Abstract
Toll like receptor 9 (TLR9) is an innate immune receptor important for recognizing DNA, both of host and foreign origin. A mechanism proposed to prevent excessive response to host DNA is the requirement for proteolytic cleavage of TLR9 in endosomes to generate a mature form of the receptor (TLR9471-1032). We previously described another cleavage event in the juxtamembrane region of the ectodomain that generated a dominant negative form of TLR9. Thus, there are at least two independent cleavage events that regulate TLR9. Here we investigated whether an N-terminal fragment of TLR9 could be responsible for regulation of the mature- or negative regulatory-form. We show that TLR9471-1032, corresponding to the proteolytically cleaved form, does not function on its own. Furthermore, activity is not rescued by co-expression of the N-terminal fragment (TLR91-440), inclusion of the hinge region (TLR9441-1032) or overexpression of UNC93B1, the latter of which is critical for trafficking and cleavage of TLR9. TLR91-440 co-immunoprecipitates with full-length TLR9 and TLR9471-1032, but does not rescue the native glycosylation pattern, thus inappropriate trafficking likely explains why TLR9471-1032 is nonfunctional. Lastly, we show that TLR9471-1032 is also a dominant negative regulator of TLR9 signaling. Together these data provide a new perspective on the complexity of TLR9 regulation by proteolytic cleavage and offer potential ways to inhibit activity through this receptor, which may dampen autoimmune inflammation.
Introduction
Innate immune receptors are positioned at various places within and on cells to allow broad detection of microbial patterns. Since DNA and RNA are encapsulated within microbes and only released upon microbial internalization into endosomes, nucleic acid sensors are positioned on host endosomal membranes or in the cytosol (1, 2). For example, nucleic acid-sensing TLR7, TLR8 and TLR9 are primarily localized in the endoplasmic reticulum (ER) until they exit to survey endosomal compartments (3–10). The unique localization and trafficking of these TLRs has been proposed as a major regulatory mechanism to limit response to host DNA and RNA (11). Indeed, artificial localization of TLR9 to the cell surface promotes response to host DNA and causes autoinflammation in mice (12). Multiple mechanisms maintain appropriate localization of TLR9 including the presence of motifs in the cytoplasmic tail that retain TLR9 in the ER prior to release to endosomal compartments (3–7, 13) and phosphorylation of TLR9 (14). Release of TLR9 to the endosomal compartment requires the ER protein UNC93B1 (15, 16). Other proteins important for trafficking and signaling of TLR9 include adapter protein 3 (AP-3), a protein associated with TLR4 (PRAT4A), Slc15a4 and glycoprotein 96 (gp96, also known as glucose regulated protein 94 [grp94]) (13, 17–21).
Upon exit from the ER and trafficking to the endosomal compartment, TLR9 and other nucleic acid-sensing TLRs are proteolytically processed. The TLR9 ectodomain contains 26 leucine rich repeat structures (LRR), interrupted by a disordered domain that forms a loop (22). This loop, also referred to as the hinge region or z-loop, is located between LRR1-14 and LRR15-26 and is the region where proteolytic processing to a mature form occurs in a processive manner (23–25). The proteolytically cleaved form of TLR9 contains amino acids 471-1032 and thus consists of LRR15 to the end of the C-terminus. Both full-length and proteolytically cleaved TLR9 bind to ligand, and both are predominantly monomeric in the absence of ligand. However, while the full-length ectodomain of TLR9 remains primarily monomeric, even upon ligand addition, the ectodomain fragment corresponding to proteolytically cleaved TLR9 forms more dimers in response to ligand (23, 24).
We previously described an alternative cleavage event in endogenous TLR9 (all other studies were performed with epitope-tagged TLR9) that generated a soluble ectodomain (26). This soluble ectodomain bound to CpG DNA ligand and was a negative regulator of signaling (26). Similarly, Lee and colleagues showed that attaching a transmembrane lysosomal targeting domain on an N-terminal fragment (LRR1-14) inhibited the ability of full-length TLR9 to signal (27). These data suggest that the cleavage events and their regulation are complex. Here we investigated the ability of a fragment of TLR9 corresponding to proteolytically cleaved TLR9 (amino acids 471-1032, TLR9471-1032) to signal and interact with the N-terminal fragment (TLR91-440) and with full-length TLR9. Our results show that expression of TLR9471-1032 in macrophages or dendritic cells is not sufficient for response to CpG DNA. Furthermore, TLR9 is a highly glycosylated protein, but our data show that the glycosylation pattern of transfected TLR9471-1032 does not recapitulate that observed for the naturally generated form TLR9471-1032. Our data support a model where defects in post-translational modification (and trafficking) of TLR9471-1032 arise if cleavage is not coincident with intracellular trafficking. However, co-expression of TLR91-440 with TLR9471-1032 neither rescues proper glycosylation nor restores signaling. In fact, both TLR91-440 and TLR9471-1032 could be co-immunoprecipitated with full-length TLR9, and both inhibited signaling by the full-length receptor. Together these data suggest that, if the naturally occurring TLR9471-1032 is indeed the active signaling form, it is likely generated from full-length TLR9 in endosomes where ligand will also be present.
Materials and Methods
Reagents and Plasmids
The following reagents were used: CpG oligodeoxynucleotides (5′-TCGTCGTTTCGTCGTTTTGTCGTT-3′, Eurofins MWG Operon, Huntsville, AL), ultrapure LPS 0111:B4 (Sigma), flagellin (Invivogen), and TNF-α ELISA kit (Biolegend). The following antibodies were used: hemaglutinin tag (HA, ABM and Roche), flag tag (Sigma), tubulin (eBioscience), green fluorescent protein (GFP, life technologies), and horseradish peroxidase-labeled secondary antibodies (Southern Biotech). TLR9471-1032-HA and TLR9441-1032-HA were generated by PCR sewing using mouse TLR9-HA (Fig 1), and the mouse IgκB leader sequence from pDisplay (Invitrogen). Two fragments were generated with primers 1 and 2, and 3 and 4 (Table 1). Those products and primers 1 and 4 were included in a second PCR reaction to generate the sewed product, which was then cloned into pcDNA3.1+(see Fig. 1 for various TLR9 mutants).
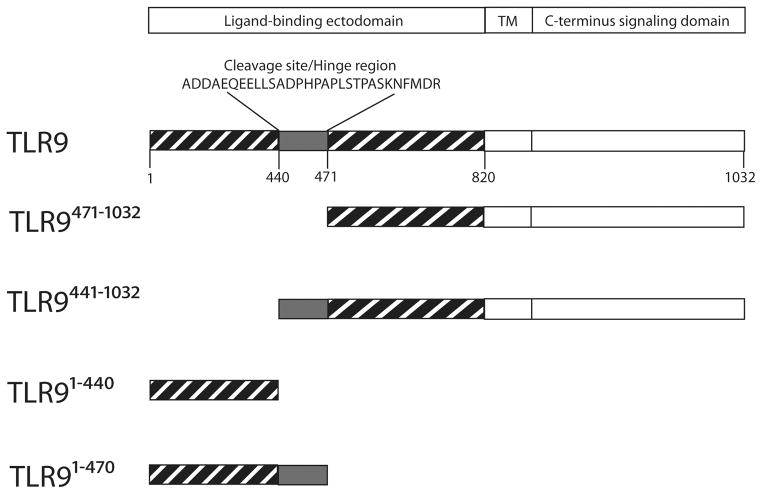
Figure 1.
Schematic for TLR9 constructs. Full-length TLR9 has 26 leucine rich repeats, a transmembrane domain and a C-terminus containing the Toll-IL-1 resistance domain. TLR9 is proteolytically cleaved between LRR14 and LRR15 resulting in TLR9471-1032 and TLR91-440. We have generated two additional forms. One form includes the hinge region (aa 441-470) in the C-terminal proteolytic cleavage product (TLR9441-1032 ) and the other includes the hinge region on the N-terminal cleavage product (TLR91-471).
Table 1.
PCR primers used in the generation of TLR9471-1032-HA and TLR9441-1032-HA
| Construct | Forward | Reverse |
|---|---|---|
| mTLR9471-1032-HA | Primer1 5′-GGCCCGCCTGGCATTATGCCCAG-3′ |
Primer 2 5′-CAGGTCCATGGTGAACTTGAAGTTCTTACAGTCACCAGTGGAACCTGGAACCCAGAGCAGCAG-3′ |
| Primer 3 5′-CTGCTGCTCTGGGTTCCAGGTTCCACTGGTGACTGTAAGAACTTCAAGTTCACCATGGAGCTG-3′ |
Primer 4 5′-TATCTCGAGCTAAGCGTAGTCTGGGACGTCGTATGGG-3′ |
|
|
| ||
| mTLR9441-1032-HA | Primer 1 5′-GGCCCGCCTGGCATTATGCCCAG-3′ |
Primer 2 5′-CAACAGCTCCTCCTGCTCTGCATCATCTGCGTCACCAGTGGAACCTGGAACCCAGAGCAGCAG-3′ |
| Primer 3 5′-CTGCTGCTCTGGGTTCCAGGTTCCACTGGTGACGCAGATGATGCAGAGCAGGAGGAGCTGTTG-3′ |
Primer 4 5′-TATCTCGAGCTAAGCGTAGTCTGGGACGTCGTATGGG-3′ |
|
Cell culture and virus transduction
The macrophage cell line derived from TLR9 knockout mice (BEI Resources, NIAID, NIH) was cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% (v/v) heat-inactivated low endotoxin fetal calf serum (FCS), 2 mM L-glutamine, 10 mM HEPES and 1 mM sodium pyruvate with the addition of 50 U/ml penicillin, 50 μg/ml streptomycin, and 10 μg/ml ciprofloxacin. The cells were cultured at 37°C with 5% CO2 and routinely tested negative for mycoplasma by PCR.
To generate transduced bone marrow derived macrophages (BMMs), femurs and tibias from TLR9 deficient mice were collected from 8- to 10-week-old mice. Bone marrow was flushed from the bones with cold DMEM supplemented with 20% L-929 cell-conditioned medium, 10% (v/v) heat-inactivated FCS, 2 mM L-glutamine, 10 mM HEPES and 1 mM sodium pyruvate, 100 U/ml penicillin and 100 mg/ml streptomycin. Bone marrow cells were cultured in 10 cm petri dishes (10 ml volume) at 1X106 cells per ml 37°C, 5% CO2 for 7 days. On days one and three, 1 ml of the media was removed and replaced with 1 ml of retrovirus-containing supernatant mixed with polybrene (final concentration 8 μg/ml). Plates were then centrifuged at 1811 × g at 32°C for 90 minutes. Post centrifugation, 1 ml of the media was removed and replaced with fresh media. At the end of 7 days, adherent cells were collected. Differentiation was confirmed by flow cytometry after staining the cells with anti-F4/80 (PE) antibody. Cultures were routinely >95% F4/80+.
A similar procedure was used to generate bone marrow derived dendritic cells (BM-DCs) except cells were cultured in 6 well plates at 1X106 cells per ml (2 ml volume) of RPMI media supplemented with 2 mM L-glutamine, 50 U/ml penicillin, 50 mg/ml streptomycin, 10% low endotoxin FBS, 20 ng/ml GM-CSF and 50 nM 2-mercaptoethanol. On days two and four, 1 ml of media was removed and replaced with 1.5 mL of retrovirus-containing supernatant mixed with polybrene (final concentration 8 μg/ml). Plates were centrifuged at 1811 × g, 32C°C for 90 minutes. Post centrifugation, 1 ml of media was removed and replaced with fresh high 2-mercaptoethanol media (same as the original media, but with 50 μM 2-mercaptoethanol). Lightly adherent and floating cells were harvested on day 7. Differentiation was confirmed by flow cytometry with anti-CD11c (PE) antibody. Cultures were routinely 80% CD11c+, and only CD11c+ cells were included in the analysis.
Retroviral transduction of macrophages
Retroviral supernatants were generated using Lipofectamine 2000 (Invitrogen) transfected ØNX-Ampho cells. TLR9−/− macrophages were spin transduced with retroviral supernatants plus polybrene (8 μg/ml final concentration) and cultured at 37°C for 44–48 hours prior to stimulation.
Flow cytometry for intracellular HA and TNF
TLR9−/− macrophage, BMM, or BM-DC were retrovirally transduced with constructs encoding mTLR9-HA or mTLR9471-1032-HA. Cells were then stimulated with 3μM CpG 10104 for 6 hours with the addition of 10μg/ml BFA for the last 4 hours. Cells were fixed with 3% PFA for 15 minutes on ice, then stained with anti-HA FITC and anti-TNF-α APC in 0.1% saponin in PBS with 10% mouse serum for 1 hour on ice. Fluorescence intensity was measured with a FACS CantoII flow cytometer. Data were collected with FACSDiva (BD Biosciences) and analyzed with FlowJo.
TNF ELISA
TNF-α in the supernatants was measured by ELISA according to the manufacturer’s recommendations (Biolegend, San Diego, CA).
Protein deglycosylation
The deglycosylation assay was performed as previously described (3). Immunoprecipitates on beads were suspended in 100 μl of deglycosidase buffer and divided into three equal portions and were either untreated or treated with endo H or PNGase F overnight at 37°C. The reactions were stopped by adding 6× SDS-PAGE reduced sample buffer and boiling at 95°C for 5 min prior to SDS-PAGE.
Immunoblotting and (co) immunoprecipitation
Immunoblotting was performed as previously described (4, 5, 26, 28). Briefly, stimulated cells were washed with ice-cold HBSS, and lysed for direct immunoblotting with 1X SDS-PAGE reduced sample buffer (62.5 mM Tris [pH 6.8], 12.5% glycerol, 1% SDS, 0.005% bromophenol blue, 1.7% beta-mercaptoethanol), or for immunoprecipitation or co-immunoprecipitation with lysis buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 10% (w/v) glycerol, 1 mM EDTA, and protease inhibitors). Lysates were incubated at 95 _C for 5 minutes prior to resolving by 10% SDS-PAGE. Proteins were transferred to nitrocellulose membranes and immunoblotted with the indicated antibodies. Membranes were incubated with a Supersignal West Pico chemiluminescence Western blotting detection system (Thermo Scientific) and exposed to x-ray film. Films were scanned and images were assembled in Photoshop. For immunoprecipitation experiments, total protein was determined in clarified lysates using the BCA protein assay (Bio-Rad) and 5μg of indicated antibody was used for immunoprecipitation. For co-immunoprecipitations, one SDS-PAGE gel was run and transferred to nitrocellulose. The blots were sequentially probed with the indicated antibodies.
Luciferase assay
Luciferase assay was performed as previously described (6, 26, 29). Briefly, HEK293 cells were transfected using TransIT (Mirus) with the indicated plasmids plus 5x-NF-kB-luciferase reporter and empty vector to total 200ng of DNA per well of a 96 well plate. After 24 hours, cells were stimulated with CpG DNA (3 μM), ultrapure LPS (100 ng/ml) or flagellin (100 ng/ml) for an additional 24 hours. Cells were lysed in Reporter Lysis Buffer (Promega) and assayed using a Veritas luminometer with luciferase substrate (20 nM tricine, 2.67 mM MgSO4·7 H2O, 33.3 mM DTT, 100 μM EDTA, 530 μM ATP, 270 μM Acetyl CoA, 12 μg/ml luciferin, 5 mM NaOH, 265 μM magnesium carbonate hydroxide).
Statistical Analysis
Data were collected in quadruplicate. Comparisons were analyzed using one-way ANOVA with Tukey’s post-hoc correction, assuming a Gaussian distribution (GraphPad Prism).
Results
Expression of a fragment corresponding to proteolytically cleaved TLR9 does not support response to CpG DNA
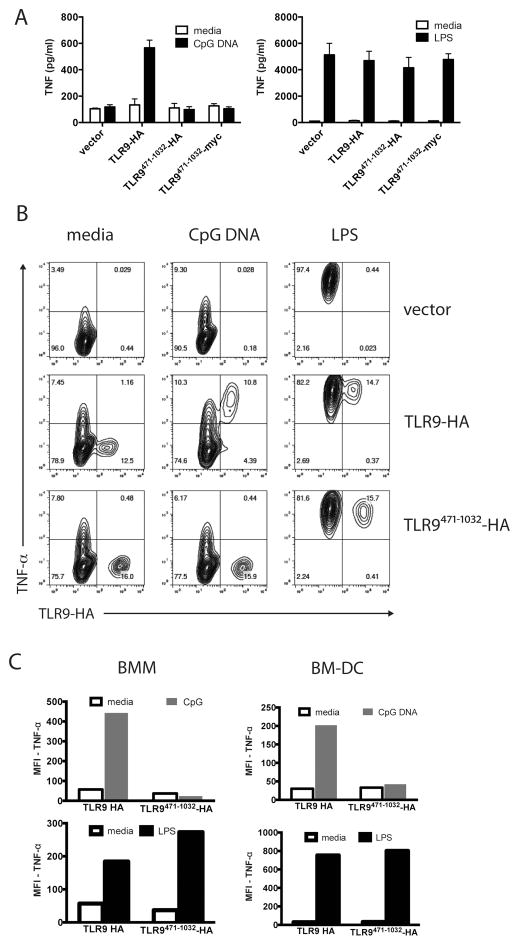
TLR9 is proteolytically cleaved between amino acids 441 and 470 to generate a mature form (Fig. 1, TLR9471-1032) that is proposed to be the active form of the receptor (23, 24). To determine whether TLR9471-1032 supported response to CpG DNA, we reconstituted a mouse macrophage cell line derived from TLR9 deficient mice on a C57BL/6 background. Reconstitution of TLR9-deficient macrophages with empty vector did not support secretion of TNF-α in response to CpG DNA (Fig 2A). As expected, expression of full-length wild-type TLR9-HA in the TLR9 deficient macrophages restored response to CpG DNA (Fig 2A). However, expression of either HA- or myc-tagged TLR9471-1032 did not support CpG DNA response (Fig 2A). Untransduced macrophages, and those expressing any of the TLR9 constructs responded similarly to LPS (Fig 2A). The level of TNF-α production by the same number of wild type, unmanipulated, BMM to 1 μM CpG DNA and 100 ng/ml LPS was similar to what we observed with retroviral reconstitution of TLR9-deficient macrophages stimulated with 3μM CpG DNA or 100 ng/ml LPS (Supplementary Fig 1). We next investigated TNF-α production by intracellular cytokine staining, thus differentiating transduced and untransduced macrophages. Retroviral transduction resulted in TLR9 or mutant TLR9 expression in around 15% of the cells (Fig 2B). Intracellular cytokine staining demonstrated that all cells responded to LPS regardless of transduction status (Fig 2B). Despite similar levels of transduction and expression of full-length and the TLR9471-1032 form, only cells expressing the full-length TLR9-HA responded to CpG DNA stimulation to produce TNF-α (Fig 2B). This was not unique to the macrophage cell line since reconstitution during in vitro differentiation of primary TLR9 deficient bone marrow derived macrophages (BMM, 15% transduction efficiency, >95% viability) or bone marrow derived dendritic cells (BM-DC, 5% transduction efficiency, >95% viability) with TLR9-HA, but not TLR9471-1032-HA supported TNF-α production in response to CpG DNA, while all the cells produced TNF-α in response to LPS (Fig 2C).
Figure 2. TLR9471-1032 does not respond to CpG DNA.
A) A TLR9-deficient macrophage cell line was retrovirally transduced with empty vector, full-length TLR9-HA, or TLR9471-1032 tagged with either HA or myc at the C-terminus. Transduced cells were replated at 2X10^5 cells per well in 24 well plates than treated with media, 3 μM CpG DNA, or 100 ng/ml LPS for six hours in quadruplicate. Supernatants were assayed for TNF-α by ELISA. This experiment was performed 3 times. B) A TLR9-deficient macrophage cell line was retrovirally transduced with empty vector, TLR9, or TLR9471-1032 for 24 hours. Cells were stimulated with 3 μM CpG DNA, or 100 ng/ml LPS for six hours and 10 μg/ml Brefeldin A was added for the final four hours of stimulation. Cells were fixed, permeabilized, and stained for intracellular TNF-α and the HA tag on TLR9. This experiment was performed 3 times. C) Primary mouse bone marrow-derived macrophages (BMM) and dendritic cells (BM-DC) were similarly reconstituted, stimulated and stained as in (B). This experiment was performed 2 times.
Inclusion of the hinge region or overexpression of UNC93B1 does not restore signaling
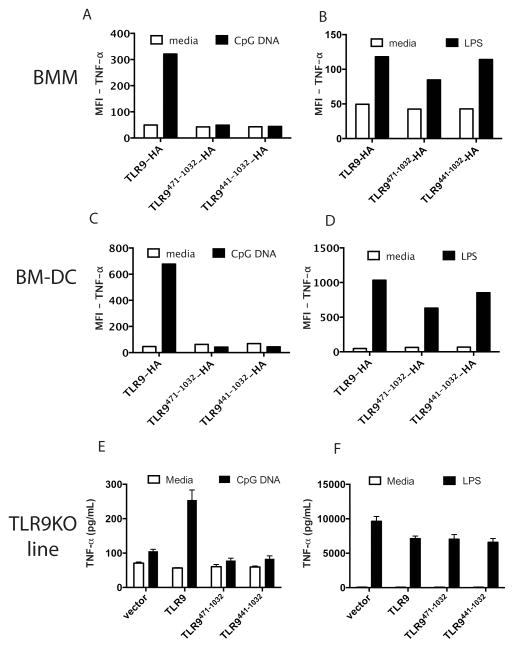
We next asked if inclusion of the hinge region (a.a.: 440-470), and thus the potential to be proteolytically processed, would restore signaling. Reconstitution of BMM or BM-DC with either TLR9471-1032-HA (without the hinge) or with TLR9441-1032-HA (with the hinge) (Fig 1) failed to support TNF-α production in response to CpG DNA (Fig 3 A, C). Similarly, reconstitution of the TLR9 deficient macrophage cell line with untagged TLR9471-1032 (without the hinge) or with TLR9441-1032 (with the hinge) also failed to support TNF-α secretion (Fig 3E). However, cells transduced with full-length TLR9-HA (Fig 3A, C) or untagged TLR9 (Fig 3E) responded to CpG DNA. All cells also responded to LPS (Fig 3 B, D, F). These findings suggest that TLR91-440 is structurally required for appropriate trafficking and cleavage, or that it is required for signaling despite being proteolytically removed.
Figure 3. Inclusion of the hinge region TLR9441-470 does not restore signaling by TLR9471-1032.
A-B) Primary bone marrow-derived macrophages (BMM) were retrovirally reconstituted with TLR9-HA, TLR9471-1032, or TLR9441-1032 and stimulated with media, 5 μM CpG DNA (A) or 100 ng/ml LPS (B) for six hours. Brefeldin A was added for the final four hours of stimulation. Cells were fixed, permeabilized, and stained for intracellular TNF-α and the HA tag on TLR9. MFI of the HA positive cells is shown. C-D) As in A-B except primary bone marrow derived dendritic cells (BM-DC) were reconstituted by retroviral transduction. E-F) A TLR9 deficient macrophage cell line (TLR9KO line) was reconstituted with empty vector, TLR9, TLR9471-1032, or TLR9441-1032 and stimulated with media, 3 μM CpG DNA (E) or 100 ng/ml LPS (F) for six hours. Supernatants were assayed for TNF-α by ELISA. Each experiment was performed 2 times.
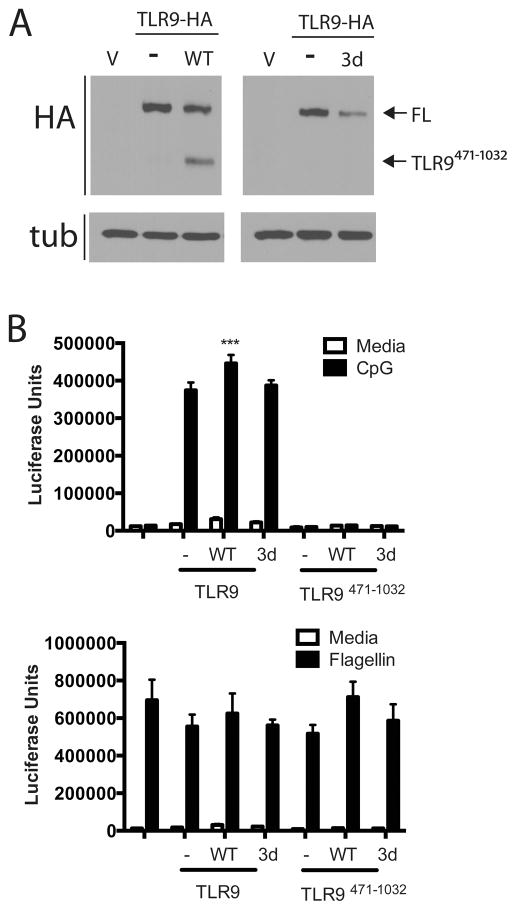
We, and others, have routinely used HEK293 cells to assay for TLR9 activity, yet TLR9 is poorly proteolytically processed to TLR9471-1032 in these cells (Fig 4A). These data raise the possibility that full-length TLR9 is capable of inducing signal transduction, which is consistent with its ability to bind ligand (22). The TLR9 trafficking chaperone UNC93B1 is required for TLR9 trafficking to endosomes (16, 30), and the mutation H412R on UNC93B1 (referred to as the UNC93B1 3d) fails to support trafficking and signaling of TLR9 (16, 31). In HEK293 cells, overexpression of wild type UNC93B1, but not the UNC93B1 3d mutant, enhanced TLR9 proteolytic cleavage (Fig 4A). Expression of wild type UNC93B1 correlated with a slight, but statistically significant, increase in TLR9 signaling (Fig 4B). The 3d mutant did not enhance signaling (Fig 4B). Expression of UNC93B1 or the 3d mutant had no effect on TLR5 response to flagellin (Fig 4B) although cells transfected with either form of UNC93B1 had an unexplained, but reproducible, increase in control β-galactosidase activity (Supplementary Fig 2A). Importantly, overexpression of UNC93B1, or the 3d mutant, did not rescue signaling by TLR9471-1032 (Fig 4B). Together, these data demonstrate that TLR9 is cleaved, and that cleavage correlates with a slight increase in signaling; however, expression of TLR9471-1032, which correlates with the cleaved form, is not sufficient for signaling, and overexpression of UNC93B1 does not rescue TLR9471-1032.
Figure 4. Increased expression of UNC93B1 enhances proteolytic cleavage of TLR9, but does not restore signaling by TLR9471-1032.
A) HEK293 cells were transfected with TLR9-HA (1.3 μg/well of a 6-well plate, total DNA 4μg/well) with or without UNC93B1 wild type (WT) or 3d mutant (3d) (0.065 μg per well) overnight. Lysates were resolved by SDS-PAGE and analyzed by immunoblotting for HA and tubulin (tub). This experiment was performed 2 times. B) HEK293 cells were transfected with empty vector, TLR9-HA or TLR9471-1032 (5ng/well of a total of 200ng per well of a 96 well plate) without or with UNC93B1 wild type (WT) or 3d mutant (3d) (0.25ng/well) plus an NF-κB–luciferase reporter overnight. Cells were stimulated with 3μM CpG DNA (top) or 100 ng/ml flagellin (bottom) for 18 hours, lysed and assayed for luciferase activity. ***p<.01. This experiment was performed 3 times.
The TLR9471-1032 is not properly glycosylated and the N-terminal fragment does not restore the native glycosylation pattern
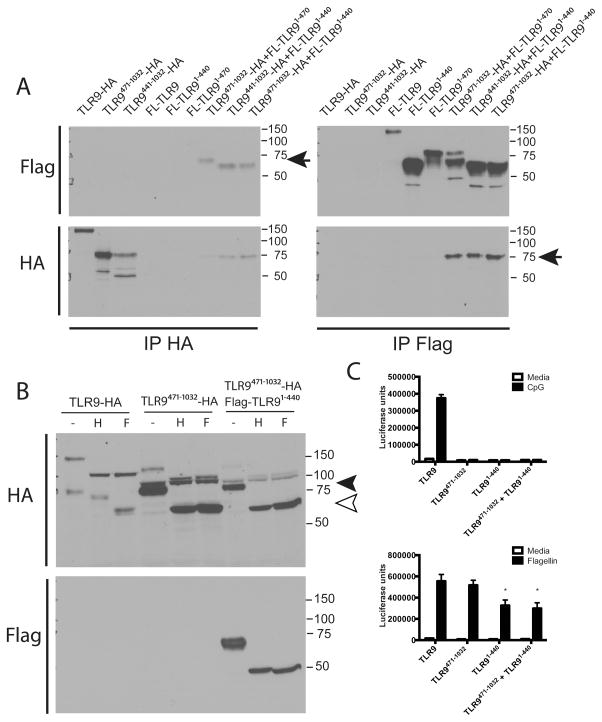
A previous study demonstrated that an N-terminal fragment of TLR9 associates with TLR9471-1032 (32). Furthermore, the hinge region is processively cleaved and may play a role in localization and trafficking of TLR9 to yeast-containing phagosomes (33, 34). Thus, we next asked whether the hinge was required for association between TLR9471-1032 and the TLR91-440. When expressed in macrophages, TLR9471-1032 co-immunoprecipitated with the N-terminus, regardless of whether the hinge region was present (TLR91-440 or TLR91-470) (Fig 5A). TLR9441-1032 (including the hinge region) also co-immunoprecipitated with the N-terminal fragment TLR91-440 (Fig 5A). Thus, N- and C- terminal fragments associate whether or not they contain the hinge region.
Figure 5. Glycosylation of TLR9471-1032 does not mimic the glycosylation pattern of naturally generated TLR9471-1032 and addition of the N-terminal fragment TLR91-470 in trans does not rescue glycosylation or signaling of TLR9471-1032.
A) HEK293 cells were transfected with (1.3 μg of a total of 4ug DNA per 6 well) TLR9-HA, TLR9471-1032-HA, TLR9441-1032-HA, Flag-TLR9 (FL-TLR9), Flag-TLR91-470, Flag-TLR91-440, or 1.3 μg of each of the combinations of the indicated plasmids. Cells were lysed and immunoprecipitated with antibody to HA or Flag, resolved by SDS-PAGE and analyzed by immunoblotting for HA or Flag. The arrows indicate co-immunoprecipitated proteins. This experiment was performed 3 times. B) HEK293 cells were transfected with TLR9-HA, TLR9471-1032, or TLR9471-1032 plus Flag-tagged TLR91-440 (1μg/well plus 1μg/well of UNC93B1 and 2 μg/well empty vector for a total of 4μg/well of a 6 well plate). Cells were lysed, and lysates were deglycosylated with endoH or PNGaseF and resolved by SDS-PAGE. Following transfer to nitrocellulose, the bots were sequentially probed for HA and Flag. This experiment was performed 2 times. C) HEK293 cells were transfected with TLR9-HA, TLR9471-1032, Flag-TLR91-440, or TLR9471-1032 plus Flag-TLR91-440 (5ng/well of a total of 200ng per well of a 96 well plate) for 24 hours. Cells were stimulated with media, 3 μM CpG DNA or 100 ng/ml flagellin (cells endogenously express TLR5) for 18 hours, lysed and assayed for luciferase activity. *p<.05. This experiment was performed 4 times.
Since TLR9471-1032 did not support signaling, and glycosylation is important for proper trafficking and signaling of TLR9, we next investigated whether the glycosylation pattern on TLR9471-1032 was the same as the glycosylation pattern generated naturally for TLR9. When full-length TLR9 is proteolytically cleaved, the full-length form of the receptor is glycosylated, but is completely sensitive to endoglycosidase H (endoH) suggesting that the glycans contain high mannose, typically associated with immature gycosylation. The naturally generated TLR9471-1032 has a molecular weight of about 80 kDa (Fig 5B, filled arrowhead) and is partially sensitive to endoH digestion. When fully deglycosylated with peptide N glycosidase F (PNGaseF), TLR9471-1032 resolves at around 65 kDa (Fig 5B, open arrowhead, and (23)). In contrast, TLR9471-1032 expressed on its own had an unusual banding pattern. We observed a highly glycosylated form (125 kDa) that was completely sensitive to endoH (105 kDa) and an intermediate glycosylated form (100kDa) that was resistant to endoH (resolves just above the black arrowhead, Fig 5B). We also observed a fragment that ran at the same molecular weight as the naturally generated mature form (80 kDa), but was completely sensitive to endoH and PNGaseF digestion (open arrowhead, Fig 5B). These data suggest that expression of a fragment of TLR9 corresponding to the proteolytically cleaved form, TLR9471-1032, is biochemically distinct than the naturally generated mature form. We next asked whether addition of TLR91-440 could restore appropriate glycosylation of TLR9471-1032. When TLR91-440 was expressed with TLR9471-1032, we detected the same fragments and glycosylation pattern observed when TLR9471-1032 was expressed by itself. Thus, intracellular processing, and therefore post-translational modifications of expressed TLR9471-1032 is distinct from naturally generated TLR9471-1032 in cells. Furthermore, supplying the N-terminal fragment in trans does not rescue post-translational modification.
We next investigated whether the N-terminal fragment could rescue the lack of signaling by TLR9471-1032. Transfection of TLR9 into HEK293 cells supported signaling in response to CpG DNA (Fig 5C). However, neither TLR9471-1032 alone, nor TLR9471-1032 co-expressed with TLR91-440 supported signaling in response to CpG DNA (Fig 5C). Low expression of TLR9471-1032 and TLR91-440 did not account for lack of activity since both protein fragments were expressed at higher levels than full-length wild type TLR9 (Fig 5A and Fig 5B). Furthermore, cells expressing TLR9, TLR9471-1032, or TLR9471-1032 and TLR91-440 supported similar signaling in response to the TLR5 ligand flagellin (HEK293 cells endogenously express TLR5) (Fig 5C). However, for unknown reasons, we consistently observed a reduction in TLR5 response to flagellin when cells overexpressed TLR91-440, which was not due to differences in transfection since control β-galactosidase activity was not significantly different among transfected cells (Supplementary Fig 2B). Thus, we conclude that supplying the two independent cleavage products is not sufficient to support proper post-translational modification and signaling of TLR9. These data would not exclude the possibility that cleavage must occur at a unique time and place within the cell to allow for appropriate generation of a functional receptor.
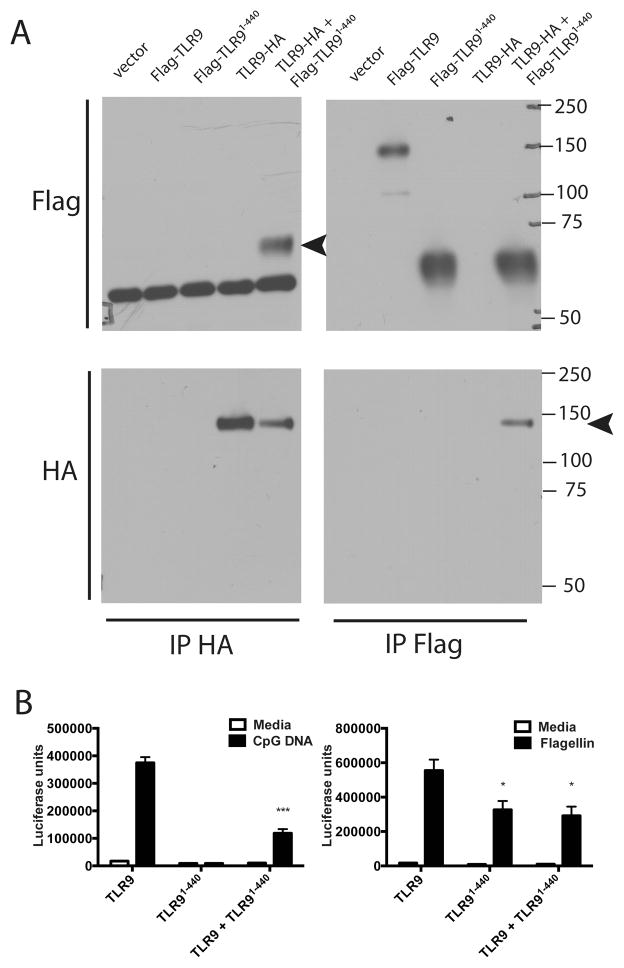
The N-terminal fragment binds directly to full-length TLR9 and inhibits signaling
We previously showed that endogenous human TLR9 is proteolytically cleaved in the ectodomain near the transmembrane domain. This proteolytic event generates a soluble form of the receptor that is capable of binding CpG DNA and inhibiting signaling through the full-length receptor. Additionally, when TLR91-440 was fused to a transmembrane domain, it acted as a dominant negative inhibitor of signaling (27). Thus, we next investigated whether the N-terminal fragment, without a transmembrane domain, could associate with full-length TLR9 and negatively regulate TLR9 signaling. TLR91-440 co-immunoprecipitated full-length TLR9-HA and full-length TLR9 co-immunoprecipitated TLR91-440 suggesting that they directly interact (Fig 6A). It is important to note that this occurs in the absence of UNC93B1 and thus in the absence of TLR9 proteolytic cleavage to TLR9471-1032 (Fig 6A). TLR9 signaling in response to CpG DNA was significantly reduced when TLR91-440 was co-expressed (Fig 6B). Interestingly, we consistently observed a reduction in TLR5 response to flagellin when TLR91-440 was expressed (Fig 6B), but control β-galactosidase activity was unaffected suggesting that the cells were equally transfected (Supplementary Fig 2C). Thus, the N-terminal fragment can negatively regulate TLR9 signaling, and may also associate with flagellin and influence TLR5 signaling.
Figure 6. The N-terminal fragment TLR91-470 binds directly to full-length TLR9 and inhibits signaling.
A) HEK293 cells were transfected with (1.3 ug of a total of 4ug DNA per 6-well plate) empty vector, Flag-TLR9, Flag-TLR91-440, TLR9-HA, or TLR9-HA plus Flag-TLR91-440. Cells were lysed and immunoprecipitated with antibody to HA or Flag, resolved by SDS-PAGE and analyzed by immunoblotting for HA or Flag. Arrows indicate coimmunoprecipitated proteins. Since the same species antibody was used for HA IP and Flag blot, heavy chain bands are observed on the HA IP, Flag blot. These are not seen in the Flag IP, Flag blot since an HRP conjugated anti-Flag was used to develop this blot. This experiment was performed 3 times. B) HEK293 cells were transfected with (5ng/well of a total of 200ng/well of a 96-well plate) TLR9, TLR91-440, TLR9 plus TLR91-440 for 24 hours. Cells were stimulated with media, 3 μM CpG DNA or 100ng/ml flagellin for 18 hours, lysed and assayed for luciferase activity. ***p<.01, *p<.05. This experiment was performed 4 times.
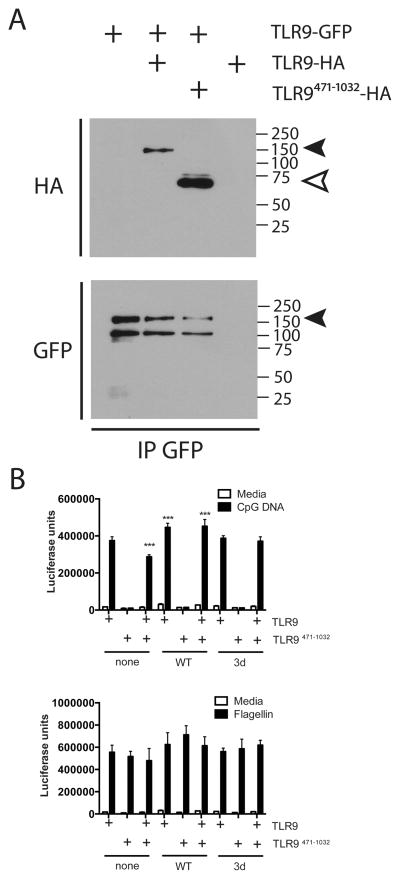
TLR9471-1032 binds to full-length TLR9 and is a dominant negative regulator of TLR9 signaling
The crystal structure of TLR9 shows that protein-protein interactions occur at multiple places within the leucine rich repeats from LRR2 to LRR26 and also in the LRR C-terminal cap region (22). Since a significant number of amino acids in the mature form are involved in these associations, we asked whether TLR9471-1032 could interact with full-length TLR9. Both full-length TLR9-HA and TLR9471-1032 co-immunoprecipitated with full-length TLR9-GFP (Fig 7A). When TLR9471-1032 was co-expressed with full-length TLR9, signaling was reduced (Fig 7B). Interestingly, this inhibitory activity was lost if UNC93B1 (WT or 3d mutant) was co-expressed (Fig 7B). Response to flagellin was unaffected (Fig 7B), but we again observed an increase in control β-galactosidase activity when either WT or 3d mutant of UNC93B1 was expressed (Supplementary Fig 2D). Thus, full-length TLR9 forms dimers with TLR9471-1032, but when UNC93B1 is overexpressed, there may be preferential trafficking of full-length TLR9, and thus a reduced impact of TLR9471-1032 on signaling. Together, these data suggest that TLR9 must be proteolytically cleaved from the full-length form to generate a functional form of the receptor and that expression of TLR9471-1032 by itself results in improper glycosylation and trafficking that interfere with proper function of full-length TLR9.
Figure 7. TLR9471-1032 is a dominant negative regulator of TLR9 signaling.
A) HEK293 cells were transfected with TLR9-GFP alone (2 ug of a total of 4ug DNA per 6-well plate) or with TLR9-HA or TLR9471-1032-HA. Cells were lysed and immunoprecipitated with antibody to GFP, resolved by SDS-PAGE and analyzed by immunoblotting for HA and GFP. This experiment was performed 2 times. B) HEK293 cells were transfected with TLR9, TLR9471-1032, or TLR9 plus TLR9471-1032 (5ng/well of a total of 200ng/well of a 96-well plate) with or without UNC93B1 (0.25ng/well) for 24 hours. Cells were stimulated with media, 3 μM CpG DNA, or 100ng/ml flagellin for 18 hours, lysed and assayed for luciferase activity. ***p<.01 compared to none-TLR9. This experiment was performed 4 times.
Discussion
Proteolytic processing of nucleic acid sensing TLRs has been proposed to be a major regulatory mechanism that occurs in endosomes (23, 24, 26). Since microbes are phagocytosed, killed, and digested in phagosomes, the phagosome is the logical place to localize nucleic acid sensing receptors. Forced expression of TLR9 to the cell surface by fusion with an ist2 domain, or fusion to the transmembrane and cytoplasmic tail of TLR4 prevents signaling in response to viral infection, reinforcing the importance of correct localization for microbial nucleic acid recognition by TLRs (12, 23).
Proteolytic cleavage in the phagosome proceeds in a stepwise fashion and requires cathepsins and asparaginyl endopeptidase (25, 34). Addition of multiple cathepsin inhibitors in combination was required to prevent cleavage and signaling, suggesting that there may be some redundancy in the enzymes required for TLR9 processing (24). We recently showed that endogenous TLR9 is proteolytically cleaved at a site near the transmembrane domain to generate a soluble negative regulator of signaling (26), and that this proteolytic event was not dependent on low pH, unlike the other cleavage event. Thus, proteolytic processing of TLR9 is complex and involves multiple steps and multiple enzymes.
It has been suggested that TLR9 processing is a required to permit signaling, yet there are several pieces of data that fail to support this model. Some studies, including this study, show that a fragment of TLR9 corresponding to the proteolytically cleaved form, when expressed in cells, does not support signaling in response to CpG DNA (32). Furthermore, in HEK293 cells, little to no proteolytic processing of TLR9 occurs in the absence of exogenously expressed UNC93B1, yet these cells support TLR9 signaling (Fig 4A, C), suggesting that processing is not absolutely required for signaling. Even more striking is the observation that forced surface expression of TLR9 by mutation of amino acids in the transmembrane domain results in poorly cleaved TLR9 that responds well to CpG DNA and host DNA, and initiates an autoinflammatory syndrome in mice (12). Thus, TLR9 appears to be capable of signaling in the absence of cleavage.
A recent structural analysis of TLR9 showed that full-length ectodomain of TLR9 bound both inhibitory and active DNA. Interestingly, when cocrystallized, TLR9 formed dimers in association with active CpG DNA but remained monomeric when in association with inhibitory DNA (22). When the mature processed form of TLR9 was generated using V8 protease and compared to the full-length ectodomain, the processed TLR9 form dimerizes more than the full-length protein (22). However, mapping of protein-protein interaction domains on the crystals revealed many amino acids involved in the interface were located throughout the N-terminal LRRs1-14, the C-terminal LRRs 15–26 and the LRR-CT, yet the agonist-binding DNA interface was primarily confined to the first 12 LRRs (22). Thus, it is unclear why some studies have found that the TLR9471-1032 directly associates with CpG DNA (23). Our studies suggest that if TLR9471-1032 is not generated from full-length TLR9, it is not properly glycosylated and can act as a dominant negative inhibitor of signaling (Fig 5B, 7B). Thus, although proteolytic cleavage is likely required, we propose that cleavage must occur at the same time and in the same location as association with ligand. Furthermore, TLR91-440 remains associated with the complex (24) for signaling to occur (32). The exact biochemical processing and involvement in signaling remains unclear and will require additional follow-up study.
TLR9-deficient mice express some partial mRNA species that results in production of the first 96 amino acids fused to the neomycin gene (35). Since there are agonist-ligand binding amino acids located in this region (22), it remains possible that a small N-terminal fragment is generated in TLR9 deficient mice that could associate at low affinity with CpG DNA. Thus it is interesting that two different studies obtained conflicting results regarding the function of the N-terminal fragment. Onji et al suggest it is required for signaling (32), while Lee et al suggested that the N-terminal fragment associates with full-length TLR9 and is a negative regulator (27). We have observed that our soluble TLR9 proteolytic fragment can be additionally cleaved to generate the same N-terminal fragment (data not shown and (26)). Thus it is possible that the negative regulatory function of soluble TLR9 could be due to generation of this fragment. We, too, saw that co-expression of the N-terminal fragment with TLR9 was inhibitory for signaling, and that when expressed in cells the N-terminal fragment bound to full-length TLR9 (Fig 6). However, we also detect an interaction between the expressed mature proteolytic form and full-length TLR9, and found that the mature TLR9 can negatively regulate signaling (Fig 7). Thus there are multiple proteolytic cleavage events generating multiple different fragments of TLR9 involved in regulating signaling. Much remains to be investigated, but it is clear that elaborate regulatory mechanisms have developed to control TLR9.
Supplementary Material
Acknowledgments
We would like to thank Bruce Beutler for the UNC93B1 plasmids, Greg Barton for the TLR9471-1032 -HA plasmid, and Boyoun Park for the TLR9471-1032 -myc plasmid. The following reagent was obtained through BEI Resources, NIAID, NIH: Macrophage Cell Line Derived from TLR9 Knockout Mice, NR-9569. We would also like to thank E. Gruber and C. Heyward for critical reading of the manuscript and helpful suggestions.
References
- 1.Hornung V. SnapShot: Nucleic acid immune sensors, part 2. Immunity. 2014;41:1066–1066. e1061. doi: 10.1016/j.immuni.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Hornung V. SnapShot: nucleic acid immune sensors, part 1. Immunity. 2014;41:868, 868 e861. doi: 10.1016/j.immuni.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Chockalingam A, Brooks JC, Cameron JL, Blum LK, Leifer CA. TLR9 traffics through the Golgi complex to localize to endolysosomes and respond to CpG DNA. Immunol Cell Biol. 2009;87:209–217. doi: 10.1038/icb.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chockalingam A, Rose WA, 2nd, Hasan M, Ju CH, Leifer CA. Cutting edge: a TLR9 cytoplasmic tyrosine motif is selectively required for proinflammatory cytokine production. J Immunol. 2012;188:527–530. doi: 10.4049/jimmunol.1102713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 6.Leifer CA, Brooks JC, Hoelzer K, Lopez J, Kennedy MN, Mazzoni A, Segal DM. Cytoplasmic targeting motifs control localization of toll-like receptor 9. J Biol Chem. 2006;281:35585–35592. doi: 10.1074/jbc.M607511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leifer CA, Kennedy MN, Mazzoni A, Lee C, Kruhlak MJ, Segal DM. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J Immunol. 2004;173:1179–1183. doi: 10.4049/jimmunol.173.2.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh H, Tatematsu M, Watanabe A, Iwano K, Funami K, Seya T, Matsumoto M. UNC93B1 physically associates with human TLR8 and regulates TLR8-mediated signaling. PLoS One. 2011;6:e28500. doi: 10.1371/journal.pone.0028500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishii N, Funami K, Tatematsu M, Seya T, Matsumoto M. Endosomal localization of TLR8 confers distinctive proteolytic processing on human myeloid cells. J Immunol. 2014;193:5118–5128. doi: 10.4049/jimmunol.1401375. [DOI] [PubMed] [Google Scholar]
- 10.Iavarone C, Ramsauer K, Kubarenko AV, Debasitis JC, Leykin I, Weber AN, Siggs OM, Beutler B, Zhang P, Otten G, D’Oro U, Valiante NM, Mbow ML, Visintin A. A point mutation in the amino terminus of TLR7 abolishes signaling without affecting ligand binding. J Immunol. 2011;186:4213–4222. doi: 10.4049/jimmunol.1003585. [DOI] [PubMed] [Google Scholar]
- 11.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 12.Mouchess ML, Arpaia N, Souza G, Barbalat R, Ewald SE, Lau L, Barton GM. Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation. Immunity. 2011;35:721–732. doi: 10.1016/j.immuni.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks JC, Sun W, Chiosis G, Leifer CA. Heat shock protein gp96 regulates Toll-like receptor 9 proteolytic processing and conformational stability. Biochem Biophys Res Commun. 2012 doi: 10.1016/j.bbrc.2012.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasan M, Gruber E, Cameron J, Leifer CA. TLR9 stability and signaling are regulated by phosphorylation and cell stress. J Leukoc Biol. 2016 doi: 10.1189/jlb.2A0815-337R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blasius AL, Arnold CN, Georgel P, Rutschmann S, Xia Y, Lin P, Ross C, Li X, Smart NG, Beutler B. Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:19973–19978. doi: 10.1073/pnas.1014051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, Shibata T, Akashi-Takamura S, Kiyokawa T, Wakabayashi Y, Tanimura N, Kobayashi T, Matsumoto F, Fukui R, Kouro T, Nagai Y, Takatsu K, Saitoh S, Miyake K. A protein associated with Toll-like receptor (TLR) 4 (PRAT4A) is required for TLR-dependent immune responses. J Exp Med. 2007;204:2963–2976. doi: 10.1084/jem.20071132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasai M, Linehan MM, Iwasaki A. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329:1530–1534. doi: 10.1126/science.1187029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Li Z. Endoplasmic reticulum HSP90b1 (gp96, grp94) optimizes B-cell function via chaperoning integrin and TLR but not immunoglobulin. Blood. 2008;112:1223–1230. doi: 10.1182/blood-2008-03-143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Yang Y, Qiu Z, Staron M, Hong F, Li Y, Wu S, Hao B, Bona R, Han D, Li Z. Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat Commun. 2010;1:79. doi: 10.1038/ncomms1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Randow F, Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat Cell Biol. 2001;3:891–896. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- 22.Ohto U, Shibata T, Tanji H, Ishida H, Krayukhina E, Uchiyama S, Miyake K, Shimizu T. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature. 2015;520:702–705. doi: 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- 23.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, Lennon-Dumenil AM, Amigorena S, Cabanie L, Manoury B. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Chockalingam A, Cameron JL, Brooks JC, Leifer CA. Negative Regulation of Signaling by a Soluble Form of Toll-Like Receptor 9. Eur J Immunol. 2011;41:2176–2184. doi: 10.1002/eji.201041034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Kang D, Ra EA, Lee TA, Ploegh HL, Park B. Negative self-regulation of TLR9 signaling by its N-terminal proteolytic cleavage product. J Immunol. 2014;193:3726–3735. doi: 10.4049/jimmunol.1400210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasan M, Ruksnis C, Wang Y, Leifer CA. Negative Regulation of TLR3 siganling by an antimicrobial peptide. J Immunol. 2011;187:5653–5659. doi: 10.4049/jimmunol.1102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ju CH, Chockalingam A, Leifer CA. Early response of mucosal epithelial cells during Toxoplasma gondii infection. J Immunol. 2009;183:7420–7427. doi: 10.4049/jimmunol.0900640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, Shamel L, Herskovits AA, Portnoy DA, Cooke M, Tarantino LM, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 32.Onji M, Kanno A, Saitoh S, Fukui R, Motoi Y, Shibata T, Matsumoto F, Lamichhane A, Sato S, Kiyono H, Yamamoto K, Miyake K. An essential role for the N-terminal fragment of Toll-like receptor 9 in DNA sensing. Nat Commun. 2013;4:1949. doi: 10.1038/ncomms2949. [DOI] [PubMed] [Google Scholar]
- 33.Kasperkovitz PV, Cardenas ML, Vyas JM. TLR9 is actively recruited to Aspergillus fumigatus phagosomes and requires the N-terminal proteolytic cleavage domain for proper intracellular trafficking. J Immunol. 2010;185:7614–7622. doi: 10.4049/jimmunol.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208:643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.