefly, samples containing equal amounts of protein were subjected to enzymatic reaction with the ubiquitin vinyl sulfone HA-tagged probe (Ub-VS-HA; Boston Biochem, USA) at a ratio of ~1:200 for 45 min at 37°C. Treated lysates were separated by SDS-PAGE and analyzed by anti-HA western blotting to visualize the active deubiquitinases. For immunoprecipitation, 8 mg of protein per sample (obtained as described in the section above) was diluted in NET buffer (50 mM Tris, 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, pH 7.4) to a protein concentration of 1 mg/ml, pre-cleared with agarose-coupled Protein A beads (Sigma-Aldrich) for 1 h at 4°C, and immunoprecipitated with 60 μl of anti-HA Agarose Conjugate (Sigma-Aldrich) for 14 h at 4°C. The resin was washed four times with NET buffer and eluted for 20 min with 0.3 ml of 100 mM glycine pH 2.5.
Sample preparation and mass spectrometric analysis
The immunoprecipitated proteins were eluted and then precipitated using chloroform and methanol: 200 μl total volume of sample, 600 μl methanol, and 150 μl chloroform were vortexed, followed by addition of 450 μl MilliQ-H20 and centrifugation at room temperature for 1 min at 14,000 rpm. The upper aqueous phase was discarded. Methanol (450 μl) was added to the lower phase and interphase, vortexed, and centrifuged at room temperature for 1 min at 14,000 rpm. The supernatants were removed and the pellets were resuspended in 0.1 M Tris pH 7.8 containing 6 M Urea and digested by trypsin digestion. Alternatively, samples were resolved on SDS-PAGE and silver stained using the Pierce Silver Stain for Mass Spectrometry kit (Fisher Scientific), followed by excision of gel bands and tryptic digestion. We then performed on-line 2D nano-LC Q Exactive MS and HCD MS/MS to analyze immunoprecipitated proteins to identify post-transcriptional modification sites of RORγt, RORγt binding proteins or active DUBs. Especially, to detect the ubiquitination of RORγt, immunoprecipitation was first subject to trypsin digestion. Digested ubiquitinated lysine (K) will leave GG isopeptide linked to K, which can be easily detected by MS due to an increased mass of 114.102 dalton. Detection of Ub footprints (GG isopeptide linked to K) at high resolution and a series of fragmentation ions from HCD allowed the accurate sequencing and assignment of the site of modification to only one site. Integrating MS and HCD MS/MS results. Protein Discoverer 1.4 automatically assigned potential modification sites with high confidence.
Enrichment of IL17-producing cells
IL17-producing EL-4 cells were enriched by using a Cell Enrichment and Detection Kit (Miltenyi Biotec Inc, Bergisch-Gladbach, Germany). Briefly, 108 cells were harvested and incubated with IL-17 Catch Reagent on ice for 5 min and at 37°C for 45 min under slow continuous rotation. Cells were washed and then stained with IL-17 detection Antibody. IL17-producing cells were sorted by BD FACSAria II cell sorter.
Enrichment of ubiquintinated proteins by Agarose-TUBE2
1.5×106 Th17 cells, differentiated in vitro as described above, were lysed in 500 μl cell lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 10% glycerol, 1 mM phenylmethanesulfonyl fluoride (PMSF)). Cell lysates were incubated with 20 μl equilibrated Agarose-TUBE2 (Lifesensors Inc., PA, USA) at 4°C for 4 hours. Agarose-TUBE2 was then washed with 1 ml TBS-T (20 mM Tirs-HCl pH8.0, 150 mM NaCl, 0.1% Tween-20) and suspended in SDS reducing sample buffer. Eluted samples were analyzed by western blotting.
Statistical analysis
Statistical analysis was performed using the unpaired, two-tailed Student’s t test. A p-value <0.05 was considered significant. Flow cytometry as well as immunoblot data shown are representative of at least three independent experiments
Results
K446 of RORγt is a ubiquitination site
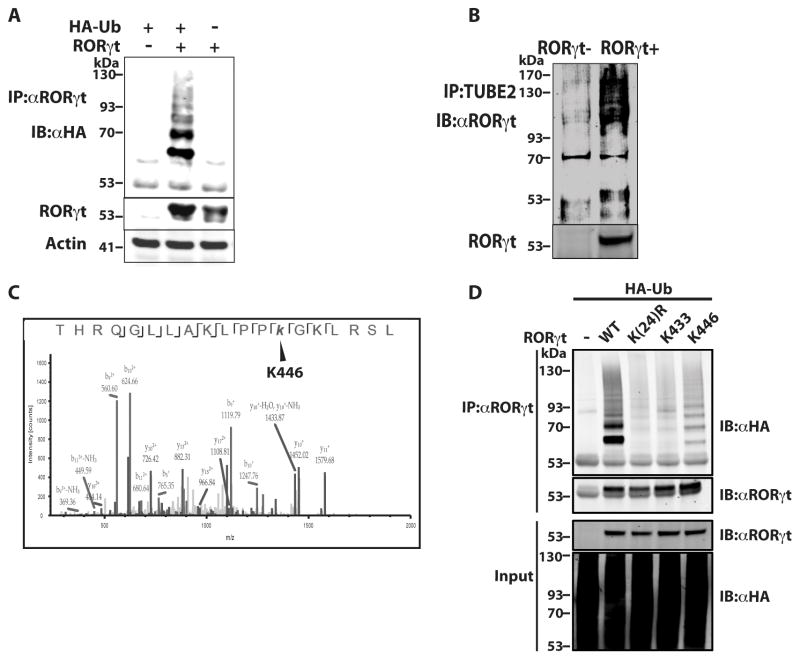
To study post-transcriptional regulation of RORγt, we first determined whether RORγt is ubiquitinated. To overcome the difficulty of detecting ubiquitinated proteins, we first used an in vitro system to overexpress both RORγt and HA-tagged ubiquitin (HA-Ub) in 293T cells (Fig. 1A). A ladder pattern typical of ubiquitinated RORγt was detected in the presence, but not absence, of HA-Ub. In T cells differentiated under Th17 conditions (TGFβ and IL-6), TUBEs (tandem ubiquitin binding entities) (40) were used to enrich ubiquitinated proteins (Fig. 1B). Ubiquitinated RORγt was detected in wild-type (WT) but not RORγt−/− cells, clearly suggesting that endogenous RORγt is ubiquitinated in Th17 cells. To identify potential ubiquitination site(s), we performed mass spectrometric (MS) analysis of RORγt immunoprecipitated from RORγt-expressing 293T cells and differentiated Th17 cells treated with deubiquitinase inhibitor. Immunoprecipitated RORγt was first digested with trypsin. Digesting ubiquitinated lysine (K) leaves GG isopeptide linked to K, which can be easily detected by MS due to an increased mass of 114.102 daltons (41). In this way, K446 of RORγt was identified as an apparent ubiquitination site in both Th17 (Fig.1C) and 293T cells (data not shown) by its MS/MS peptide spectrum. To confirm K446 ubiquitination, we mutated K446 to arginine (K446R) to prevent ubiquitination. Ubiquitination of RORγt WT and K446R was then compared in the presence of HA-Ub, but we did not observe obvious differences (data not shown), likely due to presence of other ubiquitination sites. We thus mutated all 24 lysines in RORγt to arginines [K(24)R], except K446 (K446) or K433 (K433); K433 was not a ubiquitination site in our MS analysis and was therefore used as a negative control. As expected, when expression plasmids encoding RORγt or mutants and HA-Ub were transfected into 293T cells, no ubiquitination was detected in the K(24)R mutant or K433 control, whereas obvious ubiquitination signals were detected in K446. This confirms that K446 of RORγt is a ubiquitination site.
Figure 1.
K446 of RORγt is a ubiquitination site. (A) Expression plasmids encoding RORγt and HA-Ub were transfected into 293T cells for 48 h. RORγt was then immunoprecipitated (IP) with anti-RORγt antibody (αRORγt) and resolved by SDS-PAGE. Ubiquitinated RORγt was detected by immunoblot (IB) with anti-HA antibody (αHA). Actin was used as a loading control. (B) Purified naïve CD4+ T cells were differentiated under Th17 priming conditions (TGFβ and IL-6) for three days. Ubiquitinated proteins in the purified cell lysates were enriched by IP with TUBE beads and resolved on an SDS-PAGE gel. RORγt in the immunoprecipitated ubiquitinated proteins was then detected by IB with anti-RORγt antibody. (C) RORγt immunoprecipitated from differentiated Th17 cells was digested by trypsin and analyzed by MS. Shown are the MS/MS peptide spectra that identified K446 as a ubiquitination site. (D) Expression plasmids encoding RORγt or indicated RORγt mutants and HA-Ub were transfected into 293T cells for 48 h. RORγt was then IP with αRORγt and resolved by SDS-PAGE. Ubiquitinated RORγt was detected by IB with αHA. Data shown except (C) are the representatives of at least three independent experiments.
RORγt K446 ubiquitination negatively regulates Th17 differentiation
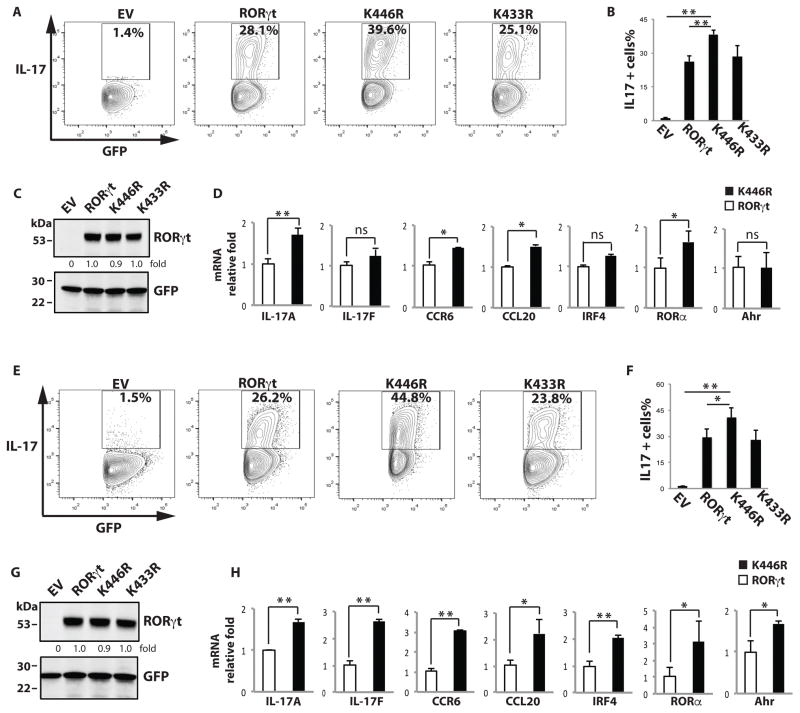
To determine the function of K446 ubiquitination in Th17 differentiation, K446 or K433 (control) were mutated to arginine (K446R and K433R). We developed an assay to evaluate K446R function using retrovirus-reconstituted RORγt−/− T cells which cannot differentiate into Th17 cells due to lack of endogenous RORγt. This system is a powerful tool, as any Th17 differentiation activity in the reconstituted RORγt−/− T cells solely depends on retrovirus-mediated expression of exogenous RORγt. T cells obtained from RORγt−/− mice (38) were transduced with retrovirus expressing GFP only or together with RORγt WT, K446R, or K433R, and differentiated under Th17 differentiation conditions. IL-17+/GFP+ cells were enumerated by flow cytometry (Fig. 2A and 2B), and RORγt and mutants expression were monitored (Fig. 2C). Consistent with the role of RORγt in Th17 differentiation, RORγt−/− T cells expressing RORγt differentiated into IL-17 producing cells, but GFP-only control cells did not. Relative to both RORγt WT and control mutant K433R, the K446R mutant greatly potentiated Th17 differentiation. We then compared the expression of critical Th17 genes between cells expressing RORγt WT and K446R (Fig. 2D). IL-17A, CCR6, CCL20 and RORα but not IL-17F, IRF4 and Ahr were significantly up-regulated in Th17 cells expressing K446R, confirming the potentiation of Th17 differentiation by K446R. To study Th17 differentiation in re-stimulated cells, the differentiated Th17 cells described above were rested in IL-2 for 48 hrs, and then placed in Th17 priming conditions again (Fig. 2E and 2F). Expression of RORγt and mutants were monitored (Fig.2G). The differences between RORγt WT- and K446R-mediated Th17 differentiation (Fig. 2E and Fig. 2F) and expression of critical Th17 genes (Fig. 2H) were further increased. Therefore, our results illustrate that the K446R mutant enhanced Th17 differentiation, demonstrating the negative role of RORγt K446 ubiquitination in the regulation of Th17 differentiation.
Figure 2.
K446 ubiquitination negatively regulates Th17 differentiation. (A–C) T cells obtained from RORγt/− mice were transduced with retrovirus expressing GFP only (empty virus, EV) or together with RORγt WT, K446R, or K433R, and differentiated under Th17-differentiating conditions. Three days later, intracellular IL-17 was detected by flow cytometry. Percentages of IL-17+ cells among GFP+ cells are indicated. B is the summary of three independent experiments. RORγt and GFP expression was detected by westernblot analysis in C, and expression levels relative to wild type RORγt was calculated. (D) RNA was prepared from RORγt (open bars)- or K446R (black bars)-reconstituted RORγt−/− T cells differentiated under Th17 conditions. qPCR was performed to quantitate expression of the indicated genes. * P < 0.05, ** P<0.01. ns stands for not significant. (E–G) Differentiated Th17 cells described in (A) were rested in IL-2 for 48 h, placed in Th17 differentiation conditions for 3 more days, and analyzed as described in (A). F is the summary of three independent experiments. RORγt and GFP expression was detected by westernblot analysis in G, and expression levels relative to wild type RORγt was calculated. (H) Critical Th17 gene expression in RORγt (open bars)- or K446R (black bars)-reconstituted RORγt−/− T cells rested and redifferentiated as described in (E) was analyzed by qPCR. Unless specified, data shown are the representative of at least three independent experiments.
RORγt K446 ubiquitination inhibits RORγt activity by preventing the recruitment of co-activator SRC1
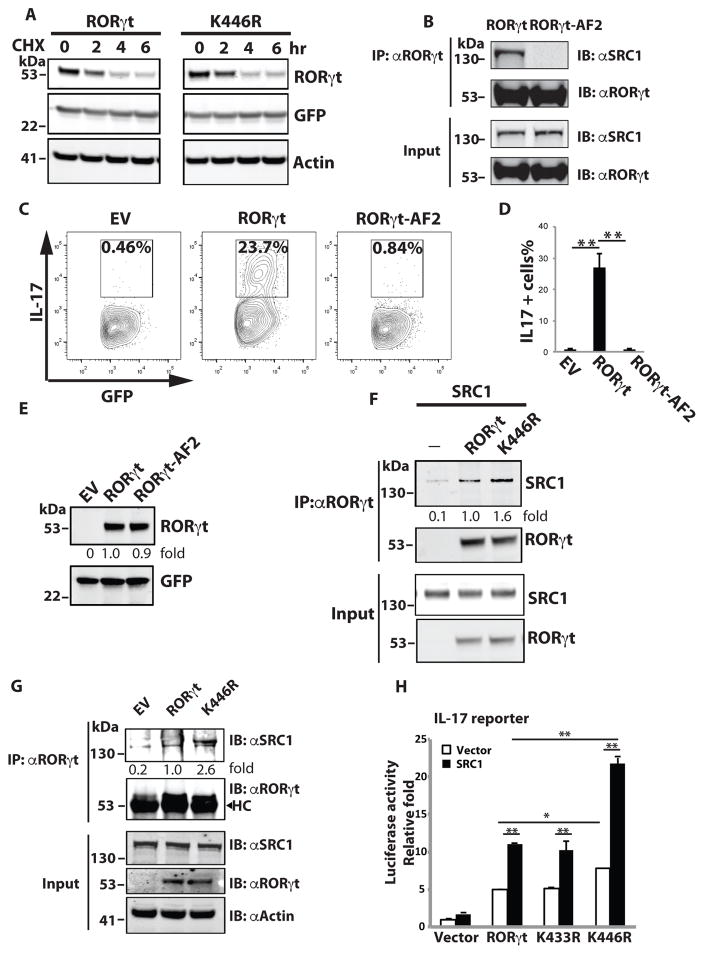
An important function of ubiquitination is to regulate protein degradation (32). Reports have suggested a role for ubiquitination in the regulation of RORγt stability (34–37). However, in our studies, RORγt WT and the K446R mutant expressed at similar levels (Fig.3A, 3F and 3G). To confirm that K446 ubiquitination does not regulate RORγt protein stability, we compared the degradation rates of RORγt WT and K446R in the presence of the protein synthesis inhibitor cycloheximide (CHX) (Fig. 3A). The rates were similar, suggesting K446 ubiquitination does not regulate RORγt protein stability. Another function of ubiquitination is to regulate protein-protein interactions. We previously identified an activation function two (AF2) motif that is located 33 amino acids downstream of K446 (28). The AF2 motif mediates the recruitment of steroid co-activator one (SRC1), which stimulates RORγt-mediated transcriptional activity. Mutation of a critical AF2 tyrosine (479) to phenylalanine (RORγt-AF2) disrupted the RORγt-SRC1 interaction (Fig. 3B). Expression of RORγt-AF2 but not RORγt WT (Fig. 3E) failed to rescue Th17 differentiation in RORγt−/− T cells (Fig. 3C and 3D), strongly supporting the important function of RORγt-recruited SRC1 in Th17 differentiation. To determine whether K446 ubiquitination affects RORγt-SRC1 interaction, we expressed RORγt WT or K446R with SRC1 in 293T cells (Fig. 3F) and RORγt−/− Th17 cells (Fig. 3G). More SRC1 was immunoprecipitated in cells expressing K446R than WT RORγt, suggesting that K446 ubiquitination inhibits SRC1 binding. Transcription activity of RORγt depends on the recruitment of co-activator SRC1 (28). To determine the effects of K446R-enhanced SRC1 recruitment on RORγt-mediated transactivation, we used an IL-17 promoter-luciferase reporter (Fig. 3H), as RORγt directly binds to IL-17 promoter to stimulate its expression (42). In the presence of SRC1, RORγt WT and K433R mutant stimulated the IL-17 reporter, consistent with the function of SRC1 as a co-activator for RORγt. The K446R mutant activated the IL-17 reporter more stronger than that of the RORγt WT. Taken together, our results suggest that K446 ubiquitination inhibits RORγt activity by preventing SRC1 recruitment.
Figure 3.
K446 ubiquitination impairs RORγt activity by reducing the recruitment of co-activator SRC1. (A) Expression plasmids for GFP together with RORγt or K446R were transfected into 293T cells for 48 h. Cells were then treated with protein synthesis inhibitor cycloheximide (CHX) for the indicated time (hr) and RORγt and GFP were detected by IB. (B) RORγt WT or AF2 mutant was expressed with SRC1 in 293T cells for 48 h. IP was performed to detect RORγt-SRC1 interactions. (C–E) RORγt−/− T cells were transduced with retrovirus expressing GFP only (empty virus, EV) or together with RORγt WT or AF2 mutant, and differentiated under Th17 conditions. Three days later, intracellular IL-17 was detected by flow cytometry. Percentages of IL-17 positive cells among GFP+ cells are indicated. D is the summary of three independent experiments. RORγt and GFP expression was detected by westernblot analysis in E. (F) RORγt WT or K446R was expressed with SRC1 in 293T cells for 48 h. IP was performed to detect RORγt-SRC1 interactions. Immunoprecipitated SRC1 levels relative to that immunoprecipiated by wild type RORγt was calculated. (G) RORγt WT or K446R was transduced into RORγt−/− T cells differentiated under Th17 conditions for three days. IP was performed to detect RORγt-SRC1 interactions. Immunoprecipitated SRC1 levels relative to that immunoprecipiated by wild type RORγt was calculated. HC indicates heavy chain. (H) IL-17 promoter luciferase reporter was transfected into 293T cells with control or SRC1 plasmid and indicated RORγt mutants for 24 h. Luciferase assay was performed to quantitate relative reporter activity. Data represent mean ± SE from three independent experiments with at least two replicas in each experiment. *<0.05 and **<0.01. Unless specified, data shown are the representative of at least three independent experiments.
USP15 interacts with RORγt and deubiquitinates K446
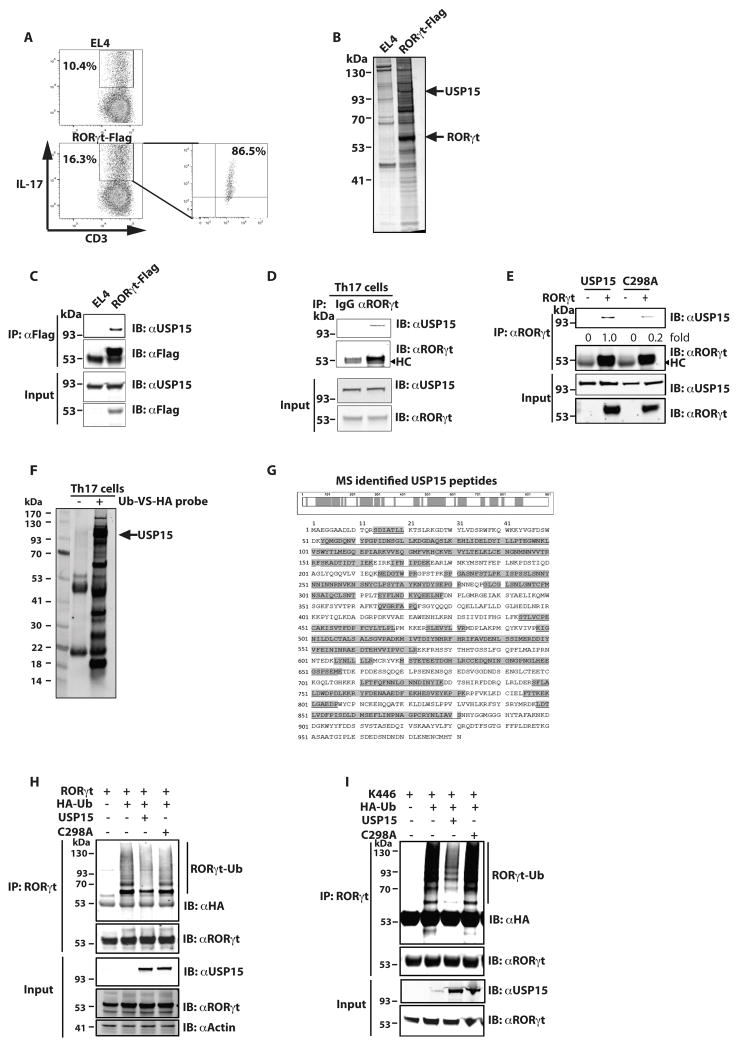
Addition of ubiquitin to lysine residues requires ubiquitin ligase enzymes, whereas removal of ubiquitin is catalyzed by deubiquitinases (32). To determine the enzyme(s) responsible for modifying K446, we used MS to identify RORγt-interacting proteins. To obtain the large amount of specific RORγt-interacting protein complexes needed for successful MS analysis, we generated a stable EL-4 cell line expressing 3xflag-tagged RORγt. EL-4 cells were used because they express RORγt and produce IL-17 (43), and thus have the components required for IL-17 production. Flag-tag avoids disguising the epitopes critical for interacting with co-factors and nonspecific binding due to cross-reactivity with anti-RORγt antibodies. We confirmed that EL-4 cells produced IL-17 which was further enhanced by overexpressing flag-tagged RORγt (Fig. 4A), indicating that flag-tag does not affect RORγt function in stimulating IL-17 production. To enrich the protein components critical for regulating IL-17 production, IL-17-producing cells were further enriched to almost 90% purity using IL-17-catching beads (Fig. 4A, right panel). Immunoprecipitation using anti-flag antibody was performed on EL-4 WT and RORγt-flag-expressing cell lysates and immunoprecipitated complexes were then resolved on a protein gel (Fig. 4B). As expected, an intense band corresponding to RORγt was observed in RORγt-flag-expressing, but not WT, EL-4 cells (Fig. 4B). MS analysis of a band observed in RORγt-flag-expressing but not WT EL-4 cells revealed a deubiquitinase, ubiquitin specific protease 15 (USP15). RORγt-USP15 interaction in the RORγt-flag EL4 cells was confirmed by immunoprecipitation analysis (Fig. 4C). We also confirmed the RORγt-USP15 interaction in differentiated Th17 cells (Fig. 4D). Interestingly, a catalytically inactive USP15 with a cysteine 298 to alanine mutation (C298A) (44), had reduced interaction with RORγt (Fig. 4E), suggesting important function of enzyme activity in RORγt-USP15 interaction. To determine whether USP15 is active in Th17 cells, we used a ubiquitin vinyl sulfone HA-tagged probe (Ub-VS-HA) that crosslinks active deubiquitinases (39). Active ubiquitinases were crosslinked to the Ub-VS-HA probe by incubating with differentiated Th17 cell lysates, followed by immunoprecipitation by anti-HA antibody and analyzed by MS (Fig. 4F). Indeed, USP15 was detected in immunoprecipitated active ubiquitinases in differentiated Th17 cells (Fig. 4F) with more than 50% peptide coverage (Fig. 4G). To determine whether USP15 can deubiquitinate RORγt, RORγt was co-expressed with USP15 WT or C298A and HA-Ub in 293T cells. WT, but not C298A, USP15 reduced ubiquitination of RORγt (Fig. 4H). To determine whether USP15 can deubiquitinate RORγt K446 specifically, K446 that has all lysines mutated to arginines except K446 was co-expressed with USP15 WT or C298A and HA-Ub in 293T cells, as we showed that K446 can be ubiquitinated (Fig. 1D). USP15 WT, but not C298A, removed ubiquitin from K446 (Fig. 4I). Taken together, our results demonstrate that USP15 is active and associated with RORγt in Th17 cells. More importantly, USP15 is able to deubiquitinate RORγt at K446 specifically.
Figure 4.
USP15 interacts with RORγt and deubiquitinates K446. (A) Control or RORγt-flag expressing EL-4 cells were established and intracellular IL-17 was detected by flow cytometry. IL-17-producing cells in gated area were enriched by IL-17 catching beads and analyzed for intracellular IL-17 (right panel). (B) Anti-flag antibody was used to IP lysates from control or RORγt-flag-expressing EL-4 cells and IP complexes were resolved on a protein gel. Arrows indicate the bands containing USP15 and RORγt on the silver stained gel. (C) RORγt-SRC1 interaction was detected by IB of anti-flag antibody IP cell lysates of control or RORγt-flag expressing EL-4 cells. (D) Naïve T cells were differentiated into Th17 cells for three days. Control IgG or αRORγt was used to perform IP to detect RORγt-USP15 interactions. HC indicates heavy chain. (E) WT USP15 or C298A was expressed with RORγt in 293T cells for 48 h. IP was performed to detect RORγt-USP15 interactions. Immunoprecipitated USP15 levels relative to that immunoprecipiated by wild type RORγt was calculated. HC indicates heavy chain. (F) Lysates of differentiated Th17 cells were incubated with or without Ub-HS-HA probe to covalently crosslink active deubiquitinases, which were then IP with αHA and resolved by SDS-PAGE. Arrow indicates the band containing USP15. (G) MS-identified USP15 peptides are highlighted by shadows. (H) RORγt was expressed alone or together with USP15 WT or inactive C298A mutant in the presence of HA-Ub in 293T cells for 48 h. RORγt was then IP and ubiquitinated RORγt (RORγt-Ub) was detected by IB with αHA. (I) K446 (all other Ks are mutated to Rs except K446) was expressed alone or together with USP15 WT or inactive C298A mutant in the presence of HA-Ub in 293T cells for 48 h. RORγt was then IP and ubiquitinated RORγt (RORγt-Ub) was detected by IB with αHA. Unless specified, data shown are the representative of at least three independent experiments.
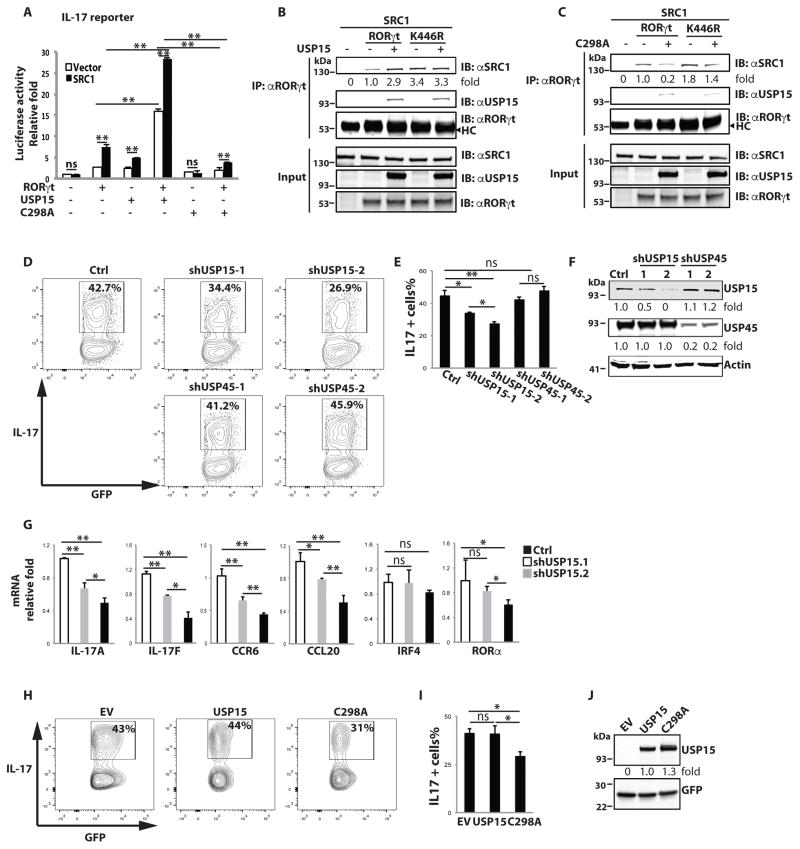
USP15 stimulates RORγt activity and Th17 differentiation by enhancing SRC1 recruitment
To determine the role of USP15 in RORγt and SRC1-regulated transactivation, we again used the IL-17 promoter reporter. USP15 WT greatly potentiated, whereas inactive C298A rather inhibited, reporter activity (Fig. 5A). Correspondingly, USP15 enhanced (Fig. 5B), whereas inactive C298A inhibited (Fig. 5C), SRC1 recruitment to RORγt. This suggests that USP15 stimulates RORγt activity via enhancing SRC1 recruitment. Consistent with our previous observation that K446R enhanced SRC1 recruitment (Fig. 5B). Furthermore, K446R-enhanced SRC1 recruitment was independent of the presence of USP15 (Fig. 5B) or C298A (Fig. 5C), strongly suggesting that USP15 regulates SRC1 recruitment via deubiquitinating K446. We next determined whether USP15 is important for Th17 differentiation using short hairpin RNA (shRNA) knockdown. Knockdown of USP15 with two different shRNAs markedly inhibited Th17 differentiation (Fig. 5D, top panels and 5E) compared to knockdown of a close member of USP15, USP45 (Fig. 5D, bottom panels and 5E). shUSP15-2 was more potent in inhibiting Th17 differentiation than shUSP15-1, which correlated well with the greater efficacy of shUSP15-2 to knock down USP15 expression (Fig. 5F). Analysis of critical Th17 genes also confirmed knockdown of USP15 impaired Th17 differentiation (Fig. 5G). In addition, retrovirus expressing USP15 WT or C298A transduced T cells to determine their function in Th17 differentiation. Overexpression of USP15 did not affect Th17 differentiation relative to empty virus expressing GFP alone (EV) (Fig. 5H, 5I and 5J), likely due to already much active USP15 detected in Th17 cells (Fig. 4F). In contrast, the inactive USP15 C298A mutant did inhibit Th17 differentiation. Impaired Th17 differentiation by both inactive C298A and knockdown of USP15 supports the positive role of USP15 in Th17 differentiation. Altogether, our results indicate that USP15 stimulates RORγt activity via enhancing SRC1 recruitment, ultimately leading to promoting Th17 differentiation.
Figure 5.
USP15 regulates RORγt activity and Th17 differentiation via enhancing SRC recruitment. (A) IL-17 reporter was transfected into 293T cells together with vector (open bars) or SRC1 expression plasmid (black bars) and indicated plasmids for 24 h. Luciferase activity was then measured. Data represent mean ± SE from three independent experiments with at least two replicas in each experiment. **<0.01. ns stands for not significant. (B) RORγt or K446R was expressed together with SRC1 in the presence or absence of USP15 in 293T cells for 48 h. IP with αRORγt was performed to detect RORγt-SRC1 interactions. Immunoprecipitated SRC1 relative to that immunoprecipitated by wild type RORγt was calculated. (C) RORγt or K446R was expressed with SRC1 in the presence or absence of inactive USP15 C298A mutant in 293T cells for 48 h. IP with αRORγt was performed to detect RORγt-SRC1 interaction. Immunoprecipitated SRC1 relative to that immunoprecipitated by wild type RORγt was calculated. (D–F) Retrovirus expressing GFP with scrambled shRNA (Ctrl) or shRNAs specific for knockdown of USP15 or USP45 was transduced into CD4+ T cells and differentiated under Th17 conditions for three days. Intracellular IL-17 was detected by flow cytometer. E is the summary of three independent experiments. Knockdown of USP15 and USP45 were monitored by IB in F. (G) RNA was prepared from control (open bars), shUSP15-1 (grey bars)- or shUSP15-2 (black bars)-transduced T cells that differentiated under Th17 conditions. qPCR was performed to quantitate expression of the indicated genes. * P < 0.05, ** P<0.01. ns stands for not significant. (H–J) Retrovirus expressing GFP alone (EV) or together with USP15 WT or inactive C298A mutant were transduced into CD4+ T cells and differentiated under Th17 conditions for three days. Intracellular IL-17 was detected by flow cytometry. (I) is the summary of three independent experiments. USP15 and GFP expression was detected by westernblot analysis in J, and expression levels relative to wild type RORγt was calculated. Unless specified, data shown are the representative of at least three independent experiments.
Discussion
The generation of robust Th17 immune responses necessary for clearance of pathogens, including fungus and Citrobacter rodentium, depends on stimulating the differentiation of Th17 cells (1–3, 45, 46). Conversely, effective prevention of Th17-mediated autoimmune diseases, such as multiple sclerosis and psoriasis, depends on inhibiting Th17 differentiation (4, 17, 18, 21, 47). RORγt is the transcription factor that instructs Th17 differentiation (6, 31), making this molecule an attractive drug target for precisely controlling Th17-mediated immunity (22, 24). It is thus important to understand the mechanisms regulating RORγt activity. Previous studies have focused on transcriptional regulation of RORγt and identification of RORγt target genes (6, 7, 14, 15). Less is known about the mechanisms of post-transcriptional regulation that is responsible for regulating RORγt function. Reports that RORγt stability is regulated by ubiquitination-mediated degradation mechanisms (34–37) suggested a role for ubiquitination in RORγt-mediated differentiation. In this study, we identified RORγt K446 as a ubiquitination site that limits Th17 differentiation by interfering with the recruitment of co-activator SRC1. Therefore, ubiquitination of RORγt has additional role in the regulation of RORγt interaction with its co-factor to regulate Th17 differentiation. This study also reveals the therapeutic potentials for clinically boosting the Th17 immunity against pathogens or preventing pathological Th17-mediated autoimmunity via controlling the RORγt ubiquitination pathways.
Like other nuclear receptors, RORγt cannot regulate gene transcription by itself; it has to recruit other cofactors such as SRC1 for activation of target genes (29). SRC1 brings in transcriptional regulators, including histone acetyltransferases and methyltransferases, to open the gene locus for active transcription. Recruitment of SRC1 is therefore a critical checkpoint for controlling RORγt activity. On the other hand, SRC1 does not directly bind to DNA and depends on transcription factors to bring it to the specific DNA targets on chromatin to regulate gene expression. The fact that SRC1 services as co-activators for many different transcription factors (48) suggests that the transcription factors associated with SRC1 determines the specificity of SRC1 in the regulation of gene expression. Based on the traditional model, ligand binding is considered a critical step for triggering the recruitment of co-activator by nuclear receptors (29). However, RORγt ligands, cholesterol intermediate metabolites, are usually abundantly available in cells (30). The question is whether there are other mechanisms regulating RORγt to recruit SRC1. We identified K446 of RORγt as a ubiquitination site in limiting SRC1 recruitment, and thus suggesting its function in controlling Th17 differentiation. Indeed, prevention of K446 ubiquitination (K446R mutation) promoted Th17 differentiation. Therefore, in addition to transcription regulation of RORγt expression and ligand-induced recruitment of SRC1, our results demonstrated additional layer of regulation mechanisms responsible for controlling RORγt activity via post-transcripitional ubiquitination. However, it is still not clear how K446 ubiquitination prevents SRC1 binding. Because K446 is close to the critical tyrosine (Y479) in the AF2 motif responsible for recruiting SRC1, it remains to be determined whether K446 ubiquitination physically blocks SRC1 binding. There are several other lysines close to K446 including K433, K442 and K448. Our study does not exclude the possibility that these neighboring lysines can be ubiqutinated and that K446 may even affect ubiquitination of the neighboring lysines. Moreover, the physiological signals regulating K446 ubiquitination are not known. Identification of such signals will facilitate understanding the function of K446 ubiquitination in the regulation of in vivo immune responses both in disease and normal conditions.
Another effort in this study was to identify the enzymes responsible for modifying K446 ubiquitination. Since K446 ubiquitination negatively regulates Th17 differentiation, K446 must be deubiquitinated to promote Th17 differentiation under Th17 priming conditions. Our efforts thus focused on identifying the deubiquitinases. Our following strong evidence supports that USP15 deubiquitinates K446 to promote Th17 differentiation by enhancing SRC1 recruitment: 1) USP15 is the active deubiquitinase associated with RORγt in Th17 cells and IL-17-producing EL-4 cells; 2) WT, but not inactive (C298A mutation), USP15 is able to remove ubiquitin from K446 of RORγt and stimulate RORγt-SRC1 interaction and RORγt activity; 3) knockdown of USP15 or forced expression of inactive USP15 impaired Th17 differentiation. USP15 has been reported to regulate multiple functions from oncogenesis to bone formation via deubiquitinating and stabilizing protein substrates (49–53), i.e. reversing ubiquitination-induced protein degradation. We report here that USP15 regulates RORγt activity via controlling SRC1 recruitment and thus reveal a new mode of action for USP15-regulated function, controlling protein-protein interaction. However, another report has shown that mice with a germline deletion of USP15 did not show defects in Th17 function (49). An obvious explanation for this discrepancy is redundancy, as other deubiquitinases may replace the function of USP15. It is also possible that a germline deletion gives mice enough time during development to adapt to the absence of USP15, for example, through up-regulation of other USPs to compensate for the lack of USP15. In fact, our demonstration that acute knockdown of USP15 affected Th17 differentiation supports the hypothesis that compensation mechanisms are likely developed in USP15 knockout mice. A conditional knockout of USP15 in Th17 cells specifically will facilitate determination of the function of USP15 in Th17 differentiation.
Th17 cells produce effector cytokines IL-17A, IL-17F, IL-22 and GM-CSF to mediate the pathological inflammation responsible for many types of autoimmune diseases; targeting Th17 cells is thus a potentially valuable treatment for these diseases (19). Indeed, inhibiting the Th17 pathway is effective in the treatment of psoriasis and multiple sclerosis (54, 55). For example, ustekinumab, a human monoclonal antibody that inhibits Th17 responses by blocking the IL-23 receptor, has been approved by the FDA for treatment of severe plaque psoriasis (56). Given the essential function of RORγt in Th17 cells, pharmaceutical and academic scientists are developing RORγt inhibitors for treatment of Th17-dependent autoimmunity (20–24). Such inhibitors are typically screened for the ability to disrupt the interaction between RORγt and co-activator SRC1 (20). We demonstrated here that recruitment of SRC1 is regulated by ubiquitination of K446, suggesting that drugs targeting this RORγt ubiquitination pathway can inhibit RORγt activity by limiting co-activator SRC1 recruitment. Interestingly, a recent report shows that USP15 is up-regulated in psoriasis, supporting the possible role of USP15 in this Th17-mediated autoimmune disease (57). We expect that USP15 inhibitors will alleviate Th17-mediated autoimmunity including psoriasis via blocking the recruitment of SRC1. Although we identified USP15 as the deubiquitinase for K446, it remains unknown about the ubiquitin ligases responsible for K446 ubiquitination. Our future work is to search for the K446 E3 ligases which are also the potential drug target for controlling Th17 immunity. Therefore, in addition to revealing a new mechanism for RORγt-regulated Th17 differentiation, our results may facilitate the development of a new category of RORγt-based drugs that treat Th17-mediated autoimmunity by controlling RORγt ubiquitination-regulated recruitment of SRC1.
Acknowledgments
This work was supported by grants from NIH R01-AI053147, NIH R01-AI109644 and institutional pilot funding. In addition, research reported in this publication was also supported by the National Cancer Institute of the National Institutes of Health under award number P30CA33572, which includes work performed in the animal, genomic, flow cytometer and mass spectrometric cores supported by this grant.
We appreciate the help by city of hope supported cores including animal, genomic, flow cytometer and mass spectrometric cores. We thank Dr. Chen Dong (Tsinghuan University) for sharing the IL-17 reporter. In addition, we thank Sarah T. Wikinson and Keely Walker for their careful proof/editing work on the manuscript.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Wang Z, Friedrich C, Hagemann SC, Korte WH, Goharani N, Cording S, Eberl G, Sparwasser T, Lochner M. Regulatory T cells promote a protective Th17-associated immune response to intestinal bacterial infection with C. rodentium. Mucosal immunology. 2014;7:1290–1301. doi: 10.1038/mi.2014.17. [DOI] [PubMed] [Google Scholar]
- 2.Basu R, Whitley SK, Bhaumik S, Zindl CL, Schoeb TR, Benveniste EN, Pear WS, Hatton RD, Weaver CT. IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the TH17 cell-iTreg cell balance. Nat Immunol. 2015;16:286–295. doi: 10.1038/ni.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O’Connor W, Jr, Rongvaux A, Van Rooijen N, Haberman AM, Iwakura Y, Kuchroo VK, Kolls JK, Bluestone JA, Herold KC, Flavell RA. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elloso MM, Gomez-Angelats M, Fourie AM. Targeting the Th17 pathway in psoriasis. Journal of leukocyte biology. 2012;92:1187–1197. doi: 10.1189/jlb.0212101. [DOI] [PubMed] [Google Scholar]
- 5.Johnson-Huang LM, Lowes MA, Krueger JG. Putting together the psoriasis puzzle: an update on developing targeted therapies. Disease models & mechanisms. 2012;5:423–433. doi: 10.1242/dmm.009092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov BS, II, McKenzie, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 7.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 9.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, Kang HS, Ma L, Watowich SS, Jetten AM, Tian Q, Dong C. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–587. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutton C, Brereton C, Keogh B, Mills KH, Lavelle EC. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J Exp Med. 2006;203:1685–1691. doi: 10.1084/jem.20060285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, Alzahrani M, Al-Muhsen S, Halwani R, Ma CS, Wong N, Soudais C, Henderson LA, Marzouqa H, Shamma J, Gonzalez M, Martinez-Barricarte R, Okada C, Avery DT, Latorre D, Deswarte C, Jabot-Hanin F, Torrado E, Fountain J, Belkadi A, Itan Y, Boisson B, Migaud M, Arlehamn CS, Sette A, Breton S, McCluskey J, Rossjohn J, de Villartay JP, Moshous D, Hambleton S, Latour S, Arkwright PD, Picard C, Lantz O, Engelhard D, Kobayashi M, Abel L, Cooper AM, Notarangelo LD, Boisson-Dupuis S, Puel A, Sallusto F, Bustamante J, Tangye SG, Casanova JL. IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349:606–613. doi: 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratajewski M, Walczak-Drzewiecka A, Salkowska A, Dastych J. Upstream stimulating factors regulate the expression of RORgammaT in human lymphocytes. J Immunol. 2012;189:3034–3042. doi: 10.4049/jimmunol.1200519. [DOI] [PubMed] [Google Scholar]
- 15.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annual review of immunology. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 16.Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nature reviews Immunology. 2014;14:585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, Becher B. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 18.El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang GX, Dittel BN, Rostami A. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12:568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Sundrud MS, Skepner J, Yamagata T. Targeting Th17 cells in autoimmune diseases. Trends in pharmacological sciences. 2014;35:493–500. doi: 10.1016/j.tips.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Huh JR, Littman DR. Small molecule inhibitors of RORgammat: targeting Th17 cells and other applications. Eur J Immunol. 2012;42:2232–2237. doi: 10.1002/eji.201242740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skepner J, Ramesh R, Trocha M, Schmidt D, Baloglu E, Lobera M, Carlson T, Hill J, Orband-Miller LA, Barnes A, Boudjelal M, Sundrud M, Ghosh S, Yang J. Pharmacologic inhibition of RORgammat regulates Th17 signature gene expression and suppresses cutaneous inflammation in vivo. J Immunol. 2014;192:2564–2575. doi: 10.4049/jimmunol.1302190. [DOI] [PubMed] [Google Scholar]
- 22.Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, Wu C, Baloglu E, Schmidt D, Ramesh R, Lobera M, Sundrud MS, Tsai PY, Xiang Z, Wang J, Xu Y, Lin X, Kretschmer K, Rahl PB, Young RA, Zhong Z, Hafler DA, Regev A, Ghosh S, Marson A, Kuchroo VK. Small-molecule RORgammat antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40:477–489. doi: 10.1016/j.immuni.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheridan C. Footrace to clinic heats up for T-cell nuclear receptor inhibitors. Nature biotechnology. 2013;31:370. doi: 10.1038/nbt0513-370. [DOI] [PubMed] [Google Scholar]
- 24.Huang Z, Xie H, Wang R, Sun Z. Retinoid-related orphan receptor gamma t is a potential therapeutic target for controlling inflammatory autoimmunity. Expert opinion on therapeutic targets. 2007;11:737–743. doi: 10.1517/14728222.11.6.737. [DOI] [PubMed] [Google Scholar]
- 25.He YW, Deftos ML, Ojala EW, Bevan MJ. RORgamma t, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity. 1998;9:797–806. doi: 10.1016/s1074-7613(00)80645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortiz MA, Piedrafita FJ, Pfahl M, Maki R. TOR: a new orphan receptor expressed in the thymus that can modulate retinoid and thyroid hormone signals. Molecular endocrinology. 1995;9:1679–1691. doi: 10.1210/mend.9.12.8614404. [DOI] [PubMed] [Google Scholar]
- 27.Xie H, Huang Z, Wang R, Sun Z. Regulation of thymocyte survival by transcriptional coactivators. Critical reviews in immunology. 2006;26:475–486. doi: 10.1615/critrevimmunol.v26.i6.10. [DOI] [PubMed] [Google Scholar]
- 28.Xie H, Sadim MS, Sun Z. RORgammat recruits steroid receptor coactivators to ensure thymocyte survival. J Immunol. 2005;175:3800–3809. doi: 10.4049/jimmunol.175.6.3800. [DOI] [PubMed] [Google Scholar]
- 29.Walsh CA, Qin L, Tien JC, Young LS, Xu J. The function of steroid receptor coactivator-1 in normal tissues and cancer. International journal of biological sciences. 2012;8:470–485. doi: 10.7150/ijbs.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santori FR, Huang P, van de Pavert SA, Douglass EF, Jr, Leaver DJ, Haubrich BA, Keber R, Lorbek G, Konijn T, Rosales BN, Rozman D, Horvat S, Rahier A, Mebius RE, Rastinejad F, Nes WD, Littman DR. Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell metabolism. 2015;21:286–297. doi: 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, Gennert D, Satija R, Shakya A, Lu DY, Trombetta JJ, Pillai MR, Ratcliffe PJ, Coleman ML, Bix M, Tantin D, Park H, Kuchroo VK, Regev A. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komander D, Rape M. The ubiquitin code. Annual review of biochemistry. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 33.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual review of biochemistry. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutz S, Kayagaki N, Phung QT, Eidenschenk C, Noubade R, Wang X, Lesch J, Lu R, Newton K, Huang OW, Cochran AG, Vasser M, Fauber BP, DeVoss J, Webster J, Diehl L, Modrusan Z, Kirkpatrick DS, Lill JR, Ouyang W, Dixit VM. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature. 2015;518:417–421. doi: 10.1038/nature13979. [DOI] [PubMed] [Google Scholar]
- 35.Han L, Yang J, Wang X, Wu Q, Yin S, Li Z, Zhang J, Xing Y, Chen Z, Tsun A, Li D, Piccioni M, Zhang Y, Guo Q, Jiang L, Bao L, Lv L, Li B. The E3 deubiquitinase USP17 is a positive regulator of retinoic acid-related orphan nuclear receptor gammat (RORgammat) in Th17 cells. J Biol Chem. 2014;289:25546–25555. doi: 10.1074/jbc.M114.565291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Yang J, Han L, Zhao K, Wu Q, Bao L, Li Z, Lv L, Li B. TRAF5-mediated Lys-63-linked Polyubiquitination Plays an Essential Role in Positive Regulation of RORgammat in Promoting IL-17A Expression. J Biol Chem. 2015;290:29086–29094. doi: 10.1074/jbc.M115.664573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, Xu P, Han L, Guo Z, Wang X, Chen Z, Nie J, Yin S, Piccioni M, Tsun A, Lv L, Ge S, Li B. Cutting edge: Ubiquitin-specific protease 4 promotes Th17 cell function under inflammation by deubiquitinating and stabilizing RORgammat. J Immunol. 2015;194:4094–4097. doi: 10.4049/jimmunol.1401451. [DOI] [PubMed] [Google Scholar]
- 38.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 39.Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chemistry & biology. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 40.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO reports. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Zhang Y, Yang XO, Nurieva RI, Chang SH, Ojeda SS, Kang HS, Schluns KS, Gui J, Jetten AM, Dong C. Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity. 2012;36:23–31. doi: 10.1016/j.immuni.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ichiyama K, Yoshida H, Wakabayashi Y, Chinen T, Saeki K, Nakaya M, Takaesu G, Hori S, Yoshimura A, Kobayashi T. Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J Biol Chem. 2008;283:17003–17008. doi: 10.1074/jbc.M801286200. [DOI] [PubMed] [Google Scholar]
- 44.Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, Guterman A, Glickman M, Schade R, Kloetzel PM, Dubiel W. The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Current biology : CB. 2005;15:1217–1221. doi: 10.1016/j.cub.2005.05.059. [DOI] [PubMed] [Google Scholar]
- 45.Zelante T, De Luca A, D’Angelo C, Moretti S, Romani L. IL-17/Th17 in anti-fungal immunity: what’s new? Eur J Immunol. 2009;39:645–648. doi: 10.1002/eji.200839102. [DOI] [PubMed] [Google Scholar]
- 46.van de Veerdonk FL, Marijnissen RJ, Kullberg BJ, Koenen HJ, Cheng SC, Joosten I, van den Berg WB, Williams DL, van der Meer JW, Joosten LA, Netea MG. The macrophage mannose receptor induces IL-17 in response to Candida albicans. Cell host & microbe. 2009;5:329–340. doi: 10.1016/j.chom.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, Kuchroo VK. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dasgupta S, Lonard DM, O’Malley BW. Nuclear receptor coactivators: master regulators of human health and disease. Annual review of medicine. 2014;65:279–292. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou Q, Jin J, Hu H, Li HS, Romano S, Xiao Y, Nakaya M, Zhou X, Cheng X, Yang P, Lozano G, Zhu C, Watowich SS, Ullrich SE, Sun SC. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol. 2014;15:562–570. doi: 10.1038/ni.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herhaus L, Al-Salihi MA, Dingwell KS, Cummins TD, Wasmus L, Vogt J, Ewan R, Bruce D, Macartney T, Weidlich S, Smith JC, Sapkota GP. USP15 targets ALK3/BMPR1A for deubiquitylation to enhance bone morphogenetic protein signalling. Open biology. 2014;4:140065. doi: 10.1098/rsob.140065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vos RM, Altreuter J, White EA, Howley PM. The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. Journal of virology. 2009;83:8885–8892. doi: 10.1128/JVI.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Faronato M, Patel V, Darling S, Dearden L, Clague MJ, Urbe S, Coulson JM. The deubiquitylase USP15 stabilizes newly synthesized REST and rescues its expression at mitotic exit. Cell cycle. 2013;12:1964–1977. doi: 10.4161/cc.25035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eichhorn PJ, Rodon L, Gonzalez-Junca A, Dirac A, Gili M, Martinez-Saez E, Aura C, Barba I, Peg V, Prat A, Cuartas I, Jimenez J, Garcia-Dorado D, Sahuquillo J, Bernards R, Baselga J, Seoane J. USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nature medicine. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- 54.Tonel G, Conrad C, Laggner U, Di Meglio P, Grys K, McClanahan TK, Blumenschein WM, Qin JZ, Xin H, Oldham E, Kastelein R, Nickoloff BJ, Nestle FO. Cutting edge: A critical functional role for IL-23 in psoriasis. J Immunol. 2010;185:5688–5691. doi: 10.4049/jimmunol.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segal BM, Constantinescu CS, Raychaudhuri A, Kim L, Fidelus-Gort R, Kasper LH, Ustekinumab MSI. Repeated subcutaneous injections of IL12/23 p40 neutralising antibody, ustekinumab, in patients with relapsing-remitting multiple sclerosis: a phase II, double-blind, placebo-controlled, randomised, dose-ranging study. The Lancet Neurology. 2008;7:796–804. doi: 10.1016/S1474-4422(08)70173-X. [DOI] [PubMed] [Google Scholar]
- 56.Benson JM, Peritt D, Scallon BJ, Heavner GA, Shealy DJ, Giles-Komar JM, Mascelli MA. Discovery and mechanism of ustekinumab: a human monoclonal antibody targeting interleukin-12 and interleukin-23 for treatment of immune-mediated disorders. mAbs. 2011;3:535–545. doi: 10.4161/mabs.3.6.17815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng AP, He YM, Liu XX, Li JW, Tu YT, Hu F, Chen SJ. Expression of USP15, TbetaR-I and Smad7 in psoriasis. Journal of Huazhong University of Science and Technology Medical sciences = Hua zhong ke ji da xue xue bao Yi xue Ying De wen ban = Huazhong keji daxue xuebao Yixue Yingdewen ban. 2014;34:415–419. doi: 10.1007/s11596-014-1293-1. [DOI] [PubMed] [Google Scholar]