Abstract
Expression of the transcription factor Zbtb1 is required for normal lymphoid development. We report here that Zbtb1 maintains genome integrity in immune progenitors without which cells undergo increased DNA damage and p53-mediated apoptosis during replication and differentiation. Increased DNA damage in Zbtb1-mutant (ScanT) progenitors was due to increased sensitivity to replication stress, which was consequence of inefficient activation of the S-phase checkpoint response. Increased p53-mediated apoptosis affected not only lymphoid but also myeloid development in competitive bone marrow chimeras and prevention of apoptosis by transgenic Bcl2 expression and p53-deficiency rescued lymphoid as well as myeloid development from Zbtb1-mutant progenitors. Interestingly, however, protection from apoptosis rescued only the early stages of T-cell development and thymocytes remained arrested at the DN3 developmental stage, indicating a strict requirement of Zbtb1 at later T-cell developmental stages. Altogether, these results indicate that Zbtb1 prevents DNA damage in replicating immune progenitors, allowing the generation of B-cell, T-cell and myeloid cells.
Keywords: Zbtb1, lymphoid, development, replication stress, DNA damage
INTRODUCTION
A prominent family of transcription factors, BTB-ZF (Broad complex, tramtrack, and Bric a brac-zinc finger), is well represented at lineage decision points during immune cell development. Among members of this family, the transcription factor Miz1 (Zbtb17) is required for B-cell development (1); Lrf (Zbtb7a), is a master regulator of B-cell differentiation (2) and it also controls erythroid development (3); Th-POK (Zbtb7b) controls CD4+ versus CD8+ T-cell differentiation (4); Bcl6 (Zbtb26) controls Tfh differentiation (5–7) and the development of CD8+ memory T-cells (8, 9); and PLZF (Zbtb16) controls the effector maturation of iNKT cells and IL-17+γδT-cells (10–13).
It was recently shown that another member of this family, Zbtb1, is expressed in hematopoietic cells and it is essential for T-cell development while it is partially required for B-cell and NK-cell development (14, 15). A point mutation in the BTB domain of Zbtb1 (ScanT mutant), as well as Zbtb1 deficiency leads to complete absence of T-cells. Under competitive conditions, however, only myeloid cells develop in ScanT mutant mice, suggesting a specific function of Zbtb1 during lymphoid but not myeloid differentiation (14). However, how Zbtb1 exerts these functions has not been explored.
More recently, using epithelial cell lines as model system, Zbtb1 was identified as a regulator of a DNA damage tolerance mechanism that occurs during DNA synthesis and it is triggered in response to stalling of the replication fork (16). At stalled replication forks, the continuous action of helicases leads to the formation of single stranded DNA, to the recruitment and activation of the ATR kinase, phosphorylation of Chk1 and inhibition of DNA synthesis (s-phase checkpoint). Zbtb1 interacts with Kap1, allowing the accessibility of Rad18 to DNA damage sites and to the recruitment of low fidelity DNA polymerases that can bypass the DNA lesion, preventing DNA damage. This process is called translesion DNA synthesis (17). Absence of the s-phase checkpoint response as it occurs in ATR-deficient or Chk1-deficient cells leads to extensive DNA damage, chromosomal abnormalities, and consequently to cell death (18) (19, 20).
In this work we have identified that Zbtb1 is required for the efficient activation of the S-phase checkpoint response in proliferating immune progenitors, preventing DNA damage and p53-mediated cell death. This function of Zbtb1 was required for efficient lymphoid as well as myeloid development. Interestingly, protection from apoptosis and p53-deficiency rescued myeloid and B-cell development but only the early stages of T-cell development in ScanT mice and thymocytes didn’t progress beyond the DN3 and DP developmental stages respectively, indicating that Zbtb1 also controls the later stages of T-cell development independently of activation of the pro-apoptotic p53 pathway.
MATERIALS AND METHODS
Animals
Experiments with animals were approved by the IACUC committee. The Zbtb1-GFP transgenic strain was kindly donated by Dr. Derek Sant’Angelo. Zbtb1-GFP mice were generated in a C57BL/6 background and were maintained by breeding with C57BL6/J mice from Jackson Laboratory (Bar Harbor, Maine 04609). Zbtb1-GFP mice grow, behave and breed normally. There is no disparity in phenotypes displayed between males and females. ScanT mice were kindly provided by Dr. Bruce Beutler and were previously described (14). Vav-Bcl2 transgenic mice were previously described (21) and p53-deficient mice were obtained from the Jackson Laboratory. All strains are in a C57BL/6 background and animals were used between 4 to 8 weeks of age. As controls for the experiments with ScanT mice we used littermates when available or C57BL/6 mice of similar age and sex.
BAC modification and generation of Zbtb1-GFP transgenic mouse
About 185 kb-long bacteria artificial chromosome (BAC) DNA containing the full length of mouse Zbtb1 gene were used for modification. Starting from translation initiation site ATG, the 1133bp long DNA fragment was replaced by GFPCre fusion protein coding sequence (1767bp), using RED/ET BAC Modification Kit (Gene Bridges GmbH, Heidelberg). Briefly, about 500bp homologous arm A and B, and GFPCre sequence in between, was subcloned and sequencing-verified. The targeted DNA sequence in the original BAC was first replaced by rpsL-neo counter-selection marker, then rpsL-neo was replaced with GFPCre by homologous recombination. The DNA integrity of the recombined BAC was checked by restriction enzyme foot-printing.
The modified BAC DNA was purified by Cesium Chloride density-gradient centrifugation. After getting rid of cesium chloride, the DNA was dialyzed in injection buffer overnight. The diluted DNA was then injected into fertilized egg to generate Zbtb1-GFP transgenic mouse.
Cell culture
OP9 and OP9-DL1 cells were cultured in αMEM containing 10% FBS (vol/vol), 100 U/ml Penicilin, 100 μg/ml Streptomycin, 2.2 g/l Sodium bicarbonate, 50 μM β-mercaptoethanol. For most cultures, bone marrow progenitor cells were sorted and seeded on OP9 stromal cells in αMEM medium supplemented with 5 ng/ml Flt3L and 1 ng/ml IL-7. Lin-Sca-1+c-kit+ cells were cultured in X-VIVOTM 10 (Lonza) medium containing 20 ng/ml IL-3, 50 ng/ml IL-6 and 50 ng/ml SCF. NIH3T3 cells were cultured in DMEM ontaining 10% FBS (vol/vol), 100 U/ml Penicilin, 100 μg/ml Streptomycin, 2.2 g/l Sodium bicarbonate.
Flow Cytometry and Cell Sorting
Single-cell suspensions were prepared at the time of autopsy from thymus, bone marrow, or spleen in PBS containing 0.1% BSA and 0.08% sodium azide (staining buffer). Antibody incubation was performed at 4°C for 20 min in staining buffer. For LMPPs and CLPs, freshly isolated bone marrow cells were incubated with biotinylated antibodies to CD3ε (145-2C11), B220 (RA3-6B2), TER-119 (TER-119), CD11b (M1/70), Gr-1 (RB6-8C5). Lineage-positive (Lin+) cells were depleted by MACS separation system according to manufacturer’s recommendations. After enrichment, Lin− cells were stained with surface markers. LMPPs were sorted by flow cytometry as Lin−Sca-1+c-Kit+Flt3hi, and CLPs were sorted as Lin−IL-7Rα+AA4.1+Flt3+. Antibodies used for flow cytometry are listed below:
CD45.1 (A20), CD45.2 (104), Sca-1 (E13-161.7), c-Kit (2B8), Flt3 (A2F10), IL-7Rα (A7R34), CD93 (AA4.1), B220 (RA3-6B2), CD19 (6D5), CD11b (M1/70), Gr-1 (RB6-8C5), CD44 (IM7), CD25 (PC61), Thy1.2 (30-H12), CD4 (RM4-5), CD8α (53-6.7), TCR-β (H57), P53 (1C12).
Sorting and FACS analysis was performed using an LSRII and FACSaria cell sorter and post analysis of data was performed was done using the Flojo software.
Tunel and Annexin staining
Anexin staining was performed according to the manufacturer instructions (eBioscience). After staining of cells with antibodies, cells were washed in PBS and resuspended in annexin binding buffer containing FITC-conjugated annexin V. After incubation at room temperature for 15 min, cells were diluted in annexin V binding buffer and analyzed by flow cytometry. For Tunel assays we used the in situ cell death detection kit-FITC (Roche) following the manufacturer’s instructions. Briefly, cells were stained with antibodies, and labeled with the fixable viable cell dye (ebioscience), cells were washed, fixed with PFA 2% for 30 min, permeabilized with 0.1% triton buffer and later incubated with the kit reagent for 60 min, washed and analyzed by FACS.
Metaphase spreads
Primary B cells were stimulated with 10 μg/ml LPS, 0.5 μg/ml anti-CD180 (RP105) and 10 ng/ml IL-4 for two days. Different concentration of ATR inhibitor VE-822 was added and cells were incubated for another 24 hours. Colcemid (Roche) was added to a final concentration of 100 ng/ml to the culture and cells were incubated 37°C for 1 to 1.5 hours. Cells were then resuspended in prewarmed (37°C) 75 mM KCl and incubated for 20 min. Cells were harvested and resuspended in freshly prepared fixative (3 parts methanol, 1 part acetic acid). After incubation at room temperature for 30 min, cells were harvested and resuspended in freshly prepared fixative at −20°C overnight. Metaphase spread were prepared by Dropping into slides using a Thermotron at about 52% humidity.
Immunofluorescence labeling, confocal microscopy and image analysis
Cells labeled using standard procedures. Briefly, cells were cytospined into slides, fixed with 4 % formaldehyde, permeabilized with 0.5 % Triton X-100 in PBS for 5 min at room temperature, blocked with 4 % normal goat serum in PBS for 15–60 min and stained with the primary antibody for 1 hour at room temperature. After washing, slides were incubated 1 hour in the presence of secondary antibody, rinsed once with 0.1 % Triton X-100 in PBS and 3 times in PBS. Slides were prepared for microscopy after staining with 100 nM DAPI in PBS for 20 min at room temperature. Images were acquired in a Zeiss Zen confocal microscope and post image analysis was done using Zeiss Zen image software, cells were segmented based on the DAPI stain. The MFI of γH2AX as well as the number of γH2AX foci was measured only in the DAPI+ region. The plotted histograms were constructed using the Countif function in Excel of the raw data extracted from the image analysis.
Western Blot Analysis
Primary B cells with different treatment were lysed with 1% NP40 lysis buffer (1% NP40, 300 mM NaCl, 0.1 mM EDTA, 50 mM Tris pH 7.5) containing protease inhibitor cocktail (Roche) and phosphatase inhibitor (Sigma), resolved by NuPAGE (Invitrogen) gel electrophoresis and transferred to a PVDF membrane. After incubation with antibodies, the positive signal was detected by enhanced chemiluminescence method. The blots were developed by LAS 3000 Imaging System (GE Healthcare Life Sciences).
In vitro labeling and staining of BrdU
Dividing Cells were in vitro labeled with 10 μM BrdU for 30 min at 37°C. Stained with Fixable Viability Dye (FVD) to label dead cells before fixation. Cells were then stained with antibodies and fixed with freshly prepared 1×BrdU Staining Buffer solution for 15 min at room temperature in the dark. After washing and resuspension, cells were digested with DNase I working solution for 1 hour at 37°C in the dark and stained with anti-BrdU antibody for 20–30 min at room temperature in the dark.
Cell Proliferation Assay by CellTrace™ Violet Labeling
Cells were incubated with PBS dye solution containing 5 μM CellTrace™ Violet for 20 min at 37°C in the dark. Free dye remaining in the solution was removed by washing with culture medium. Cells were cultured for the indicated time period before flow cytometry analysis.
RNA isolation and real time PCR
RNA was extracted using RNeasy Plus Micro Kit (QIAGEN) and reverse transcribed using iScript™ cDNA Synthesis Kit (BIO-RAD). Real time PCR was performed in a 10 μl reaction volume with iTaq™ Universal SYBR® Green Supermix (BIO-RAD). Reaction was performed on 7900HT Fast Real-Time PCR System (Applied Biosystems). Expression of the gene of interest was calculated relative to HPRT mRNA levels.
Generation of Lhx2 cell lines by retroviral transduction
We generated a retrovirus expressing Lhx2 under the control of the TRE promoter by deleting the dsRED-miR30 fragment from TrmpVIR plasmid generated by Dr. Scott Lowe (Addgene plasmid#27994) (22) with BamH1-EcoRI restriction enzymes and inserting the Lhx2 cDNA. Retrovirus were generated by transfection of 293T cells with packaging plasmids pMD2G (from Dr. Trono Addgene #12259) and pCL-Eco from (Dr. Inder Verma Addgene 12371) and the generated virus was collected in the supernatants 2 days after transfection. Sca1+ bone marrow cells were isolated with magnetic beads (Miltenyi Biotech) and cells were stimulated to proliferate with hematopoietic cytokines IL-3, IL-6 and SCF overnight in serum-free ExVivo media. Cells were then transduced by spin infection in the presence of 8μM of polybrene for 2 hours. Transduced immune progenitors were then washed and cultured in ExVivo media for additional 2 days before sorting of GFP+ cells. Cells were maintained in IMDM media supplemented with antibobiotics and 10%FBS in the presence of SCF, IL6 and Doxycycline (10ng/ml) to induce Lhx2 expression.
Generation of Flag-Zbtb1-IRES-Thy1.1 and Flag-ScanT-IRES-Thy1.1 constructs
The Flag-Zbtb1 and Flag-ScanT constructs were cloned into a Thy1.1+ expressing retrovirus (23) using EcoRI and XhoI restriction enzymes. The generated virus from these constructs were used to transduce NIH3T3 cells and transduced Thy1.1+ cells were selected by positive selection with anti-biotin MACs beads (Miltenyi Biotech). Stably transduced Thy1.1+NIH3T3 were maintained in DMEM media supplemented with antibiotics and 10%FBS.
Generation of competitive BM chimeras
For the generation of competitive bone marrow chimeras, recipient F1 mice (CD45.1+CD45.2+) of over 2 month of age were lethally irradiated with two doses of 500 rads separated by 5 hours before i.v. injection of a mixture of bone marrow cells from donor mice (2–5×106 cells/recipient). Mice were analyzed 2–4 months after reconstitution.
RESULTS
Zbtb1 is required for efficient development of immune cells
Zbtb1 was previously reported to be expressed in hematopoietic stem cells and progenitors and to control lymphoid development (14, 15). To better understand the expression pattern of Zbtb1 during immune cell development, we analyzed GFP levels in a BAC transgenic Zbtb1-GFP reporter mouse which was generated and kindly provided by Dr. Derek Sant’Angelo. FACs analysis of bone marrow cells from Zbtb1-GFP mice revealed that lineage+ cells were GFPlo while lineage− cells were homogenously GFPhi. We then validated the reporter model by RT-PCR of endogenous Zbtb1 transcripts on sorted GFP+ and GFP− cells from Bone Barrow (Supplementary Figure 1a). Using this reporter model we analyzed Zbtb1 levels in different immune populations. As reported previously by RT-PCR, we identified that hematopoietic progenitors in bone marrow were already Zbtb1 expressing cells. Thymocytes at different stages of development as well as mature T-cells, B-cells and myeloid cells were also Zbtb1-expressing cells (Supplementary Figure 1b). In summary, the high Zbtb1 expression in hematopoietic progenitors suggested a functional role at this developmental stage.
To analyze the impact of Zbtb1 expression to immune cell development, we obtained ScanT (Zbtb1-mutant) mice from Dr. Bruce Beutler. A point mutation within the BTB domain of Zbtb1, termed “ScanT”, leads to absence of T-cells similarly to Zbtb1 deficiency. As it was not reported if the ScanT point mutation affected the stability or function of Zbtb1, we set out to investigate if the ScanT mutant form of Zbtb1 protein could be detected. For this purpose we generated plasmid constructs expressing either wild type (Flag-Zbtb1) or mutant (Flag-ScanT) Zbtb1 forms upstream of an IRES-Thy1.1. Using this system we could simultaneously detect the presence of the Flag-Zbtb1 or Flag-ScanT and Thy1.1 proteins which are derived from the same mRNA in transduced NIH3T3 cells.
Although similar levels of Thy1.1 were detected by FACs analysis of NIH3T3 cells transduced with either Flag-Zbtb1 or Flag-ScanT constructs, the ScanT form could not be detected by FACs analysis with an anti-flag antibody (Supplementary Figure 2a) or by immunofluorescence (Supplementary Figure 2c). We could also not detect presence of the Zbtb1 protein by western blot of NIH3T3 cells transduced with the Flag-ScanT construct (Supplementary Figure 2b). In correlation, B-cells isolated from ScanT mice didn’t have detectable Zbtb1 protein levels (Supplementary Figure 2d). Altogether, these data indicate that the ScanT mutation in Zbtb1 severely affects its stability, validating the ScanT mouse model as a Zbtb1-deficient model.
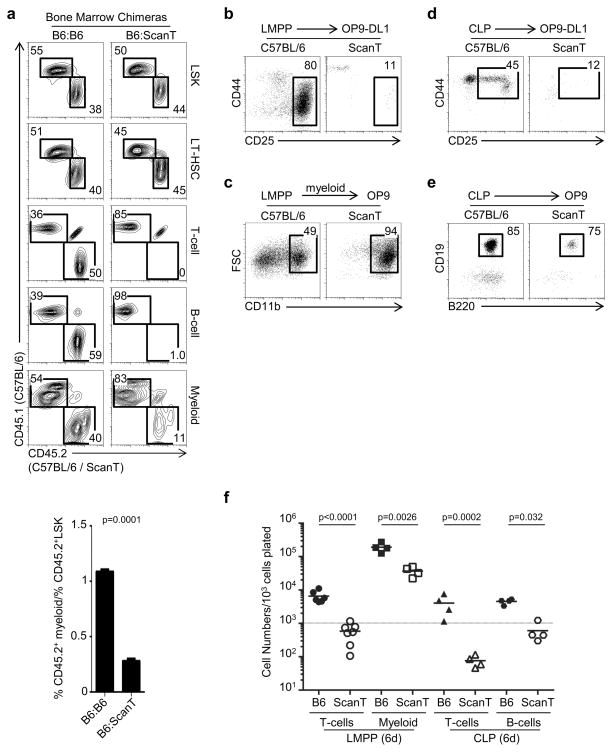
To evaluate the impact of Zbtb1 to immune cell development, we performed mixed bone marrow chimeras in which ScanT progenitor cells were in competition with wild type cells. We identified the origin of the cells by the congenic markers CD45.1 and CD45.2. We observed that ScanT progenitors could, similar to wild type cells, efficiently reconstitute LSK progenitors as well as LT-HSCs in the bone marrow under these competitive conditions. As previously described, T-cell and B-cell development from ScanT progenitors were highly affected (16). Although myeloid cells could be generated from ScanT progenitors, their development was partially affected. Analysis of the percentage of ScanT-derived myeloid cells relative to the percentage in of ScanT-derived LSK cells revealed that myeloid reconstitution was approximately 75% lower than that from wild type progenitors (Figure 1a). Interestingly, despite the impaired lymphoid and myeloid differentiation of ScanT-derived cells, lymphoid and myeloid progenitors were efficiently generated (Supplementary Figure 3).
Figure 1. Zbtb1 is required for the generation of normal numbers of lymphoid and myeloid cells.
a) FACS analysis of mixed bone marrow chimeras generated with a 1:1 mixture of CD45.1+(C57BL/6):CD45.2+(C57BL/6 or ScanT) donor cells. Donor chimerism is identified in LSK (Lin−Sca1+cKit+) and LT-HSC (Lin−CD150+Flt3− Sca1+cKit+) cells in bone marrow and T-cells (TCRβhi), B-cells (CD19+) and myeloid cells (CD11b+) in spleen. The numbers represent the proportion of cells within the indicated gates. Below is the quantitation of the proportion of myeloid/LSK cells derived from CD45.2+ cells. Data was calculated by dividing the % of CD45.2+CD11b+ in spleen by the % of CD45.2+LSK in bone marrow from each mouse and data of 4 independent chimeras for each condition is represented. Three independent experiments with 3–4 chimeras per condition were performed with similar results. b) FACS analysis of LMPP cells co-cultured with OP9-DL1 cells in the presence of lymphoid (IL-7 and Flt3l) cytokines for 15 days to initiate T-cell development. The numbers indicate the proportion of DN3 cells generated. c) FACS analysis of LMPP cells co-cultured with OP9 cells in the presence of myeloid (GM-CSF, M-CSF, IL-3) cytokines for 6 days to initiate myeloid development. The numbers indicate the proportion of myeloid cells generated. d) FACS analysis of CLP cells co-cultured with OP9-DL1 cells in the presence of lymphoid (IL-7 and Flt3l) cytokines for 6 days to initiate T-cell development. The numbers indicate the proportion of DN2 cells generated. e) FACS analysis of CLP cells co-cultured with OP9 cells in the presence of lymphoid (IL-7 and Flt3l) cytokines for 6 days to initiate B-cell development. The numbers indicate the proportion of B cells generated. f) Quantitation of cell numbers obtained during in vitro differentiation experiments outlined in (b–e) at 6 days. Each dot represents data of cells isolated from an individual mouse, horizontal bars represent the mean and the dotted line represent the original cell number plated. p values corresponding to the significance using T-test are shown.
The impaired generation of mature immune cells from ScanT progenitors was also observed in in vitro developmental systems. ScanT LMPP cells could not develop into DN3 (CD44−CD25+) thymocytes in OP9-DL1 co-cultures but could develop into myeloid (CD11b+) cells in myeloid-promoting conditions (Figure 1b and c). Similarly to LMPP cells, CLP progenitors could not develop into T-cells in OP9-DL1 co-cultures but generated B-cells (B220+CD19+) in OP9 co-cultures (Figure 1d and e). However, the number of cells generated from ScanT progenitors in culture was highly and significantly reduced in all differentiating conditions (Figure 1f).
Altogether these results indicate that Zbtb1 is not required for the generation of immune progenitors from hematopoietic stem cells but for the differentiation of immune progenitors towards mature myeloid and lymphoid cells.
Zbtb1 prevents apoptosis in replicating immune progenitors
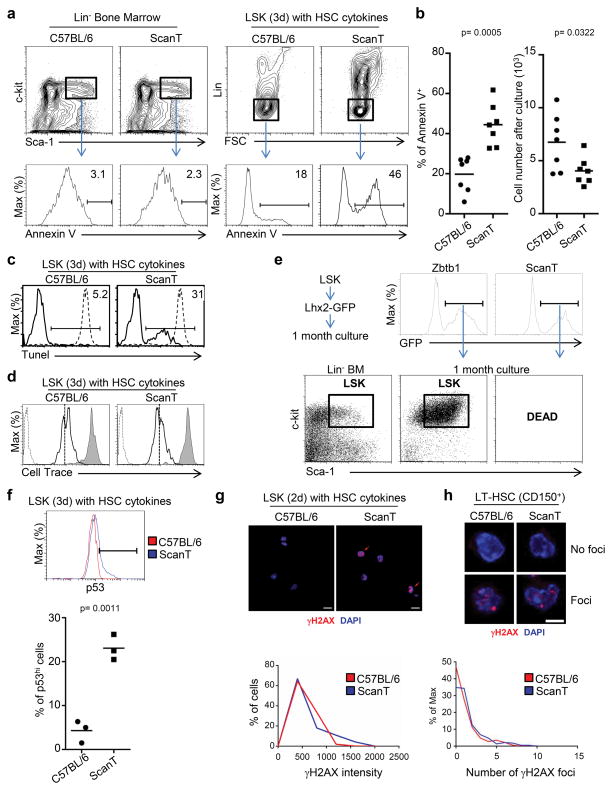
As the ScanT mutation affected development of all immune lineages, and as the generation of mature immune cells is associated with active proliferation, we postulated that impaired proliferation or survival of ScanT progenitors may underlie their impaired development. To test this, we sorted the LSK progenitor population from wild type and ScanT bone marrow and cultured them with SCF, IL-6, and IL-3 cytokines, which stimulate the proliferation of undifferentiated cells. Under these conditions, we measured the ability of the cells to proliferate by dilution of cell trace label and the proportion of apoptotic cells by annexin V staining, gating on PI negative live cells. We didn’t detect an increase of apoptotic (Annexin V+) LSK cells from ScanT mice when analyzed ex-vivo, however, when stimulated to proliferate in vitro, approximately 45% of ScanT lineage-negative cells were apoptotic as compared to approximately 20% of wild type cells as measured by Annexin V staining (Figure 2a and b). The increased apoptosis of ScanT progenitors correlated with significantly reduced cell numbers (Figure 2b). Approximately 30% of ScanT cells presented apoptotic DNA breaks after culture as measured by the Tunel assay (Figure 2c). However, despite their increased apoptosis, ScanT LSK cells diluted the cell trace label similarly to wild type cells, indicating that the ScanT mutation does not affect their ability to initiate proliferation (Figure 2d). To investigate if the apoptosis of ScanT LSK cells was unrelated to a specific differentiation pathway, we attempted to generate hematopoietic cell lines from wild type and ScanT immune progenitors. For this purpose we generated a retroviral construct expressing Lhx2 under the control of the TRE promoter (Dox induced) (pTrmpVIR-Lhx2). Overexpression of Lhx2 is sufficient to drive the differentiation of Embryonic Stem cells (ESC) towards immune progenitors and to generate hematopoietic cell lines in culture from adult bone marrow (24). Although LSK cells derived from ScanT and wild type mice could be similarly transduced with the TrmpVIR-Lhx2 construct, only wild type cells could be efficiently maintained in culture and presented an undifferentiated phenotype (Figure 2e). These results further confirm that although Zbtb1 is required for the development of immune cells from lineage-committed progenitors, its function is not related to a specific developmental pathway but to prevent the apoptosis of actively proliferating cells, presumably by protecting cells to replication stress.
Figure 2. Zbtb1 protects hematopoietic progenitors from apoptosis.
a) FACs analysis for annexin V staining in LSK (Lin−Sca1+cKit+) cells ex-vivo or after 3 day culture with SCF, IL-6 and IL-3. The gate indicates the proportion of Annexin V+ cells within the PI− live cells. b) Scatter plots representing the percentage of annexin V+ cells obtained in (a), and the number of cells obtained after 3 days in culture. Each dot corresponds to data from one mouse and horizontal bars represent the mean. c) Histogram showing the percentage of Tunel+ cells after culture. The dashed line represents T-cells treated with DNAse I before the Tunel assay as positive control. Data is representative of two mice. d) FACS analysis of cell trace dilution in cultured LSK cells as in (a), the gray histogram represents non-proliferating T-cells maintained with IL-7 for 3 days and the dashed line represents cells not stained with the cell trace dye. The vertical dotted line is placed as reference for comparison and the stripped histogram represent cells non-labeled with Cell Trace to indicate the background cell trace fluorescence. This experiment is representative of three independent experiments with two mice per condition. e) LSK cells were transduced with the TrmpVIR-Lhx2 retrovirus, transduced GFP+ cells were sorted and kept in culture for one month. The dot plot shows the LSK phenotype of generated cell lines. f) FACS analysis of intracellular p53 levels in cultured LSK cells as in (a). The scatter plot represents the proportion of p53hi cells, each dot corresponds to data from one mouse and horizontal bars represent the mean. g) Confocal microscopy to identify γH2AX fluorescence in LSK cells after 2 day culture with SCF, IL-6 and IL-3 as in (a). Red arrows indicate cells that present increased γH2AX staining. The histogram represents the Mean Fluorescence Intensity of γH2AX within the DAPI+ region corresponding to the nucleus. h) Immunofluorescence showing γH2AX foci in LT-HSCs. The histogram represents the number of γH2AX foci within the DAPI+ region corresponding to the nucleus. p values corresponding to the significance using T-test are shown. Data is representative of more than 3 independent experiments.
To test if the ScanT mutation leads to increased DNA damage and p53-mediated apoptosis in response to replication stress, we analyzed γH2AX by microscopy, a marker of DNA damage and replication stress (25), and intracellular p53 levels on LSK cells that have been stimulated to proliferate in vitro. We observed that ScanT progenitors presented a significantly higher proportion of cells expressing high p53 levels than wild type progenitors (Figure 2f). Increased p53 levels in ScanT cells correlated with an increased proportion of cells presenting high γH2AX intensity levels (MFI range 1200–2000) (Figure 2g). Interestingly, ScanT mice had the same proportion of LT-HSCs (Lin−ckit+Sca1+CD150+) than wild type mice, some of which presented γH2AX foci, in similar proportions to wild type cells, indicating that ScanT LT-HSC are not undergoing increased replication stress or DNA damage in vivo (Figure 2h). Thus, Zbtb1 prevents increased γH2AX and p53 levels, and prevents apoptosis in immune progenitors when stimulated to proliferate in vitro.
Zbtb1 prevents DNA damage in proliferating B-cells
Upon replication stress, the ATR kinase is recruited to the site of stalled replication forks, ATR phosphorylates Chk1, which blocks DNA synthesis, allowing time for the DNA lesion to be repaired (18). Inefficient ATR activation and Chk1 phosphorylation is lethal for the cell, consequence of high levels of double strand breaks (DSB) and chromosomal aberrations (26).
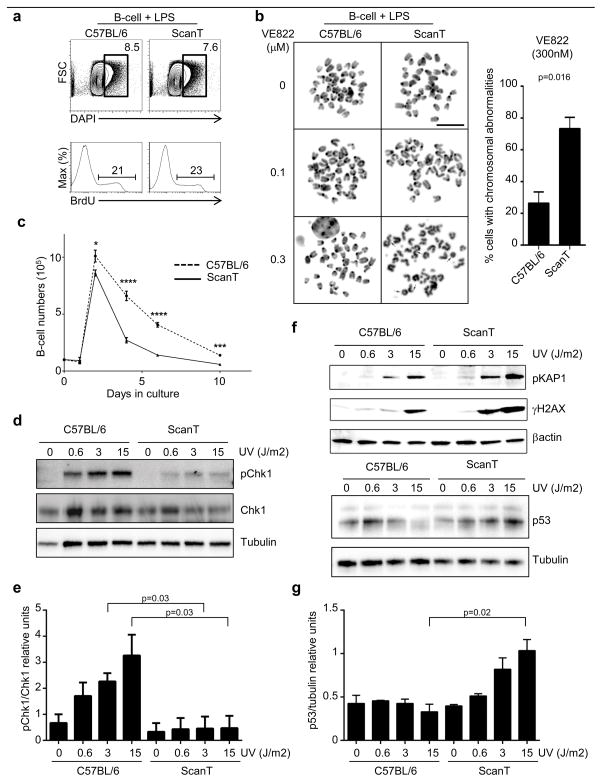
To test if ScanT cells were more sensitive to generate DNA damage upon replication stress, as some B-cells but no T-cell develop in ScanT mice, we used ScanT B-cells as a model system. We first evaluated if ScanT B-cells were able to initiate cell cycle after stimulation with LPS overnight. We analyzed the proportion of cells that were in S/G2-phase by FACs analysis of the intracellular DNA levels (DAPI) and their ability to incorporate BrdU, which occurs during DNA synthesis. ScanT B-cells initiated cell cycle similarly to wild type cells as observed by a similar proportion of cells in S/G2-phase and a similar incorporation of BrdU (Figure 3a). To directly evaluate if Zbtb1 prevents DSBs in proliferating cells we analyzed metaphase spreads from B-cells that have been treated with different concentrations of the ATR inhibitor (VE-822). Interestingly, partial inhibition of ATR function by 300nM treatment caused chromosomal aberrations in approximately 20% of wild type B-cells, but it led to a significant increase in the proportion of cells with chromosomal aberrations in ScanT B-cells (Figure 3b), indicating that ScanT cells are more sensitive to replication stress and to the generation of DNA damage and chromosomal aberrations. Increased sensitivity to replication stress in ScanT B-cells also led to a significant reduction of B-cell numbers at four days after stimulation of proliferation (Figure 3c).
Figure 3. Zbtb1 protects B-cells from DNA damage in response to replication stress.
a) FACs analysis of DNA content and BrdU incorporation in isolated B-cells after 18hs LPS treatment to induce proliferation. Number represent the proportion of cells within the gates identifying cells that have initiated DNA synthesis. b) Metaphase spreads to identify the chromosomal integrity of proliferating B-cells after treatment with suboptimal concentrations of the ATR inhibitor (VE822). Arrows indicate chromosomal aberrations (deletions or fusions). The column graph represents the percentage of cells presenting chromosomal abnormalities after treatment with 0.3μM of VE822. Data corresponds to three independent experiments c) Analysis of B-cell numbers in culture in the presence of LPS stimulation, (Mean ± SD, n=6). d) Western-blot 90 minutes after UV irradiation in proliferating B-cells. e) Quantitation of the relative intensity observed in (d) from three independent experiments. f) Western-blot of proliferating B-cells 5 hours after UV irradiation. g) Quantitation of the relative intensity of p53 observed in (f) from two independent experiments. p values corresponding to the significance using T-test are shown.
Zbtb1 is required for efficient phosphorylation of Chk1 preventing increased p53 levels in response to replication stress
As we observed that partial inhibition of ATR led to increased chromosomal abnormalities in ScanT B-cells, we postulated that Zbtb1 is required for efficient activation of the ATR-Chk1 pathway, which is necessary to avoid DNA damage and cell death. To test this hypothesis, we isolated B-cells, stimulated them to proliferate with LPS for 18hs and treated them with different UV doses to generate thymidine-thymidine dimers that block DNA synthesis and induce replication stress. 90 minutes after irradiation we measured phosphorylated (pChk1) and total Chk1 by western-blot. We observed a dose-response increase of pChk1 in wild type B-cells. Interestingly, ScanT B-cells presented significantly lower pChk1 levels, indicative of inefficient activation of this pathway (Figure 3d and e). Inefficient activation of the S-phase response in ScanT cells led to increased DNA damage evidenced as increased levels of phosphorylated Kap1, γH2AX and p53 levels (Figure 3f and g).
These data indicate that Zbtb1 is required for efficient Chk1 phosphorylation in replicating B-cells, protecting from DNA damage and increased p53 levels.
Transgenic Bcl2 expression and p53 deficiency rescue B-cell development in ScanT mice
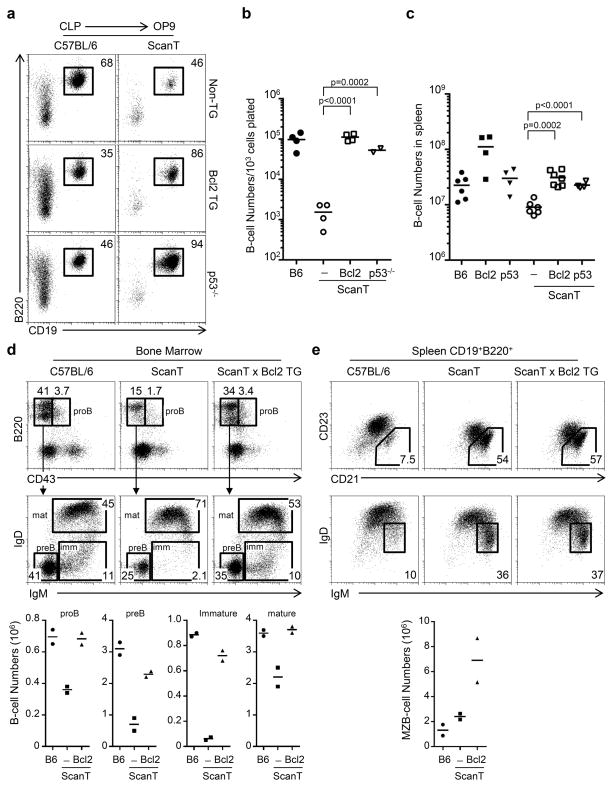
As we have observed that ScanT progenitors upregulate p53 levels and undergo apoptosis when cultured in vitro, we wanted to test if protection from apoptosis and p53 deficiency would rescue B-cell development of ScanT progenitors.
To test this, we analyzed the B-cell developmental potential of CLPs isolated from compound ScanT x Vav-Bcl2 transgenic mice, which protects cells from apoptosis (21) and from ScanT p53−/− mice. We observed that both transgenic Bcl2 expression as well as p53-deficiency significantly rescued the generation of B-cells in vitro from CLP ScanT progenitors (Figure 4a and b). Transgenic Bcl2 expression as well as p53-deficiency similarly rescued B-cell numbers in ScanT mice (Figure 4c). Analysis of B-cell development in the Bone marrow revealed lower numbers of proB and preB cells, immature and mature recirculating B cells in ScanT mice, and this deficiency was reverted by transgenic Bcl2 expression (Figure 4d). We observed a proportional increase of marginal zone B-cells in the spleens of ScanT mice as previously described (14), and this increase was not reverted by transgenic Bcl2 expression (Figure 4e), strongly suggesting that Zbtb1 may also affect the differentiation of B-cells independently of its effects at early stages of B-cell development. In summary, these results indicate that protection from apoptosis rescues the early stages of B-cell development leading to normal B-cell numbers, however, ScanT progenitors preferentially generate marginal zone B-cells.
Figure 4. Transgenic Bcl2 expression and p53-deficiency rescue B-cell development of ScanT progenitors.
a) FACs analysis of in vitro B-cell differentiation in OP9 co-cultures of sorted CLP cells from the indicated strains. The numbers represent the proportion of cells within the gates. b) Scatter plot representing the number of cells obtained under the differentiation conditions in (a). c) B-cell numbers (CD19+B220+) in spleen. Each dot represents data from a mouse. Horizontal bars represent the mean value. p values corresponding to the significance using T-test are shown. Data in (a) is representative of 4 independent experiments. d) Analysis of immature B-cell populations in Bone marrow from the indicated mice corresponding to proB, preB, immature (imm) and mature (mat) B-cells. The numbers represent the percentage of cells within the indicated gates. Below is the quantitation of cell numbers corresponding to each subpopulation in two mice from each strain. e) FACs analysis of B-cells in spleen, numbers represent the proportion of events within each gate, which correspond to Marginal Zone B-cells (CD21+CD23− and IgMhiIgDlo), data is representative of two mice from each strain. Below is the quantitation of Maginal Zone B-cell numbers corresponding to each subpopulation in two mice from each strain
Transgenic Bcl2 expression and p53 deficiency partially rescue T-cell development in ScanT mice
In order to evaluate if p53-mediated apoptosis of ScanT thymic progenitors was the main mechanism affecting T-cell development, we analyzed the thymic profile of compound ScanT x Vav-Bcl2 mice and ScanT p53−/− mice.
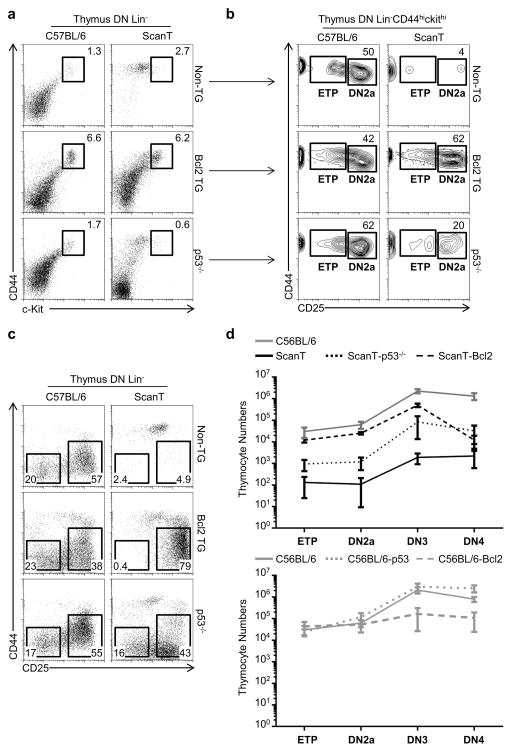
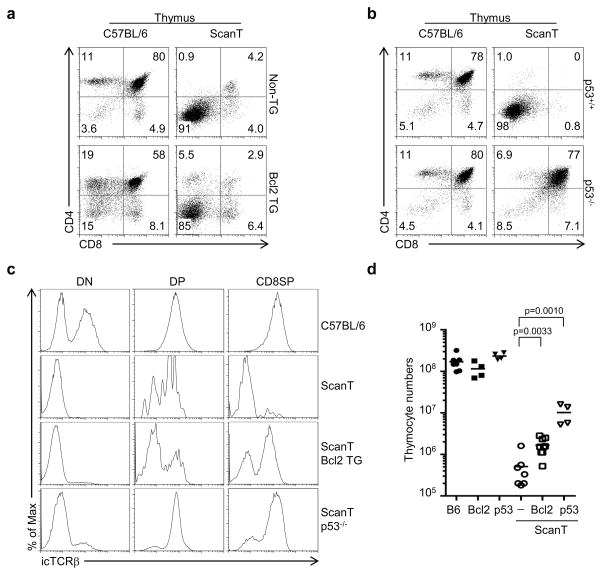
As previously reported, ETP, DN2, DN3 and DN4 thymocytes were absent in the ScanT thymus. We observed that transgenic Bcl2 expression rescued the generation of ETP, DN2a and DN3 thymocytes but not the generation of DN4 thymocytes (Figure 5a, b, c and d) or DP thymocytes (Figure 6a). p53-deficiency, however, was sufficient to rescue the generation of ETP, DN2a, DN3 and DN4 thymocytes (Figure 5a, b, c and d), as well as DP thymocytes, but not the generation of CD4 or CD8 single positive thymocytes (Figure 6b).
Figure 5. Transgenic Bcl2 expression and p53 deficiency partially rescue the early stages of T-cell development in ScanT mice.
a) FACs analysis of lineage negative thymocytes in the indicated strains showing the percentage of CD44hicKithi thymocytes. b) Analysis of ETP and DN2 proportions in cells gated in (a). c) Analysis of DN3 (CD25+CD44−) and DN4 (CD25−CD44−) proportion in the indicated strains. d) Quantitation of cell numbers at different developmental stages in the indicated strains. (Mean ± SD). Data is representative of more than 4 independent experiments.
Figure 6. Transgenic Bcl2 expression and p53 deficiency fail to rescue the late stages of T-cell development in ScanT mice.
FACs analysis of the indicated strains (a and b). Numbers indicate the percentage of cells within the gates. c) Intracellular TCRβ staining of the indicated gated thymocytes. d) Scatter plot showing thymocyte numbers in the indicated mice, each dot corresponds to a mouse, horizontal bars indicate the mean. p values corresponding to the significance using T-test are shown.
These results indicate that prevention of apoptosis in ScanT progenitors is sufficient to rescue the early but not the late stages of T-cell development, and that ScanT thymocytes cannot progress beyond the preTCR checkpoint and remain arrested at the DN3 stage of development. They also show that p53-deficiency is sufficient to bypass the preTCR developmental block of ScanT thymocytes, leading to the generation of DP thymocytes, but thymocytes cannot undergo positive selection. This partial rescue of T-cell development in ScanT mice by transgenic Bcl2 expression as well as p53-deficiency was observed as a partial increase of thymic cellularity (Figure 6d).
The T-cell developmental phenotypes of ScanT cells, when protected from apoptosis, are highly reminiscent of Rag-deficient thymocytes, in which absence of a rearranged TCRβ leads to a developmental block at the DN3 stage, which can be bypassed by p53-deficiency but not by transgenic Bcl2 expression (27, 28). Therefore, we postulated that Zbtb1 might be required for TCRβ rearrangements and absence of a rearranged TCR in ScanT thymocytes may be responsible for these phenotypes. To evaluate this, we measured intracellular TCRβ levels which can be detected in cells that underwent productive TCRβ rearrangements at the DN3 stage. Surprisingly, we observed that the few DP thymocytes that are generated in ScanT mice express a rearranged TCRβ as well as ScanT p53−/− DP thymocytes (Figure 6c), indicating that complete lack of TCR rearrangements is not the underlying cause of the developmental block.
Transgenic Bcl2 expression and p53 deficiency rescue the generation of Myeloid and B-cells but not the generation of T-cells from ScanT progenitors in bone marrow chimeras
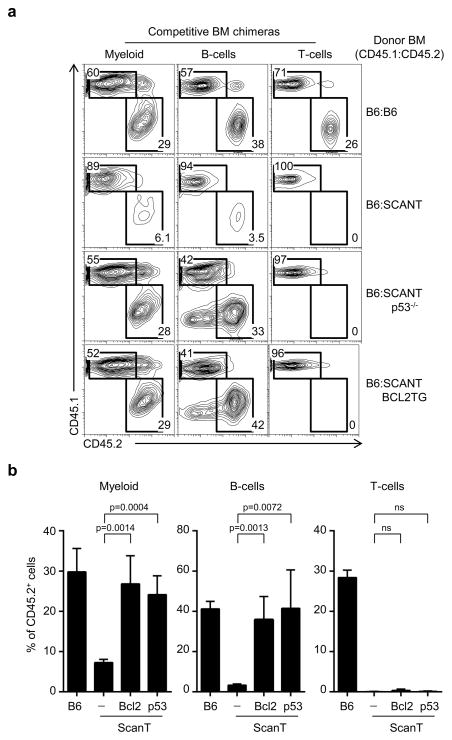
As the defect in the generation of myeloid cells from ScanT progenitors was observed only in competitive bone marrow chimeras, we set up to analyze if transgenic Bcl2 and p53-deficiency could rescue myeloid, as well as B and T-cell development from ScanT progenitors in competitive bone marrow chimeras with wild type cells as performed in Figure 1a. We observed that both the development of myeloid and B-cells from ScanT progenitors were significantly recovered by transgenic Bcl2 and p53-deficiency. As expected, however, T-cell development was not recovered (Figure 7a and b).
Figure 7. Transgenic Bcl2 expression and p53 deficiency rescue the generation of Myeloid and B-cells from ScanT progenitors in hematopoietic bone marrow chimeras.
a) FACS analysis of mixed bone marrow chimeras generated with the indicated mixture of bone marrow donor cells. Donor chimerism is identified in myeloid cells (CD11b+), B-cells (CD19+) and T-cells (TCRβhi), in spleen. The numbers represent the proportion of cells within the indicated gates. A representative plot out of four per condition is shown. b) Quantitation of the results obtained in (a), (Mean ± SD; n=4). p values corresponding to the significance using T-test are shown.
Altogether, we have identified that Zbtb1 is expressed in hematopoietic stem cells and immune progenitors before commitment to lymphoid or myeloid lineages occur, and at this stage, it prevents the generation of DNA damage and p53-mediated apoptosis by allowing the activation of the S-phase checkpoint response. Interestingly, although B-cell and myeloid development from ScanT progenitors was rescued by protection from apoptosis and p53-deficiency, abrogating the p53 pathway only rescued the earlier but not the later stages of T-cell development, indicating that p53-mediated apoptosis is not the only mechanisms by which Zbtb1 controls T-cell development.
DISCUSSION
Zbtb1 is required for lymphoid development, but its mechanism of action has not been elucidated (14, 15). We report here that the main function of Zbtb1 is to maintain genome stability in immune progenitors without which cells undergo increased DNA damage and p53-mediated apoptosis during replication and differentiation.
Two isoforms of Zbtb1 were described a 644aa and a 713aa form. The 713aa form is highly expressed in hematopoietic cells including HSC and MPP progenitors in bone marrow, T cells and B-cells. The expression pattern of the 713aa form of Zbtb1 was shown to be regulated during B and T-cell development. Interestingly, although Zbtb1 expression was shown to be high in immature B-cells and downregulated during B-cell development, the pattern of expression seemed different during T-cell development and it was reported to be upregulated in DP thymocytes (15). As previously reported, we observed an increase of relative Zbtb1 expression levels in DP thymocytes compared to DN thymocytes by RT-PCR (data not shown), however analysis of Zbtb1 expression using our reporter Zbtb1-GFP mice didn’t show this pattern and Zbtb1 levels were downregulated during T-cell development. This apparent discrepancy could be explained by differences in the housekeeping gene expression (HPRT) between DN and DP subsets. In agreement with the reported expression of Zbtb1, we identified high expression of Zbtb1 in HSCs and immune progenitors.
Replication stress occurs during normal proliferation due to either base modifications, DNA adducts or absence of other factors, which lead to stalling of the replication fork. The continuous action of helicases at stalled forks generates single stranded DNA, leading to the recruitment and activation of the ATR kinase, which in turn phosphorylates the S-phase checkpoint kinase Chk1. Phosphorylated Chk1 stimulates the degradation of Cdc25a, leading to a reversible block of DNA synthesis to allow repair of the DNA lesion before mitosis (17, 18). Once DNA synthesis stops, several mechanisms exist to repair the DNA lesion, one of them is called translesion DNA synthesis (TLS) and consists on the recruitment of low fidelity DNA polymerases that can bypass the lesion by incorporating either matched or mismatched nucleotides (19). Zbtb1 was shown to interact with Kap1, leading to relaxation of the DNA after replication stress, to the recruitment of Rad18 to the lesion and initiation of TLS. Knock-down of Zbtb1 in Hela cells leads to deficient recruitment of TLS polymerases in response to UV irradiation. However, as normal Chk1 phosphorylation was observed in these experiments, it was postulated that Zbtb1 exerts a specific function in TLS but not in initiation of the S-phase checkpoint (16). Our data differs from this previous report in that we have observed severely impaired Chk1 phosphorylation in ScanT primary cells after UV irradiation, which led to increased sensitivity to replication stress, chromosomal aberrations and p53-mediated cell death, suggesting that Zbtb1-deficiency has a more profound effect in preventing DNA damage than previously described.
A possible explanation for this discrepancy is that Zbtb1 may play specific DNA damage functions in immune cells. Alternatively, as the ATR-Chk1 pathway is essential for the normal proliferation of cells, cells with defects in this pathway would disappear from the culture in cell lines, and therefore, defects in this pathway would have not been observed in the reported manuscript (16).
Impaired Chk1 phosphorylation in ScanT cells led to increased sensitivity to replication stress and to the induction of chromosomal aberrations when the Chk1 pathway was partially inhibited. Increased DNA damage in ScanT cells in response to replication stress was also observed as increased phosphorylation of H2AX (γH2AX) and Kap1, which are downstream targets of ATM and markers of DNA damage (29–31), indicating that deficient activation of the ATR-Chk1 pathway leads to increased double strand breaks (DSB) in ScanT cells.
As activation of the ATR-Chk1 pathway is required for the survival of all replicating cells, it is intriguing that the ScanT phenotype is highly restricted to the immune lineage. Furthermore, we have observed that disruption of Zbtb1 doesn’t affect the reconstitution of HSCs and immune progenitors in competitive chimeras while the generation of mature immune cells is highly affected. Although ScanT immune progenitors can be generated in vivo in bone marrow chimeras, they become apoptotic when stimulated to proliferate in culture. A possible explanation for these observations is that compensatory mechanisms exist in hematopoietic stem cells that allow their efficient expansion and repopulation of immune progenitors in vivo and that the described function of Zbtb1 is restricted to lineage committed immune progenitors. Self-renewal of HSCs occurs in the hypoxic environment of the bone marrow and it is driven by anaerobic glycolysis, leading to a different metabolic profile from that of committed progenitors (32). It is possible that Zbtb1 effects are evidenced in committed progenitors due to different metabolic requirements. Alternatively, it is possible that the proliferative burst of differentiating immune progenitors is of higher magnitude than that of repopulating HSCs and cells are therefore more sensitive to Zbtb1 deficiency.
Our data supports a requirement of Zbtb1 for the expansion of immune progenitors in culture as we were not able to generate cell lines from ScanT immune progenitors, strongly suggesting that Zbtb1 functions are not restricted to a specific immune lineage. We have observed that mature B-cells are also susceptible to Zbtb1 deficiency when stimulated to proliferate in vitro. ScanT mice were previously shown to have reduced number of immature B-cells in bone marrow (14), and Zbtb1-deficient mice showed a proportional reduction of pre-B cells (15). Our analysis of B-cell development indicated that although we detected reduced numbers of pro-B and pre-B cells in ScanT mice, a severe developmental deficiency is observed in immature B-cells after the pre-B cell stage in correlation with the previous report (14), and this deficiency was reverted by transgenic Bcl2 expression. As previously reported (14), we have observed an increase of Marginal Zone B-cells in ScanT mice, however, this increase was not reverted by transgenic Bcl2 expression, strongly suggesting that Zbtb1 may also control the differentiation of mature B cells into Follicular B and Marginal Zone B-cells. We have also observed that T-cell deficiency of ScanT mice was correlated with a severe reduction of ETP, DN2 and DN3 thymocytes, which are generated after extensive proliferation of few thymic seeding progenitors (33).
Based on the inability of Bim-deficiency, which rescues TCR-signaled apoptosis of self-reactive thymocytes (34), to rescue the generation of T-cells in ScanT mice (14), it was postulated that the requirement of Zbtb1 for T-cell development is not due to prevention or protection from apoptosis. Contrary to this postulate, our data shows that increased p53 levels and p53-mediated apoptosis in ScanT cells are responsible for disrupted lymphoid, myeloid and T-cell development of ScanT progenitors. Protection from apoptosis by transgenic expression of Bcl2 or p53-deficiency rescued B-cell development, myeloid development in competitive chimeras, and rescued the early but not late stages of T-cell development. Although ETP, DN2a and DN3 thymocytes were recovered, T-cell development remained arrested at the DN3 developmental stage by expression of transgenic Bcl2.
Why is Zbtb1 more profoundly required for T-cell than for B-cell development?. One possible explanation is that the proliferative burst that occurs after preTCR signal would lead to extensive DNA damage in ScanT cells and to a mitotic catastrophe that cannot be rescued by protecting cells from apoptosis (35, 36). Another possible mechanism would be that Zbtb1 plays additional functions during T-cell development that are not required during B-cell development. Although the block in T-cell development that we observed at the preTCR and TCR checkpoint in compound ScanT x Vav-Bcl2 and ScanT p53−/− mice respectively could be explained by deficient TCR rearrangements, we observed that ScanT p53−/− thymocytes presented intracellular TCRβ. Although this would suggest that deficient TCR rearrangements in ScanT mice are not the main mechanism of the developmental block, it is also possible that Zbtb1 controls TCRβ rearrangements and that DN3 thymocytes that have rearranged TCRβ actively proliferated, leading to an accumulation of DP thymocytes. A requirement of Zbtb1 for productive TCRα rearrangements could be the underlying cause of the DP developmental block that we observed in ScanT p53−/− mice and deserves further characterization.
Altogether, we have identified a novel DNA damage function of Zbtb1 in controlling the activation of the ATR-Chk1 pathway in response to replication stress. This function of Zbtb1 was essential to maintain genome integrity during lymphoid development, preventing DNA damage, activation of p53 and apoptosis. We have further identified a specific requirement of Zbtb1 at the preTCR and TCR checkpoints during T-cell development, which may be independent of the DNA damage function described here and explains the severe T-cell defects in Zbtb1-mutant and -deficient mice.
Supplementary Material
Acknowledgments
We thank Dr. Bruce Beutler for providing the ScanT strain. Kangxin Jin and Dr. Derek Sant’Angelo for kindly providing us the Zbtb1-GFP reporter mice. Dr. Zuniga Pflucker for providing the OP9-DL1 cell line. Dr. Andre Nussenzweig for experimental advice in relation to DNA damage. Dr. Hyun Park, Dr. Remy Bosselut, Dr, Avinash Bhandola and Dr. Nussenzweig for reviewing a draft of the manuscript. Dr M. Kruhlak for microscopic analysis. Dr. S. Sharrow, L. Granger and T. Adams for flow cytometry and cell sorting.
Footnotes
This work was funded by NIH intramural funds (1ZIABC011429).
References
- 1.Kosan C, Saba I, Godmann M, Herold S, Herkert B, Eilers M, Moroy T. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 2010;33:917–928. doi: 10.1016/j.immuni.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 2.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, Teruya-Feldstein J, Cattoretti G, Pandolfi PP. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeda T, Ito K, Merghoub T, Poliseno L, Hobbs RM, Wang G, Dong L, Maeda M, Dore LC, Zelent A, Luzzatto L, Teruya-Feldstein J, Weiss MJ, Pandolfi PP. LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Developmental cell. 2009;17:527–540. doi: 10.1016/j.devcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He X, V, Dave P, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 5.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Ichii H, Sakamoto A, Kuroda Y, Tokuhisa T. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J Immunol. 2004;173:883–891. doi: 10.4049/jimmunol.173.2.883. [DOI] [PubMed] [Google Scholar]
- 9.Ichii H, Sakamoto A, Hatano M, Okada S, Toyama H, Taki S, Arima M, Kuroda Y, Tokuhisa T. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nature immunology. 2002;3:558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 10.Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, Sant’Angelo DB. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J Immunol. 2010;184:6746–6755. doi: 10.4049/jimmunol.1000776. [DOI] [PubMed] [Google Scholar]
- 11.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant’Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature immunology. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Y, Cao X, Zhang X, Kovalovsky D. PLZF Controls the Development of Fetal-Derived IL-17+Vgamma6+ gammadelta T Cells. J Immunol. 2015;195:4273–4281. doi: 10.4049/jimmunol.1500939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siggs OM, Li X, Xia Y, Beutler B. ZBTB1 is a determinant of lymphoid development. The Journal of experimental medicine. 2012;209:19–27. doi: 10.1084/jem.20112084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Punwani D, Simon K, Choi Y, Dutra A, Gonzalez-Espinosa D, Pak E, Naradikian M, Song CH, Zhang J, Bodine DM, Puck JM. Transcription factor zinc finger and BTB domain 1 is essential for lymphocyte development. J Immunol. 2012;189:1253–1264. doi: 10.4049/jimmunol.1200623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H, Dejsuphong D, Adelmant G, Ceccaldi R, Yang K, Marto JA, D’Andrea AD. Transcriptional repressor ZBTB1 promotes chromatin remodeling and translesion DNA synthesis. Molecular cell. 2014;54:107–118. doi: 10.1016/j.molcel.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez Besteiro MA, Gottifredi V. The fork and the kinase: a DNA replication tale from a CHK1 perspective. Mutation research Reviews in mutation research. 2015;763:168–180. doi: 10.1016/j.mrrev.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes & development. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harbor perspectives in biology. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rass U. Resolving branched DNA intermediates with structure-specific nucleases during replication in eukaryotes. Chromosoma. 2013;122:499–515. doi: 10.1007/s00412-013-0431-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogilvy S, Metcalf D, Print CG, Bath ML, Harris AW, Adams JM. Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14943–14948. doi: 10.1073/pnas.96.26.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nature biotechnology. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Godec J, Ben-Aissa K, Cui K, Zhao K, Pucsek AB, Lee YK, Weaver CT, Yagi R, Lazarevic V. The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-gamma-producing T helper 17 cells. Immunity. 2014;40:355–366. doi: 10.1016/j.immuni.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitajima K, Minehata K, Sakimura K, Nakano T, Hara T. In vitro generation of HSC-like cells from murine ESCs/iPSCs by enforced expression of LIM-homeobox transcription factor Lhx2. Blood. 2011;117:3748–3758. doi: 10.1182/blood-2010-07-298596. [DOI] [PubMed] [Google Scholar]
- 25.Gagou ME, Zuazua-Villar P, Meuth M. Enhanced H2AX phosphorylation, DNA replication fork arrest, and cell death in the absence of Chk1. Molecular biology of the cell. 2010;21:739–752. doi: 10.1091/mbc.E09-07-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jardim MJ, Wang Q, Furumai R, Wakeman T, Goodman BK, Wang XF. Reduced ATR or Chk1 expression leads to chromosome instability and chemosensitization of mismatch repair-deficient colorectal cancer cells. Molecular biology of the cell. 2009;20:3801–3809. doi: 10.1091/mbc.E09-04-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang D, Lenardo MJ, Zuniga-Pflucker JC. p53 prevents maturation to the CD4+CD8+ stage of thymocyte differentiation in the absence of T cell receptor rearrangement. The Journal of experimental medicine. 1996;183:1923–1928. doi: 10.1084/jem.183.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maraskovsky E, O’Reilly LA, Teepe M, Corcoran LM, Peschon JJ, Strasser A. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1−/− mice. Cell. 1997;89:1011–1019. doi: 10.1016/s0092-8674(00)80289-5. [DOI] [PubMed] [Google Scholar]
- 29.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. The Journal of biological chemistry. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 30.White D, I, Rafalska-Metcalf U, Ivanov AV, Corsinotti A, Peng H, Lee SC, Trono D, Janicki SM, Rauscher FJ., 3rd The ATM substrate KAP1 controls DNA repair in heterochromatin: regulation by HP1 proteins and serine 473/824 phosphorylation. Molecular cancer research: MCR. 2012;10:401–414. doi: 10.1158/1541-7786.MCR-11-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nature cell biology. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 32.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nature reviews Molecular cell biology. 2014;15:243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciofani M, Zuniga-Pflucker JC. The thymus as an inductive site for T lymphopoiesis. Annu Rev Cell Dev Biol. 2007;23:463–493. doi: 10.1146/annurev.cellbio.23.090506.123547. [DOI] [PubMed] [Google Scholar]
- 34.Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, Pellegrini M, Cory S, Adams JM, Strasser A. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 2002;415:922–926. doi: 10.1038/415922a. [DOI] [PubMed] [Google Scholar]
- 35.Eykelenboom JK, Harte EC, Canavan L, Pastor-Peidro A, Calvo-Asensio I, Llorens-Agost M, Lowndes NF. ATR activates the S-M checkpoint during unperturbed growth to ensure sufficient replication prior to mitotic onset. Cell reports. 2013;5:1095–1107. doi: 10.1016/j.celrep.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 36.Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, Lin L, Hoog J, Goiffon RJ, Prat A, Aft RL, Ellis MJ, Piwnica-Worms H. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. The Journal of clinical investigation. 2012;122:1541–1552. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.