Abstract
Staphylococcus aureus is both a commensal and a pathogen, and USA300, a strain that is usually methicillin-resistant S. aureus (MRSA) but can sometimes be methicillin-susceptible (MSSA), has been causing skin and soft tissue infections (SSTIs) in epidemic proportions among otherwise healthy individuals. Although many people are colonized with S. aureus strains, including some with USA300, few of these colonized individuals develop SSTIs. This prompts the hypothesis that infections may develop in individuals with somewhat reduced innate and/or adaptive immune responses to S. aureus, either because prior S. aureus colonization has dampened such responses selectively, or because of more globally reduced immune reactivity. Here, we analyzed the S. aureus colonization status and PBMC responses to innate and adaptive stimuli in 72 patients with SSTIs and 143 uninfected demographically matched controls. Contrary to the hypothesis formulated, PBMCs from infected patients obtained at the time of infection displayed enhanced innate cytokine production upon restimulation compared with PBMCs from controls, a difference that disappeared after infection resolution. Notably, PBMCs from patients infected with a documented USA300 SSTI displayed greater innate cytokine production than those from patients infected with documented non-USA300 genotypes. Moreover, colonization with USA300 in infected patients, regardless of their infecting strain, correlated with increased production of IL-10, IL-17A and IL-22 compared with patients colonized with non-USA300 subtypes. Thus, our results demonstrate that infected patients associated with USA300 either as an infecting, or as a colonizing strain, have systemic immune responses of greater magnitude than those associated with other S. aureus subtypes.
Introduction
The Gram-positive bacterium S. aureus is a frequent commensal of the human skin and mucosal areas, but it is also the most commonly isolated human bacterial pathogen and an important cause of SSTIs (1). Since the mid-1990s, there has been a sharp increase in the incidence of S. aureus SSTIs in community-dwelling populations in the absence of frequent exposure to the healthcare system. This increase has coincided with the identification of new, virulent S. aureus strain types that are usually resistant to nearly all β-lactam antibiotics (methicillin-resistant S. aureus, MRSA). Such community-associated (CA)-MRSA strains can be distinguished molecularly from health care-associated strains and appear to cause infections in otherwise healthy individuals. Since 2001, in the US, USA300 MRSA has been the dominant strain causing infection in the community, although methicillin-susceptible S. aureus (MSSA) strains sharing many genetic characteristics of USA300 also exist (2, 3).
It is often cited that 20% of the human population is persistently colonized with S. aureus (4), although the prevalence is likely higher (5). Aside from patients with immune genetic defects known to be associated with recurrent S. aureus infections (6), it is not clear why some seemingly healthy individuals develop S. aureus SSTIs and others do not. One possibility is that individuals with slightly lower immune responses (towards the left of the Gaussian distribution of normal immune responses) are the ones who develop SSTIs. Indeed, polymorphisms in the cytokine genes IL6, TNF, IL10, IL17A and IFNG, as well as in the innate immune modulating gene TLR10, have been linked to susceptibility to complicated SSTIs, though how these polymorphisms affect cytokine levels has not been resolved (7, 8). An alternative possibility is that colonization with S. aureus may tolerize the immune system to S. aureus antigens or microbial products, as such predisposing an individual to subsequent infections by any S. aureus strain. This is conceivable as the superantigen properties of S. aureus can result in deletion or anergy of an entire Vβ family of T cells (9), and desensitization of pattern-recognition receptors can occur in innate cells following exposure to certain microbial products (10).
S. aureus infections can activate both innate and adaptive immune cells. The cell surface Toll-like receptor 2 (TLR2) and the cytosolic nucleotide-binding oligodimerization domain 2 (NOD2) are the main microbial molecular pattern-recognition receptors known to signal immune cells during S. aureus infection (11). Activated innate cells produce inflammatory cytokines, including IL-6, that promote production of C-reactive protein (CRP), a primitive opsonin and an activator of complement (12). An association between the presence of neutralizing anti-IL-6 antibodies in the serum, low CRP and recurrent S. aureus infection has been reported (13), emphasizing the importance of this cytokine in anti-S. aureus immunity. T cell responses have received less attention, but emerging data support their role in protection against S. aureus infections. For instance, T cells, but not antibodies, were shown to mediate protection against S. aureus lethality, following vaccination in mice (14, 15). A link between T cell-IL-17 and IL-22 and protection against SSTIs caused by S. aureus has emerged both in rodents and humans (16–19) and our group has shown that Th17 cells can confer partial protection that limits the size of dermonecrotic lesions upon secondary skin infection in mice (20). Notably, patients with mutations in the STAT3 gene that signals downstream of IL-6, develop recurrent S. aureus infections of the skin and lung, but not systemic infections (21–26). Because IL-6 is important for Th17 differentiation, these patients have an absence of S. aureus-mediated IL-17 production by T cells. These observations suggest that reduced systemic Th17 responses may result in increased susceptibility to local SSTIs. By extension to otherwise healthy individuals, these reports suggest that evaluating systemic immunity to S. aureus by analyzing PBMCs may reveal some causes of susceptibility to localized skin infections.
In this study, we set out to address specific questions related to the susceptibility to S. aureus SSTIs: 1) Do people who develop SSTIs have weaker than average immune responses? 2) Does colonization with S. aureus result in dampened innate or adaptive reactivity to S. aureus restimulation? 3) Are immune responses to a USA300 infection of different magnitude or quality than immune responses to other S. aureus genotypes, explaining the increase in this specific infection? To address these questions, we took advantage of an ongoing multicenter clinical trial comparing different treatment modalities in patients presenting to the emergency department (ED) with an SSTI, in the absence of co-morbidities (27, 28). Blood was collected from patients with skin infections, and from control individuals presenting to the ED for minor non-infectious causes, and production of cytokines was compared in both serum and stimulated PBMCs. In parallel, skin swabs from 3 anatomic sites in SSTI patients (at sites distinct from the infection) and uninfected controls were cultured to identify S. aureus colonization. The infecting SSTI S. aureus isolate also underwent genotypic characterization. Our results document the systemic immune response that takes place following SSTIs, reveal stronger immune responses to USA300 than non-USA300 strain types, and argue against systemic immunosuppressive effects by colonizing S. aureus strains.
Materials and Methods
Patients and samples
The patients enrolled were otherwise healthy adults who presented to the ED at the University of Chicago Medicine with a skin infection between September 2010 and September 2012. Exclusion criteria included human or animal bite at the infection site, fever (oral temperature >38.5°C), receipt of immunosuppressive medications, presence of a co-morbid condition such as diabetes or chronic renal failure, morbid obesity (BMI >40 kg/m2), or antibacterial therapy with anti-staphylococcal activity within the prior 14 days. All inclusion/exclusion criteria were as described previously (27, 28). Patients were randomized to receive either trimethoprim/sulfamethoxazole, or clindamycin (or a placebo in cases of a single abscess <5cm in diameter). Surgical incision and drainage were performed if an abscess was present, and purulent material was cultured. Blood was drawn on the day of the infected patients’ first visit (day 0, D0) and 40 days (D40) after enrollment.
Uninfected controls enrolled were adults presenting to the ED during the same time period with minor, non-infectious complaints without co-morbidities. Controls included individuals of the same age, gender and self-reported race as the patients with an SSTI. Blood was drawn only when the control patient was identified (D0).
At enrollment of SSTI patients and controls, informed consent was obtained. Each consenting subject was cultured at nasal, oropharyngeal, and perianal/inguinal sites using separate dry rayon-tipped applicators (CultureSwab, BD Diagnostic Systems). These cultures were repeated in infected patients at the follow up visit on D40. The protocol was approved by the Institutional Review Board of The University of Chicago.
Colonization swab culture conditions and molecular diagnosis of colonization and infection isolates
Culture swabs were transported to the University of Chicago MRSA Research Laboratory and were incubated in trypticase soy broth with 7% sodium chloride overnight at 37° C. An aliquot of the broth was then spread onto BBL CHROMagar S. aureus media (BD Diagnostic Systems) and incubated for 24 hours at 37° C. One mauve colony was picked from the plate and confirmed to be S. aureus by typical colony morphology, positive catalase and agglutination with the StaphAureux test (Remel, Thermo Scientific, US). S. aureus isolates were stored at −70° C in skim milk (Difco) until further evaluation.
Genomic DNA was extracted from each isolate using the Qiagen DNeasy Blood and Tissue Kit following manufacturer’s instructions and modified by incubation with lysostaphin in resuspension buffer (at 37°C for at least 30 minutes) to facilitate S. aureus lysis (29). S. aureus speciation was confirmed using a polymerase chain reaction (PCR) assay specific for spa (encoding protein A). S. aureus isolates were characterized by multilocus sequence typing (MLST) (30) to determine the genetic background and by typing of the SCCmec element, the mobile genetic element that carries mecA (31) with type assignments using published guidelines (32). Detection of the genes encoding the Panton-Valentine leukocidin (PVL) was performed as described (33). Two S. aureus isolates were considered indistinguishable if they shared a MLST and SCCmec type and were concordant with respect to the presence or absence of the PVL genetic determinants. Based on a previous investigation demonstrating that the presence of both the PVL genes (luk-S-PV and luk-F-PV) and the arcA gene found on ACME is highly concordant with USA300 MRSA genetic background assessed by pulsed field gel electrophoresis (34), isolates with these characteristics were considered to be USA300 MRSA (Table I). USA300 isolates that lacked mecA and an SCCmec element were termed USA300 MSSA.
Table I.
Algorithm for classification of S. aureus strains.
| Strain type | mecA | PVL | arcA | ST | SCCmec type | ||
|---|---|---|---|---|---|---|---|
| USA300 | MRSA | USA300 | + | + | + | Not done | Not Done |
| probable USA300 |
+ | + | − | 8 | IV | ||
| probable USA300 |
+ | − | + | 8 | IV | ||
| MSSA | probable USA300 |
− | + | − | 8 | NA | |
| probable USA300 |
− | + | + | 8 | NA | ||
| Anything other than the above is not USA300 | |||||||
| non USA300 | MRSA | + | − | − | 8 | IV | |
| + | +/− | +/− | 8 | Not SCCmec IV | |||
| + | − | − | NOT ST8 | any | |||
| + | + | − | NOT ST8 | any | |||
| + | − | + | NOT ST8 | any | |||
| MSSA | − | +/− | +/− | NOT ST8 | NA | ||
Preparation and Isolation of serum, peripheral blood mononuclear cells (PBMC) and cytokine assays
Serum was prepared from about 3 ml of non-heparinized blood by centrifuging the tube at 3,000 RPM for 10 min at room temperature. The serum was collected and aliquots frozen at −20°C. Cytokine levels of IL12p70, TNF, IL-1β, IL-6, IL-23, IL-10, IFNγ, IL-2, IL-17, IL-17F and IL-22 in serum were measured by Multiplex Beads assay (EMD Millipore, Billerica. MA, USA).
About 12 ml of blood containing sodium heparin anticoagulant were collected from all patients and uninfected controls. PBMCs from whole blood were isolated following Ficoll gradient centrifugation to eliminate red blood cells. These fresh PBMCs (106 cells/ml/well) were cultured in 48-well plates in a final volume of 1 ml, either unstimulated or stimulated with anti-CD3 mAb (OKT3, 5 µg/ml), the TLR2 agonist Pam3CSK4 (1 µg/ml), or lysate from sonicated USA300 MRSA strain 923 (50 µl/well) for 24 hours. PBMCs from frozen aliquots from a single healthy donor were used in every plate as a positive control to verify the quality of the stimulation. The supernatants were collected into Eppendorf tubes and frozen in aliquots at −80°C. The cytokine concentration of TNF, IL-1β, IL-6, IL-23, IL-10, IFN-γ, IL-2, IL-17 and IL-22 was determined by Elisa (R&D System Inc, Minneapolis, MN) according to the manufacturer’s instructions, or, for IL-17A following USA300 MRSA sonicate restimulation, by microfluidic ELISA (Siloam Biosciences, Cincinnati, OH).
Data analysis
The numbers of infected patients and uninfected controls to enroll in the study was determined by a sample size calculation to obtain >80% power (with p<0.05) in detecting a predetermined difference in the IL-17:IFN-γ ratio between cases (median 0.25, interquartile range [IQR] 0.12–0.050) and controls (median 0.43, IQR 0.16–1.02) based on the distribution of cytokine production observed in preliminary results. The sample size was calculated forcing a ratio of cases to controls of 1:2 in order to increase power with a smaller number of cases.
Statistical analyses on cytokines from PBMCs were performed using the non-parametric Mann-Whitney test and GraphPadPrism 6.0 software. Analyses of serum cytokines were performed using Wilcoxon rank-sum test, or Wilcoxon rank-sum test for matched pairs when appropriate. The Chi-square test or the McNemar test for paired data was used to compare categorical data. A p-value <0.05 was considered to be statistically significant.
Results
Patient demographics, colonization and infection status
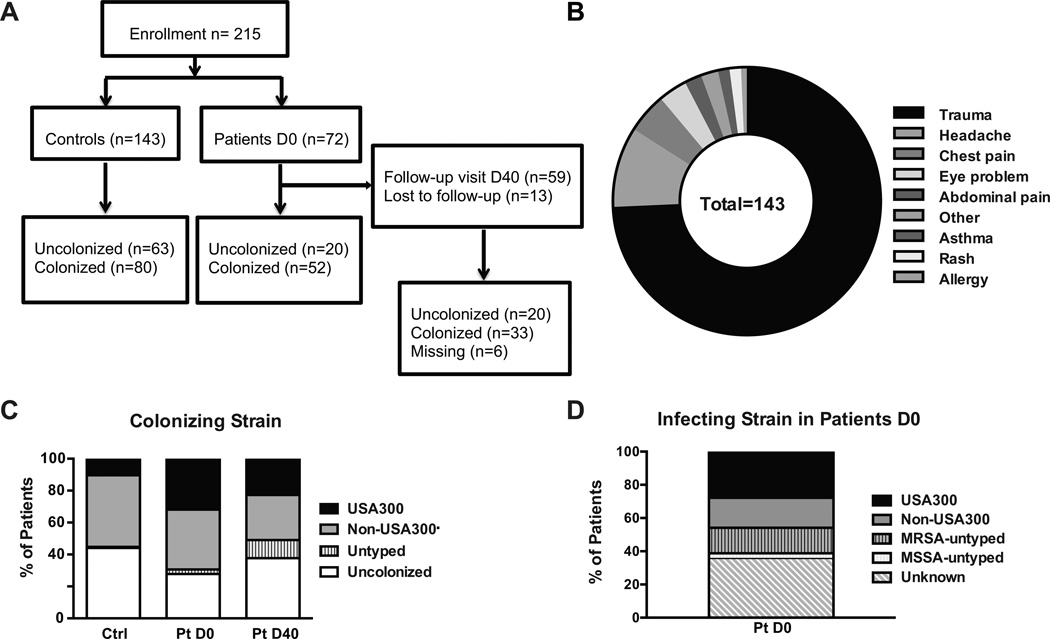
Overall, 215 adults were enrolled, including 72 patients with an SSTI and 143 controls (Figure 1A). The majority of uninfected controls presented to the ED following minor traumatic events (Figure 1B). All patients were treated with surgical incision and drainage unless they had non-purulent cellulitis. Patients were randomized to 1 of 2 antibiotics, clindamycin or trimethoprim/sulfamethoxazole, in patients with multiple or large lesions, or to these antibiotics or a placebo if the abscess was small. Patients and controls had a similar distribution of gender, age and self-reported race (Table II). The majority of individuals were African Americans, reflecting the population served by the ED at the University of Chicago, and younger than 40 years of age. Nasal, oropharyngeal and perianal/inguinal cultures were obtained to establish the colonization status of infected patients at D0 and D40 and uninfected controls at D0, and colonization was considered positive if at least 1 of the sites tested positive. Colonization was detected in similar proportions at all 3 body sites. Individuals were classified as uncolonized (with S. aureus) or as colonized with USA300 versus non-USA300 S. aureus strain types. Isolates not genotyped were designated untyped (Figure 1C). Fifty six% of controls and 72% of patients on D0 were colonized with S. aureus, and within those, 19% of controls and 46% of SSTI patients were colonized with USA300 (Figure 1C). Analysis of colonization in patients on D40 revealed a decrease in colonization prevalence that was not statistically significant (using the Chi-square test) following antibiotic treatment, with 50% of patients colonized with S. aureus, among which 44% were USA300. Among colonized individuals, 52.6%, 51.9% and 39.4% were positive at more than 1 site in controls and infected patients D0 and D40, respectively. Of note, some shifts in colonizing strains were apparent in infected patients between D0 and D40 (Table III) either because of the antibiotic treatment received by the patients or because shifts occur normally over time.
Figure 1. Enrollment, colonization and infection status.
A. Schematic representation of enrollment among controls and SSTI patients. B. Distribution of the chief complaint by uninfected controls presenting to the ED. C. Enrolled individuals were swabbed at 3 body sites and the colonization status and genotype of the S. aureus strain when at least 1 site was positive were determined at the first visit to the ED for SSTI patients and uninfected controls (D0) as well as on D40 for SSTI patients. D. Patients presenting with SSTI on D0 had their skin lesion incised and drained when possible and the obtained material sent to the laboratory for genotyping. Patients whose samples grew S. aureus MRSA or MSSA in the clinical microbiology laboratory but that were not genotyped are labeled as untyped. Patients whose samples were not obtained or whose culture was negative are labeled as unknown.
Table II.
Patient Demographics.
| Controls | Patients | |
|---|---|---|
| Total Numbers | 143 | 72 |
| Gender | ||
| Female | 56% | 54% |
| Male | 44% | 46% |
| Age | ||
| 18–25y | 30% | 38% |
| 26–40y | 41% | 33% |
| >41 | 29% | 29% |
| Race | ||
| African American | 92% | 97% |
| Caucasian | 7% | 3% |
| Asian | <1% | 0 |
Table III.
Colonizing S. aureus strains change over time.
| Patient Colonization D0 |
Patient Colonization D40 | ||||||
|---|---|---|---|---|---|---|---|
| Uncolonized | Colonized | N/D | |||||
| USA300/MRSA | Non- USA300/MRSA |
USA300/MSSA | Non- USA300/MSSA |
N/A | |||
| Uncolonized n = 20 |
10 | 1 | 3 | 6 | |||
| N/D (n=2) |
1 | 1 | |||||
| USA300/MRSA n = 18 |
1 | 4 | 1 | 3 | 2 | 7 | |
| USA300/MSSA n = 5 |
1 | 2 | 2 | ||||
| Non-USA300/MSSA n = 27 |
8 | 5 | 6 | 4 | 4 | ||
| Total = 72 | 20 | 9 | 2 | 3 | 13 | 6 | 19 |
N/A: unknown. N/D: not done
The infection isolate was identified in the clinical microbiology laboratory and later sent to the MRSA Research Laboratory for molecular typing and classification into USA300 or non-USA300 strain types. Eleven infection samples were identified as MRSA and 2 as MSSA by the clinical microbiology laboratory but were not typed by the MRSA Research Laboratory and were labeled as MRSA-untyped or MSSA-untyped in Figure 1D. Patients with non-purulent cellulitis that could not be drained (n=10), negative cultures and samples that did not undergo typing (n=16), were all combined into a category termed “unknown” for the infecting agent (Figure 1D). All positive cultures revealed S. aureus as the infecting agent, and 27.8% of SSTI patients were infected with documented USA300. Of interest, the infecting S. aureus strain was often different from the colonizing strain (Table IV), suggesting that distinct strains may colonize versus infect, or that by typing only 1 colony from each anatomic site, we missed detection of other colonizing strains that may have included the infecting strain.
Table IV.
Colonization status of SSTI patients with documented S. aureus infection.
| Infecting Strain | Colonization Status | |||||
|---|---|---|---|---|---|---|
| D0 | D40 | |||||
| Uncolonized | USA300 | Non- USA300 |
Uncolonized | USA300 | Non- USA300 |
|
| USA300 (n=20) | 7 | 8 | 5 | 8 | 5 | 2 |
| Non-USA300 (n=13) | 2 | 6 | 5 | 3 | 1 | 3 |
| Total | 9 | 14 | 10 | 11 | 6 | 5 |
Stimulated PBMCs from SSTI patients produced more innate but not adaptive cytokines
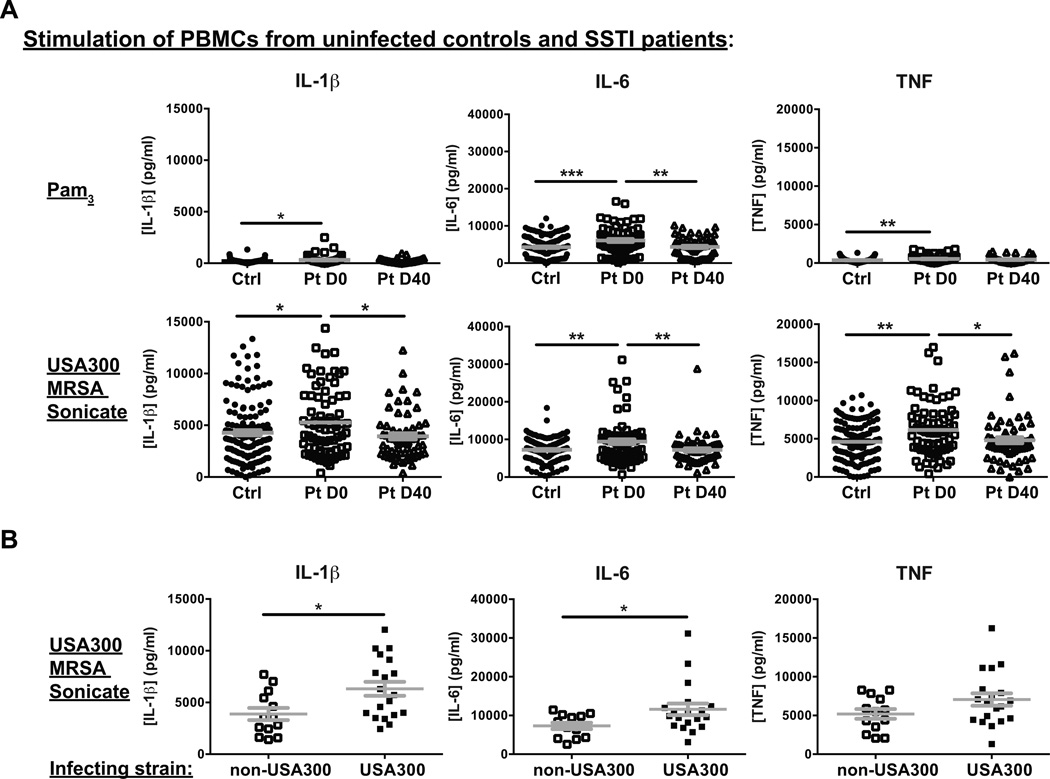
Otherwise healthy individuals who develop S. aureus SSTIs may be those with slightly lower global immune responses, which might fail to contain colonizing bacteria or emerging infections. To test this hypothesis, we compared the innate cytokines produced by PBMCs of SSTI patients during and after infection versus uninfected controls upon in vitro stimulation with the TLR2 agonist Pam3CysK4, or with a USA300 MRSA sonicate, to mimic signals elicited by S. aureus. In addition, we measured the adaptive cytokines produced following exposure to the pan-T cell activator anti-CD3 mAb, or to the USA300 MRSA sonicate. In contrast to our hypothesis, innate cytokine production by PBMCs obtained from SSTI patients on D0 was actually increased (Il-1β, IL-6 and TNF) when compared with that from control individuals, both following Pam3CysK4 and USA300 stimulation (Figure 2A). In most cases, cytokine levels were lower when the infection was resolved after treatment on D40, but not lower than from control PBMCs (Figure 2A). Of note, similar cytokine level decreases between D0 and D40 were observed in infected patients treated with trimetroprim/sulfamethoxazole or those treated with clindamycin (Supplemental Figure 1). These results support a systemic consequence of the localized infection on D0, and suggest that the SSTI results in either increased numbers of circulating cells that can respond to these stimuli, or in an increased capacity on a per cell basis to produce these cytokines.
Figure 2. Greater innate cytokines produced by stimulated PBMCs from SSTI patients than controls.
A. PBMCs from uninfected controls (Ctrl) and SSTI infected patients (Pt) were stimulated with Pam3CysK4 (Pam3, top panels) or with USA300 MRSA sonicates (bottom panels) for 24 hours. Supernatants were assayed for IL-1β, IL-6 and TNF by ELISA. Statistical significance between uninfected and infected D0 was calculated using the Chi-square test and between D0 and D40 using the McNemar test for paired data. B. Levels of cytokines induced by USA300 MRSA sonicates were compared between PBMCs from patients with documented non-USA300 infection and PBMCs from patients with documented USA300 infection. Plots represent Mean + SEM. Statistical significance was calculated using the Mann-Whitney test. * p<0.05, ** p<0.01, ***p<0.001.
To investigate whether USA300 infection elicited distinct responses compared with other S. aureus strain types, we compared the production of innate cytokines by PBMCs from patients infected with documented USA300 versus from patients infected with documented non-USA300. As shown in Figure 2B, PBMCs from patients with documented USA300 infection produced more IL-1β and IL-6 upon restimulation with S. aureus sonicates, suggestive of a more robust systemic immune response.
In contrast to the differences in innate cytokine production, no differences were observed in the adaptive cytokines (IL-2, IFN-γ, IL-17A, IL-22) detected following anti-CD3- or USA300 MRSA sonicate-mediated stimulation, either between SSTI patients and uninfected controls (Supplemental Figure 2A), or between SSTI patients infected with USA300 versus non-USA300 (Supplemental Figure 2B), although significantly higher IL-10 production was detected from USA300-infected versus non-USA300-infected patients upon S. aureus restimulation (Supplemental Figure 2B).
Together, these results argue against the hypotheses that infected patients express down-modulating polymorphisms that decrease their immune responsiveness to S. aureus strains, or that the infection dampens immune responses.
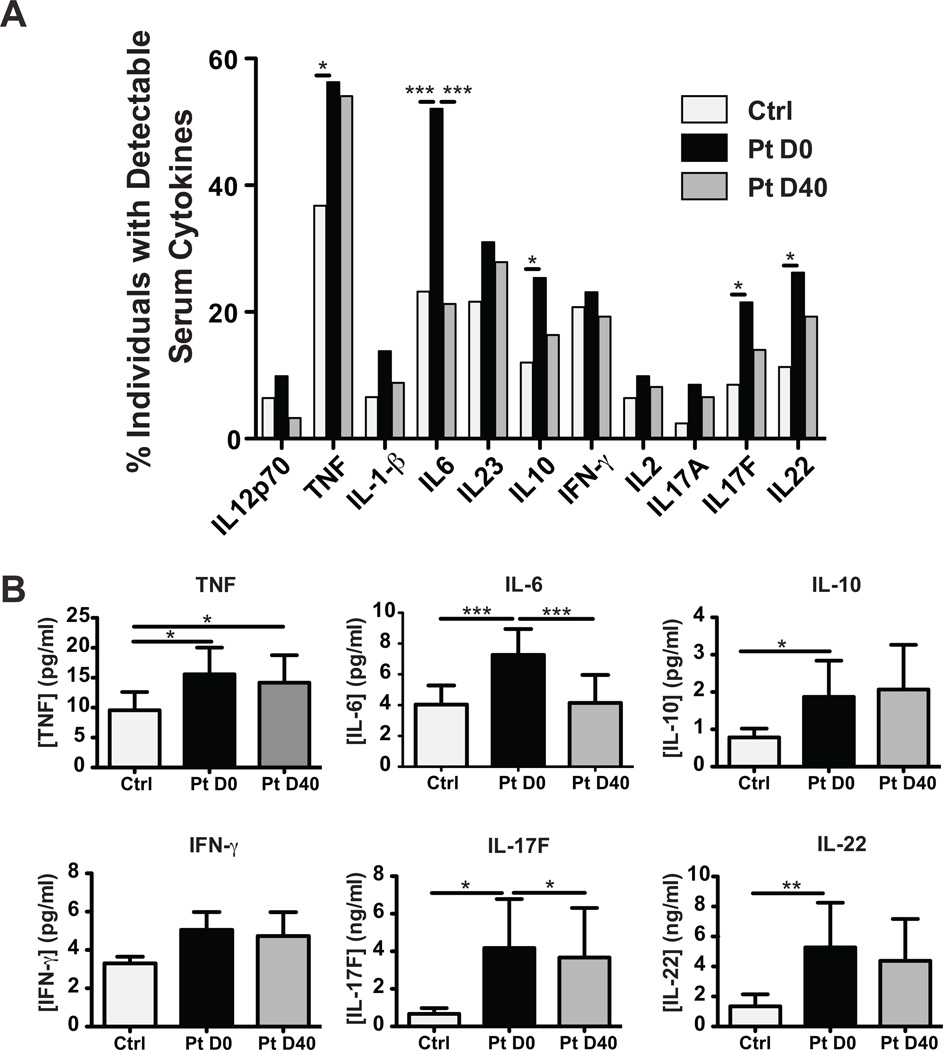
Serum cytokines were increased in infected patients
The increased production of innate cytokines by PBMCs from infected patients compared with uninfected controls suggested that a systemic immune response was taking place in response to the localized infection, at least in some patients. To further investigate possible systemic consequences of the infection, the concentration of serum cytokines was measured in uninfected controls and SSTI patients. A higher proportion of infected patients on D0 had detectable serum cytokines compared with uninfected controls (Figure 3A), and infected patients had significantly higher serum levels of TNF, IL-6, IL-10, IL-17F and IL-22 (Figure 3B). By D40, the serum IL-6 and IL-17F levels were significantly decreased compared with D0 (Figure 3B), and similarly so in patients treated with trimethoprim/sulfamethoxazole or clindamycin (Supplemental Figure 1 for IL-6 and data not shown for IL-17F). The detectable level of circulating cytokines was probably not due to systemic dissemination of the infection, as a criterion for enrollment was a body temperature below 38.5°C. In fact, all patients but 4 had a body temperature below 37°C (Supplemental Figure 3), and the 4 patients with a temperature between 37°C and 37.9°C did not have higher serum cytokine levels than the afebrile patients (data not shown). Of note SSTI patients infected with documented USA300 did not have greater serum cytokine levels than those infected with documented non-USA300 (Supplemental Figure 4), although the study was underpowered to analyze this parameter, as not all patients had measurable serum cytokines (Figure 3A).
Figure 3. Elevated serum cytokines in SSTI patients.
Serum was isolated from SSTI patients (Pt) and uninfected controls (Ctrl) and cytokine levels were measured by MultiPlex bead assay. A. Percentage of patients with detectable levels of given cytokines. B. Levels of select cytokines are shown (Mean + SEM). Statistical significance was calculated using the Two-Way ANOVA, * p<0.05, **p<0.01, ***p<0.001.
Impact of S. aureus colonization on immune responses
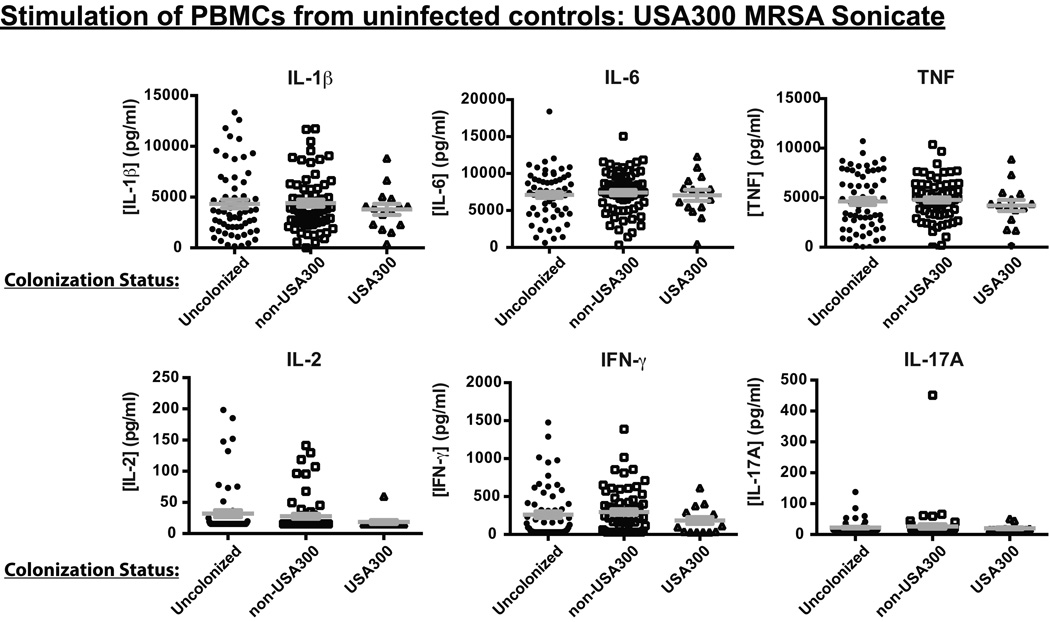
Our study design also allowed us to test the hypothesis that asymptomatic colonization in uninfected individuals may tolerize immune cells, such that immune responses to S. aureus may become blunted, and colonized individuals may thus become more susceptible to infection either by the colonizing strain itself or by a subsequent strain to which they may become exposed. To test this hypothesis, we compared levels of cytokines produced upon in vitro stimulation with USA300 MRSA sonicates by PBMCs from non-infected controls that were either not colonized at the time of ED visit, or were colonized by USA300 versus non-USA300 strain types. Neither innate nor adaptive cytokine production was significantly different in the colonized versus uncolonized control individuals (Figure 4), suggesting that colonization does not desensitize the innate immune response to S. aureus in these individuals and that a T cell repertoire capable of recognizing S. aureus antigens or superantigens is still present in the blood of colonized hosts. Colonization also did not impact responses to Pam3CysK4 or anti-CD3 restimulation, or production of other cytokines tested (data not shown).
Figure 4. Documented S. aureus colonization is not associated with reduced PBMC responses in uninfected subjects.
PBMCs from uninfected controls were stimulated with USA300 MRSA sonicates for 24h and cytokine levels in supernatants measured by ELISA. The plots compare the cytokines produced by uninfected controls classified as uncolonized versus those classified as colonized with USA300 and those classified as colonized with non-USA300 genotypes (Mean + SEM). Differences were not significant as determined by the Mann-Whitney test.
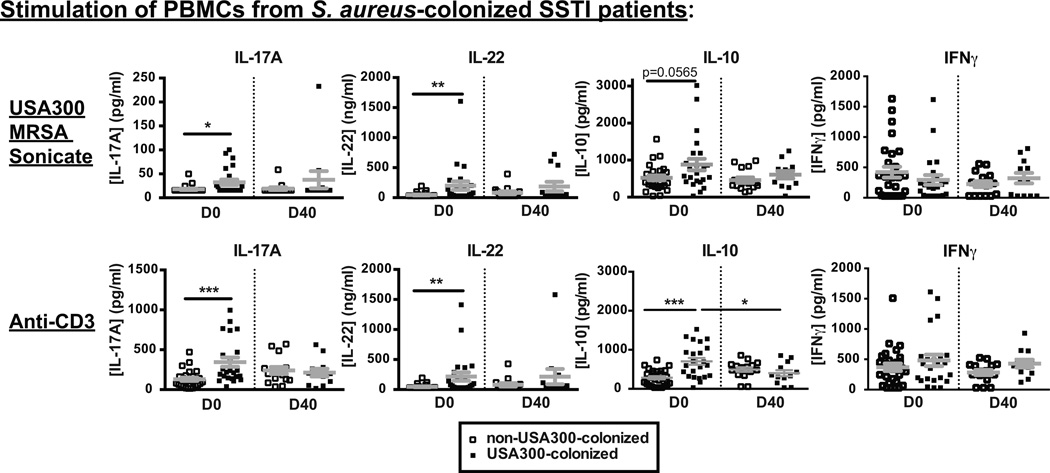
Finally, we tested whether the type of S. aureus colonization impacted the immune responses in infected patients. Surprisingly, irrespective of the infecting strain, PBMCs from infected patients who were also colonized with USA300 on D0 produced more IL-17A, IL-22 and IL-10, but not IFNγ (or IL-2, not shown) upon anti-CD3 or USA300 MRSA sonicate restimulation than those colonized with non-USA300 strains, and those differences dissipated after resolution of the infection on D40 (Figure 5). IL-17A, IL-22 and IL-10 are cytokines that have been associated with immune responses to S. aureus in vitro (35), suggesting that we may be detecting a circulating memory T cell response in infected patients who are colonized with USA300.
Figure 5. PBMCs from SSTI patients colonized with USA300 produce more IL-17, IL-22 and IL-10 cytokines than those from SSTI patients colonized with non-USA300.
PBMCs from SSTI patients were stimulated for 24h with USA300 MRSA sonicates (top panels) or anti-CD3 (bottom panels) and cytokine levels in the supernatants were determined by ELISA. Results from patients with documented colonization with USA300 were compared with those from patients with documented colonization with non-USA300 S. aureus strain types. Plots show Mean + SEM. * p<0.05, **p<0.01, ***p<0.001 as determined by the Mann-Whitney test.
Discussion
Skin infections constitute a public health concern. However, the immune responses in patients with SSTIs are not well documented and hypotheses to address why some individuals are susceptible to these infections have not been tested. In this study, we genotyped the colonizing and infecting S. aureus strains of patients with SSTIs, analyzed their immune responses at D0 and D40 post-infection and tested whether the reason why otherwise healthy individuals develop SSTIs is because they have reduced global or S. aureus-specific immune responses when compared with uninfected controls. In contrast to the hypothesis, we found that patients with SSTIs produced higher levels of innate cytokines than uninfected controls, as determined in serum measurements as well as upon PBMC restimulation. Notably, skin infection with documented USA300 elicited greater innate cytokine production by PBMCs than skin infection with documented non-USA300. Moreover, colonization with USA300, compared with non-USA300 strain types, correlated with increased recall adaptive cytokine production by PBMCs of SSTI patients at the time of infection, but not after resolution of the infection or in uninfected controls. Our results argue against systemic tolerization by colonizing strains, or inhibitory immune polymorphisms, as facilitators of SSTIs in otherwise healthy individuals and, instead, demonstrate a detectable systemic immune response in patients with localized, uncomplicated SSTIs, with greater systemic immune responses when USA300 was detected either as the pathogen in an SSTI or as a commensal in the cultures obtained to test for colonization.
The prevalence of S. aureus colonization observed in the adult population enrolled in this study, namely 56% of controls and 72% of patients, matched that obtained in the parent study, which included both adult and pediatric subjects (27), but was higher than that usually cited in the general population when only the nares are sampled (36). Because we considered patients to be colonized when S. aureus was isolated from any of 3 body sites cultured, and because analysis of only the nares swabs yielded similar percentages to previous reports (27), our data suggest that S. aureus colonization is more frequent than commonly recognized and more widespread throughout the body (27). People become colonized with limited microbiota at birth, and staphylococci have been shown to be among the first colonizing taxa (37). The demonstration that isotype-switched anti-S. aureus antibodies can be rapidly and increasingly detected during childhood, in the absence of obvious infection (38), implies rapid general exposure and generation of adaptive immunity to colonizing S. aureus strains or cross-reacting strains. This is also consistent with a recent report identifying a high level of T cell memory to S. aureus in healthy individuals, with the S. aureus-specific T cell frequency estimated between 0.2 and 5.7% of the T cell repertoire (4). Together these studies suggest that healthy individuals may in fact not be immunologically naïve to S. aureus antigens.
The epidemiologic data from our study demonstrates that USA300 colonization was more frequent in SSTI patients than controls, and that antibiotic treatment failed to clear S. aureus carriage in many patients, irrespective of its genotype and despite resolution of the skin infection in the majority of cases. Moreover, the genotype of colonizing strains changed from day 0 to day 40 in many patients. This may be due to pressure imparted by the antibiotics administered to treat the infection and/or to shifts that may occur over time even in healthy controls not deliberately exposed to antimicrobial therapy. Alternatively, people may often be colonized by multiple genotypes simultaneously, such that sampling single colonies at 2 different time points may simply reveal 2 distinct strain types that were both present throughout the time course. Indeed, even with the limited approach of genotyping only 1 bacterial colony from each of the 3 cultured sites, we previously found that 25% of individuals of the combined adult and pediatric population enrolled in the parental study were polyclonally colonized (27). Consistent with the concept of polycolonization, we also found that the infecting S. aureus isolates often differed genotypically from isolates that colonized the same individual, indicating that many patients are infected with one strain and simultaneously colonized with another. Whether the infecting strain is also present as a colonizing commensal at another site of the body in these patients, or has been present as a colonizing commensal before the infection remains to be determined.
S. aureus was the only infectious agent identified in positive cultures from SSTI lesions in our patients. Although we cannot exclude that the non-purulent cellulitis that could not be drained, or lesions with negative cultures, were caused by non-S. aureus microbial organisms, it is reasonable to speculate that the vast majority of SSTI lesions are caused by S. aureus. Thus, when analyzing the data, we first grouped all infected patients into a single category regardless of the results of cultures from their SSTI site.
Our data reveal increased innate cytokines in infected patients compared with controls. Although the differences in mean cytokine production between the groups were small, the statistically significant increase in innate cytokine levels in infected patients on D0 when compared with uninfected controls correlated with the ongoing infection, and the significant reduction on D40 compared with D0 (and the lack of statistical difference on D40 with uninfected controls) correlated with resolution of the infection, suggesting that, even if subtle, these differences were associated with a functional response. While it is unclear if the higher cytokine levels in patients infected with USA300 compared with non-USA300 are biologically meaningful, this stronger immune response may be necessary to clear the infection by such a virulent strain, or may result in more severe inflammatory symptoms that then bring more patients to the ED, which, in turn, may explain the detection of this strain type among SSTI cultures in epidemic proportions.
Our results also reveal increased recall responses in infected patients colonized with USA300. Recall memory T cell responses to S. aureus have been shown to reside in circulating CCR6+CCR4+ T cells that comprise Th17 as well as some Th1 cells, rather than in the CXCR3+ or CCR4+ subsets, which are enriched in Th1 and Th2 cells, respectively (35). Differentiation of T cells from healthy controls for 12 days in vitro with heat-inactivated S. aureus-pulsed monocytes revealed production of IL-17, IL-22 and IL-10 predominantly (35). Strikingly, IL17, IL22 and IL-10 were also the cytokines whose 24h-recall production we found to be significantly increased in PBMCs from SSTI patients colonized with USA300, irrespective of the infecting strain’s genotype. These cytokines can induce tissue-protective and immunosuppressive effects, and have been implicated at cutaneous sites (35). The increased capacity of circulating PBMCs to produce these cytokines upon in vitro recall may reflect an additive Th17-type effector/memory response to the ongoing infection and USA300-colonization. This is supported by the fact that colonization with USA300 did not result in increased T cell recall responses in uninfected controls.
We also tested whether exposure of a person’s immune system to asymptomatically colonizing S. aureus could dampen immune responses to S. aureus antigens and microbial signals, thereby enhancing susceptibility of the person to infection. However, our analysis of the uninfected control subjects with detectable but asymptomatic S. aureus colonization did not reveal any decrease in the ability of their PBMCs to produce innate or adaptive cytokines upon S. aureus restimulation when compared with uncolonized controls. This result should be interpreted with caution because it is possible, and perhaps likely, that all individuals are colonized with S. aureus either transiently or persistently, such that the uninfected subjects whom we characterized as uncolonized may have in fact been colonized and already downregulated their immune responses to S. aureus.
In conclusion, our results do not support the hypothesis that SSTIs occur in individuals with more blunted systemic immune responses when compared with people who do not develop infections, but rather that ongoing USA300 skin infections drive a stronger innate inflammatory response than non-USA300 skin infections while USA300 colonization may induce stronger Th17 memory. A limitation of our study is that only systemic immune responses were analyzed. Although our data show that there are no defects in global or S. aureus-specific T cell immunity, it remains possible that S. aureus-reactive T cells may be impaired in their migration to the skin, or that the infection milieu may directly suppress the function of these T cells following cutaneous infiltration. Local immune responses to skin commensals and pathogens are only starting to be investigated (39) and may reveal additional insights into susceptibility to SSTIs. Our study also raises additional questions for subsequent studies, including why pre-existing T cell responses do not protect from infection; what role these immune responses play in the clearing of the infection and in tissue pathology; and what genetic traits present in USA300, but not other S. aureus strain types, drive stronger immune responses.
Supplementary Material
Acknowledgments
We thank Thomas Gajewski and Chris Montgomery for helpful discussions.
This work was supported by NIH grant AR059414 to RD and MLA
Abbreviations
- BMI
body-mass index
- CRP
C-reactive protein
- ED
emergency department
- HIES
hyper IgE syndrome
- MLST
multilocus sequence typing
- MRSA
methicillin-resistant S. aureus
- MSSA
methicillin-sensitive S. aureus
- NOD2
nucleotide-binding oligodimerization domain 2
- NS
not significant
- PCR
polymerase chain reaction
- PVL
Panton-Valentine leukicidin
- SSTI
skin and soft tissue infection
- TLR2
Toll-like receptor 2
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.David MZ, Glikman D, Crawford SE, Peng J, King KJ, Hostetler MA, Boyle-Vavra S, Daum RS. What is community-associated methicillin-resistant Staphylococcus aureus? J Infect Dis. 2008;197:1235–1243. doi: 10.1086/533502. [DOI] [PubMed] [Google Scholar]
- 3.Boyle-Vavra S, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab Invest. 2007;87:3–9. doi: 10.1038/labinvest.3700501. Epub 2006 Dec 2004. [DOI] [PubMed] [Google Scholar]
- 4.Kolata JB, Kuhbandner I, Link C, Normann N, Vu CH, Steil L, Weidenmaier C, Broker BM. The Fall of a Dogma? Unexpected High T-Cell Memory Response to Staphylococcus aureus in Humans. J Infect Dis. 2015;212:830–838. doi: 10.1093/infdis/jiv128. Epub 2015 Mar 1093. [DOI] [PubMed] [Google Scholar]
- 5.Brown AF, Leech JM, Rogers TR, McLoughlin RM. Staphylococcus aureus Colonization: Modulation of Host Immune Response and Impact on Human Vaccine Design. Front Immunol. 2014;4:507. doi: 10.3389/fimmu.2013.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marodi L, Cypowyj S, Toth B, Chernyshova L, Puel A, Casanova JL. Molecular mechanisms of mucocutaneous immunity against Candida and Staphylococcus species. J Allergy Clin Immunol. 2012;130:1019–1027. doi: 10.1016/j.jaci.2012.09.011. Epub 2012 Oct 1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stappers MH, Thys Y, Oosting M, Plantinga TS, Ioana M, Reimnitz P, Mouton JW, Netea MG, Joosten LA, Gyssens IC. Polymorphisms in cytokine genes IL6, TNF, IL10, IL17A and IFNG influence susceptibility to complicated skin and skin structure infections. Eur J Clin Microbiol Infect Dis. 2014;33:2267–2274. doi: 10.1007/s10096-014-2201-0. Epub 12014 Jul 10015. [DOI] [PubMed] [Google Scholar]
- 8.Stappers MH, Oosting M, Ioana M, Reimnitz P, Mouton JW, Netea MG, Gyssens IC, Joosten LA. Genetic Variation in TLR10, an Inhibitory Toll-Like Receptor, Influences Susceptibility to Complicated Skin and Skin Structure Infections. J Infect Dis. 2015;212:1491–1499. doi: 10.1093/infdis/jiv229. Epub 2015 Apr 1420. [DOI] [PubMed] [Google Scholar]
- 9.Fleischer B. Superantigens produced by infectious pathogens: molecular mechanism of action and biological significance. Int J Clin Lab Res. 1994;24:193–197. doi: 10.1007/BF02592461. [DOI] [PubMed] [Google Scholar]
- 10.Hedl M, Li J, Cho JH, Abraham C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc Natl Acad Sci U S A. 2007;104:19440–19445. doi: 10.1073/pnas.0706097104. Epub 12007 Nov 19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hruz P, Zinkernagel AS, Jenikova G, Botwin GJ, Hugot JP, Karin M, Nizet V, Eckmann L. NOD2 contributes to cutaneous defense against Staphylococcus aureus through alpha-toxin-dependent innate immune activation. Proc Natl Acad Sci U S A. 2009;106:12873–12878. doi: 10.1073/pnas.0904958106. Epub 12009 Jun 12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison CJ. Innate immunity as a key element in host defense against methicillin resistant Staphylococcus aureus. Minerva Pediatr. 2009;61:503–514. [PubMed] [Google Scholar]
- 13.Puel A, Picard C, Lorrot M, Pons C, Chrabieh M, Lorenzo L, Mamani-Matsuda M, Jouanguy E, Gendrel D, Casanova JL. Recurrent staphylococcal cellulitis and subcutaneous abscesses in a child with autoantibodies against IL-6. J Immunol. 2008;180:647–654. doi: 10.4049/jimmunol.180.1.647. [DOI] [PubMed] [Google Scholar]
- 14.Spellberg B, Ibrahim AS, Yeaman MR, Lin L, Fu Y, Avanesian V, Bayer AS, Filler SG, Lipke P, Otoo H, Edwards JE., Jr The antifungal vaccine derived from the recombinant N terminus of Als3p protects mice against the bacterium Staphylococcus aureus. Infect Immun. 2008;76:4574–4580. doi: 10.1128/IAI.00700-08. Epub 2008 Jul 4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin L, Ibrahim AS, Baquir B, Avanesian V, Fu Y, Spellberg B. Immunological surrogate marker of rAls3p-N vaccine-induced protection against Staphylococcus aureus. FEMS Immunol Med Microbiol. 2009;55:293–295. doi: 10.1111/j.1574-695X.2008.00531.x. Epub 2009 Jan 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, Iwakura Y. Differential roles of interleukin-17A and-17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Chan LC, Chaili S, Filler SG, Barr K, Wang H, Kupferwasser D, Edwards JE, Jr, Xiong YQ, Ibrahim AS, Miller LS, Schmidt CS, Hennessey JP, Jr, Yeaman MR. Nonredundant Roles of Interleukin-17A (IL-17A) and IL-22 in Murine Host Defense against Cutaneous and Hematogenous Infection Due to Methicillin-Resistant Staphylococcus aureus. Infect Immun. 2015;83:4427–4437. doi: 10.1128/IAI.01061-15. Epub 02015 Sep 01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, Magorien JE, Blauvelt A, Kolls JK, Cheung AL, Cheng G, Modlin RL, Miller LS. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest. 2010;120:1762–1773. doi: 10.1172/JCI40891. Epub 42010 Apr 40891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolk K, Warszawska K, Hoeflich C, Witte E, Schneider-Burrus S, Witte K, Kunz S, Buss A, Roewert HJ, Krause M, Lukowsky A, Volk HD, Sterry W, Sabat R. Deficiency of IL-22 contributes to a chronic inflammatory disease: pathogenetic mechanisms in acne inversa. J Immunol. 2011;186:1228–1239. doi: 10.4049/jimmunol.0903907. Epub 0902010 Dec 0903908. [DOI] [PubMed] [Google Scholar]
- 20.Montgomery CP, Daniels M, Zhao F, Alegre ML, Chong AS, Daum RS. Protective immunity against recurrent Staphylococcus aureus skin infection requires antibody and interleukin-17A. Infect Immun. 2014;82:2125–2134. doi: 10.1128/IAI.01491-14. Epub 02014 Mar 01410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, Anderson VL, Darnell DN, Welch PA, Kuhns DB, Frucht DM, Malech HL, Gallin JI, Kobayashi SD, Whitney AR, Voyich JM, Musser JM, Woellner C, Schaffer AA, Puck JM, Grimbacher B. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–1619. doi: 10.1056/NEJMoa073687. Epub 2007 Sep 1619. [DOI] [PubMed] [Google Scholar]
- 22.Ma CS, Chew GY, Simpson N, Priyadarshi A, Wong M, Grimbacher B, Fulcher DA, Tangye SG, Cook MC. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205:1551–1557. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renner ED, Rylaarsdam S, Anover-Sombke S, Rack AL, Reichenbach J, Carey JC, Zhu Q, Jansson AF, Barboza J, Schimke LF, Leppert MF, Getz MM, Seger RA, Hill HR, Belohradsky BH, Torgerson TR, Ochs HD. Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced T(H)17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J Allergy Clin Immunol. 2008;122:181–187. doi: 10.1016/j.jaci.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, Kanno Y, Spalding C, Elloumi HZ, Paulson ML, Davis J, Hsu A, Asher AI, O'Shea J, Holland SM, Paul WE, Douek DC. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–776. doi: 10.1038/nature06764. Epub 2008 Mar 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, Metin A, Karasuyama H. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. Epub 2007 Aug 1055. [DOI] [PubMed] [Google Scholar]
- 26.Minegishi Y, Saito M, Nagasawa M, Takada H, Hara T, Tsuchiya S, Agematsu K, Yamada M, Kawamura N, Ariga T, Tsuge I, Karasuyama H. Molecular explanation for the contradiction between systemic Th17 defect and localized bacterial infection in hyper-IgE syndrome. J Exp Med. 2009;206:1291–1301. doi: 10.1084/jem.20082767. Epub 2009 Jun 1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar N, David MZ, Boyle-Vavra S, Sieth J, Daum RS. High Staphylococcus aureus colonization prevalence among patients with skin and soft tissue infections and controls in an urban emergency department. J Clin Microbiol. 2015;53:810–815. doi: 10.1128/JCM.03221-14. Epub 02014 Dec 03224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller LG, Daum RS, Creech CB, Young D, Downing MD, Eells SJ, Pettibone S, Hoagland RJ, Chambers HF. Clindamycin versus trimethoprim-sulfamethoxazole for uncomplicated skin infections. N Engl J Med. 2015;372:1093–1103. doi: 10.1056/NEJMoa1403789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. Journal of clinical microbiology. 2004;42:792–799. doi: 10.1128/JCM.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus . Journal of clinical microbiology. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(IWG-SCC)., I. W. G. o. t. C. o. S. C. C. E. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;12:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle-Vavra S, Ereshefsky B, Wang CC, Daum RS. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec(SCCmec) type VT or SCCmec type IV. Journal of clinical microbiology. 2005;43:4719–4730. doi: 10.1128/JCM.43.9.4719-4730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 34.David MZ, T. A., Lynfield R, Boxrud DJ, Short G, Zychowski D, Boyle-Vavra S, Daum RS. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for panton-valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. Medical Center. Journal of clinical microbiology. 2013;3:814–819. doi: 10.1128/JCM.02429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, Monticelli S, Lanzavecchia A, Sallusto F. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 36.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10:505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS biology. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dryla A, Prustomersky S, Gelbmann D, Hanner M, Bettinger E, Kocsis B, Kustos T, Henics T, Meinke A, Nagy E. Comparison of antibody repertoires against Staphylococcus aureus in healthy individuals and in acutely infected patients. Clin Diagn Lab Immunol. 2005;12:387–398. doi: 10.1128/CDLI.12.3.387-398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.