Abstract
Complete genes encoding the predicted nucleoprotein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), M2-1protein, M2-2protein, small hydrophobic protein (SH), and attachmentprotein (G) of seven newly isolated human metapneumoviruses (hMPVs) were analyzed and compared with previously published data for hMPV genes. Phylogenetic analysis of the nucleotide sequences indicated that there were two genetic groups, tentatively named groups 1 and 2, similar to the grouping of human respiratory syncytial virus. Although the predicted amino acid sequences of N, P, M, F, and M2 were highly conserved between the two groups (amino acid identities, 96% for N, 85% for P, 97% for M, 94% for F, 95% for M2-1, and 90% for M2-2), the amino acid identities of the SH and G proteins were low (SH, 58%; G, 33%). Furthermore, each group could be subdivided into two subgroups by phylogenetic analysis, tentatively named subgroups 1A and 1B and subgroups 2A and 2B. The predicted amino acid sequences of G within members of each subgroup were highly conserved (amino acid identities, 88% for group 1A, 93% for group 1B, and 96% for group 2B). The G of hMPV is thought to be the major antigenic determinant and to play an important role in the production of neutralizing antibodies. Clarification of the antigenic diversity of G is important for epidemiological analysis and for establishment of strategies to prevent hMPV infection.
Human metapneumovirus (hMPV), first isolated in The Netherlands in 2001, is a member of the genus Metapneumovirus of the subfamily Pneumovirinae of the family Paramyxoviridae (37). hMPV causes upper respiratory tract infections and flu-like illnesses (5, 32); but it is also associated with lower respiratory tract infections, such as wheezing bronchitis, bronchitis, bronchiolitis, and pneumonia, in very young children, elderly individuals, and immunocompromised patients (7, 10, 11, 15, 30).
The genomic organization of hMPV is 3′-N-P-M-F-M2-SH-G-L-5′ (36, 37). The M2 gene contains two open reading frames (ORFs), M2-1 and M2-2. Although the predicted hMPV proteins have not yet been identified or functionally analyzed, hMPV is thought to encode the following nine proteins: N, the nucleocapsid RNA binding protein; P, the nucleocapsid phosphoprotein; M, the nonglycosylated matrix protein; F, the fusion glycoprotein; M2-1, the transcription elongation factor; M2-2, the RNA synthesis regulatory factor; SH, the small hydrophobic surface protein; G, the major attachment protein; and L, the major polymerase subunit (4, 36, 37).
Pneumoviruses encode two major surface glycoproteins (glycoproteins F and G). F promotes fusion of the viral and cell membranes, allowing penetration of the viral ribonucleoprotein into the cell cytoplasm (39), and G mediates virus binding to the cell receptor (24). G is known to be the most variable protein in human respiratory syncytial virus (hRSV) and avian pneumovirus (APV) (8, 22). In both viruses there are at least two distinct groups. For hRSV G, there is 53% identity in amino acid sequences between groups A and B (19), whereas there is only 38% identity in the amino acid sequences between types A and B for APV G (20). Neutralizing antibodies against F and G are important for protection against hRSV infection. The antibody against F protects animals from viruses of both hRSV group A and hRSV group B (17, 18, 28). G induces an antibody response protective only against infections caused by one hRSV group (19, 40).
The sequences of several hMPV genes (the genes for N, P, M, F, and L) and gene fragments have been determined, and the results of phylogenetic analysis have shown that hMPV can be divided into two major groups (2, 3, 5, 6, 9, 25, 29, 38). However, sequence data for G are limited (4, 37). In the present study, we determined the nucleotide sequences of putative ORFs for N, P, M, F, M2-1, M2-2, SH, and G of seven newly isolated hMPVs and compared them with the previously published sequences for hMPV.
MATERIALS AND METHODS
Virus propagation.
Seven hMPV isolates (9) were propagated in LLC-MK2 cells in vitro. The isolates were from nasopharyngeal swab samples of seven Japanese patients (age range, 8 months to 3 years) with lower respiratory tract infections obtained during the winter and spring of 2003-2004 in Sapporo, Japan. The cells were cultured in Eagle's minimum essential medium for 3 to 4 weeks, with the medium changed weekly. Trypsin (Sigma-Aldrich) was added to the medium at a concentration of 1 μg/ml.
RNA extraction, cDNA synthesis, RT-PCR, and sequencing.
Virus RNA was extracted from inoculated LLC-MK2 cells by the RNAzol B (TEL-TEST, Inc., Friendswood, Tex.) method, according to the protocol of the manufacturer. Approximately 1 μg of each RNA sample was incubated in a solution containing 100 ng of random hexadeoxynucleotides and 200 U of Moloney murine leukemia virus reverse transcriptase (First-Strand cDNA Synthesis kit; Amersham Pharmacia Biotech, Piscataway, N.J.) in a final volume of 15 μl at 37°C for 1 h to synthesize cDNA. The cDNA (0.3 μl) was subjected to reverse transcription-PCR (RT-PCR). The RT-PCR mixture consisted of 100 μmol of each deoxyribonucleotide, 1.0 U of AmpliTaq Gold, 50 mmol of potassium chloride per liter, 10 mmol of Tris-HCl (pH 8.3) per liter, 1.5 mmol of magnesium chloride per liter, 0.01% (wt/vol) of gelatin, 10 pmol of each primer, and cDNA in a volume of 25 μl. The PCR conditions were as follows: 94°C for 10 min, followed by 35 cycles of 94°C for 1 min, 50°C for 2 min, and 72°C for 3 min. The PCR products were sequenced directly by using a BigDye Dye terminator cycle sequencing ready reaction kit (Perkin-Elmer Applied Biosystems, Tokyo, Japan) with an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems).
Strategy for sequencing.
Specific primers were designed on the basis of the total sequence of hMPV strain 00-1 and the partial sequence of isolate JPS02-76, which were obtained previously (data not shown). Fragments ranging from 1,000 to 3,000 bp were generated by RT-PCR. The PCR product was sequenced directly by the genome-walking method. The sequences were subsequently confirmed by generating overlapping RT-PCR fragments. The genomes of hMPV isolates were sequenced from the start of the gene for N to the noncoding sequences between the genes for G and L.
Nucleotide sequences.
The GenBank database nucleotide sequence accession numbers for the pneumovirus strains and proteins used in this study are AF371337 for strain hMPV 00-1, AY297749 for strain hMPV CAN97-83, AY297748 for strain hMPV CAN98-75, AF176590 for the APV type C (APVC) gene for N, AF176591 for the APVC gene for P, AF262571 for the APVC gene for M, AY579780 for the APVC gene for F, AF176592 for the APVC gene for M2, APN457967 for the APVC gene for SH, and APN457967 for the APVC gene for G.
Analysis of nucleotide and amino acid sequences.
Sequences were aligned by use of the CLUSTAL W program (35). The transmembrane region was predicted by the method of Stoffel et al. (33), available online at the Swiss Institute for Experimental Cancer Research (http://www.ch.embnet.org/software/TMPRED_form.html). ORFs were predicted by using ORF finder software (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). O-linked glycosylation and N-linked glycosylation were predicted by using NetOGlyc2.0 (14) and NetNGlyc1.0 (13) software, respectively; all software was obtained from the Center for Biological Sequence Analysis, Technical University of Denmark, Lyngby, Denmark (http://www.cbs.dtu.dk/services/). Local similarities between nucleotide and amino acid sequences were studied by use of the BLAST 2 algorithm (1).
Phylogeny.
The nucleotide sequences of the genes for N, P, M, F, M2-1, M2-2, SH, and G were assembled by using CLUSTAL W software. Phylogenetic trees were generated by the neighbor-joining method with the MEGA program (21).
Nucleotide sequence accession numbers.
The hMPV sequences presented in this paper have been deposited in GenBank under accession numbers AY530089 to AY530095 for strains JPS02-76, JPS03-176, JPS03-178, JPS03-180, JPS03-187, JPS03-194, and JPS03-240, respectively.
RESULTS
Phylogenetic analysis.
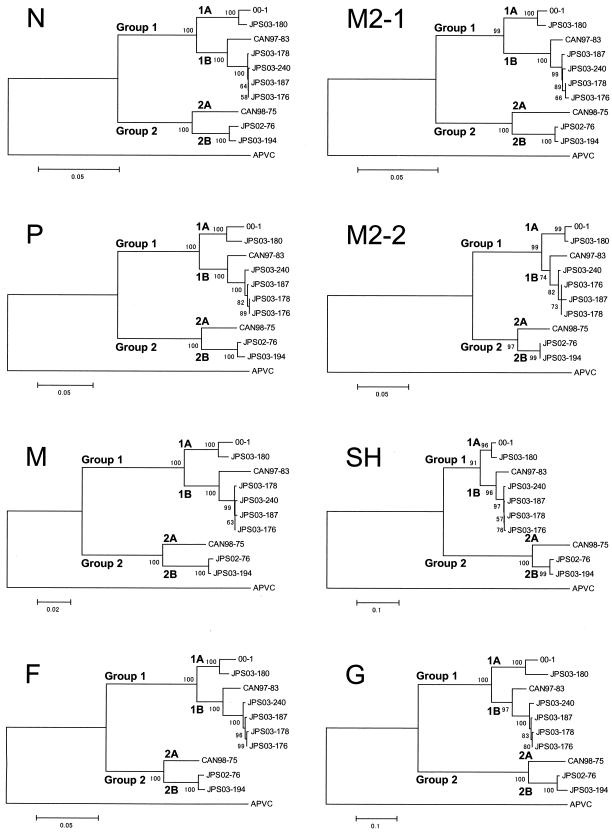
Phylogenetic analyses were based on the putative ORFs for N, P, M, F, M2-1, M2-2, SH, and G of seven hMPV isolates, three published full-length hMPV sequences (for isolates 00-1, CAN97-83, and CAN98-75), and the APVC sequence (Fig. 1). Ten hMPV strains were divided into two major genetic groups that were previously tentatively named groups 1 and 2 (2, 5). The hMPV Dutch isolate characterized by van den Hoogen et al. (37) (isolate 00-1) belongs to group 1. The results of our phylogenetic analysis are consistent with data previously reported for the partial or complete sequences of the genes for N, P, M, F, and L of hMPV isolates from The Netherlands (37), Canada (2, 5, 31), Germany (38), Italy (25), and Japan (9). Furthermore, seven hMPV strains in group 1 were subdivided into two subgroups, tentatively named subgroup 1A (strains 00-1 and JPS03-180) and subgroup 1B (strains CAN97-83, JPS03-176, JPS03-178, JPS03-187, and JPS03-240). Three hMPV strains in group 2 were also subdivided into two subgroups, tentatively named subgroup 2A (strain CAN98-75) and subgroup 2B (strains JPS02-76 and JPS03-194).
FIG. 1.
Phylogenetic analysis of hMPV isolates. The putative ORFs for N, P, M, F, M2-1, M2-2, SH, and G were analyzed. The corresponding gene sequences from APVC were also analyzed. Bootstrap proportions were plotted at the main internal branches of the phylogram to show support values. Note that the scale of each tree was different. Phylogenetic analysis was performed by use of the neighbor-joining method of the MEGA program.
ORFs and proteins of hMPV isolates.
Table 1 shows the lengths of the putative ORFs for N, P, M, F, M2-1, M2-2, SH, and G and the predicted amino acid sequences of isolates 00-1, CAN97-83, and CAN98-75 and seven hMPV isolates. The calculated molecular masses of the predicted proteins are also shown. The lengths of the coding regions of the putative genes for N, P, M, F, M2-1, and M2-2 and the predicted amino acid sequences were consistent between isolates (N, 1,185 bp and 394 amino acids [aa]; P, 885 bp and 294 aa; M, 765 bp and 254 aa; F, 1620 bp and 539 aa; M2-1, 564 bp and 187 aa; and M2-2, 216 bp and 71 aa). The lengths of the coding region of the putative gene for the SH protein and the predicted amino acid sequence were different for strains in groups 1 and 2 (group 1, 552 bp and 183 aa; group 2, 534 bp and 177 aa) except for isolate CAN97-83 (540 bp and 179 aa), which had a stop codon (TAA) at position 541 from the start of the gene for SH. The lengths of the coding region of the putative gene for G and predicted amino acid sequences were different for strains in subgroups 1A, 1B, 2A, and 2B (subgroup 1A, 711 bp and 236 aa; subgroup 1B, 660 bp and 219 aa; subgroup 2A, 711 bp and 236 aa; and subgroup 2B, 696 bp and 231 aa).
TABLE 1.
Lengths of putative ORFs and predicted amino acids of hMPV isolates
| Group, subgroup, and isolate | N
|
P
|
M
|
F
|
M2-1
|
M2-2
|
SH
|
G
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nt (aa)a | MMb | nt (aa) | MM | nt (aa) | MM | nt (aa) | MM | nt (aa) | MM | nt (aa) | MM | nt (aa) | MM | nt (aa) | MM | |
| Group 1 | ||||||||||||||||
| Subgroup 1A | ||||||||||||||||
| 00-1 | 1,185 (394) | 43.5 | 885 (294) | 32.5 | 765 (254) | 27.6 | 1,620 (539) | 58.5 | 564 (187) | 21.2 | 216 (71) | 8.2 | 552 (183) | 20.9 | 711 (236) | 25.8 |
| JPS03-180 | 1,185 (394) | 43.5 | 885 (294) | 32.6 | 765 (254) | 27.6 | 1,620 (539) | 58.5 | 564 (187) | 21.2 | 216 (71) | 8.2 | 552 (183) | 21.1 | 711 (236) | 25.7 |
| Subgroup 1B | ||||||||||||||||
| CAN97-83 | 1,185 (394) | 43.5 | 885 (294) | 32.7 | 765 (254) | 27.6 | 1,620 (539) | 58.5 | 564 (187) | 21.2 | 216 (71) | 8.2 | 540 (179) | 20.6 | 660 (219) | 23.7 |
| JPS03-176 | 1,185 (394) | 43.5 | 885 (294) | 32.7 | 765 (254) | 27.6 | 1,620 (539) | 58.5 | 564 (187) | 21.3 | 216 (71) | 8.2 | 552 (183) | 21.1 | 660 (219) | 23.8 |
| JPS03-178 | 1,185 (394) | 43.5 | 885 (294) | 32.7 | 765 (254) | 27.6 | 1,620 (539) | 58.5 | 564 (187) | 21.3 | 216 (71) | 8.2 | 552 (183) | 21.1 | 660 (219) | 23.8 |
| JPS03-187 | 1,185 (394) | 43.5 | 885 (294) | 32.7 | 765 (254) | 27.6 | 1,620 (539) | 58.6 | 564 (187) | 21.3 | 216 (71) | 8.1 | 552 (183) | 21.1 | 660 (219) | 23.8 |
| JPS03-240 | 1,185 (394) | 43.5 | 885 (294) | 32.7 | 765 (254) | 27.6 | 1,620 (539) | 58.6 | 564 (187) | 21.3 | 216 (71) | 8.2 | 552 (183) | 21.1 | 660 (219) | 23.7 |
| Group 2 | ||||||||||||||||
| Subgroup 2A, CAN98-75 | 1,185 (394) | 43.6 | 885 (294) | 32.5 | 765 (254) | 27.6 | 1,620 (539) | 58.4 | 564 (187) | 21.2 | 216 (71) | 8.2 | 534 (177) | 20.4 | 711 (236) | 25.5 |
| Subgroup 2B | ||||||||||||||||
| JPS02-76 | 1,185 (394) | 43.6 | 885 (294) | 32.5 | 765 (254) | 27.6 | 1,620 (539) | 58.5 | 564 (187) | 21.2 | 216 (71) | 8.2 | 534 (177) | 20.4 | 696 (231) | 25.4 |
| JPS03-194 | 1,185 (394) | 43.6 | 885 (294) | 32.4 | 765 (254) | 27.6 | 1,620 (539) | 58.5 | 564 (187) | 21.2 | 216 (71) | 8.2 | 534 (177) | 20.3 | 696 (231) | 25.3 |
nt, length of putative ORF (in base pairs).; aa, length of predicted amino acid sequence.
MM, estimated molecular mass (in kilodaltons) calculated from the sequences.
Table 2 shows the levels of identity of the nucleotide and amino acid sequences of the various hMPV ORFs between and within groups. Between two groups, the ORFs for N, P, M, M2-1, and M2-2 shared identities of 81 to 86%, on average, whereas the ORFs for the three surface proteins (F, SH, and G) were more divergent (F, 60%; SH, 67%; G, 58%). The predicted amino acid sequences of N, P, M, M2-1, and M2-2 were highly (85 to 97%) conserved between two groups, as expected. While the predicted amino acid sequences of SH and G were divergent between two groups (58 and 33%, respectively), as expected, those of F were surprisingly well conserved (94%).
TABLE 2.
Levels of identity of nucleotide and amino acid sequences within and between two hMPV groups
| Protein | % Nucleotide sequence identity within groupa
|
% Nucleotide sequence identity between groupsa | % Amino acid sequence identity within groupa
|
% Amino acid sequence identity between groupsa | ||
|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 1 | Group 2 | |||
| N | 94-100 (96) | 95-99 (96) | 86-87 (86) | 99-100 (99) | 98-100 (99) | 95-96 (96) |
| P | 92-100 (95) | 98-99 (95) | 80-82 (81) | 94-99 (97) | 95-99 (97) | 84-86 (85) |
| M | 94-99 (97) | 95-99 (96) | 84-86 (85) | 98-100 (99) | 99-100 (99) | 96-97 (97) |
| F | 85-100 (92) | 81-98 (87) | 55-61 (60) | 98-99 (98) | 98-99 (98) | 93-94 (94) |
| M2-1 | 94-99 (97) | 94-99 (96) | 86-87 (86) | 96-100 (98) | 97-100 (98) | 94-95 (95) |
| M2-2 | 95-100 (97) | 95-100 (97) | 85-87 (86) | 95-100 (97) | 97-100 (98) | 88-90 (90) |
| SH | 90-99 (94) | 88-98 (91) | 66-69 (67) | 83-99 (92) | 81-98 (88) | 54-60 (58) |
| G | 75-99 (82) | 78-97 (85) | 56-60 (58) | 61-99 (78) | 63-96 (75) | 31-35 (33) |
Values are ranges (average) of identity obtained for comparison of strains within each group and between groups.
Within each group, the ORFs for N, P, M, M2-1, and M2-2 shared high degrees of identity, on average, 95 to 97%; and the predicted amino acid sequences of these proteins were highly conserved (97 to 99%) (Table 2). The ORFs for F and SH were well conserved within the two groups (F, 92 and 87%, respectively; SH, 94 and 91%, respectively), and the predicted amino acid sequences of F and SH were also well conserved (F, 98% for both groups; SH, 92 and 88%, respectively). The ORFs for G were moderately identical within groups (82 and 85%, respectively), and the predicted amino acid sequences of G were marginally less conserved (78 and 75%, respectively).
Tables 3 and 4 show the levels of identity of the nucleotide and amino acid sequences of various hMPV ORFs between and within subgroups. The ORFs and the predicted amino acid sequences of G were highly identical within members of each subgroup (nucleotide identities, 92% for subgroup 1A, 96% for subgroup 1B, and 97% for subgroup 2B; amino acid identities, 88% for subgroup 1A; 93% for subgroup 1B, and 96% for subgroup 2B).
TABLE 3.
Levels of identity of nucleotide and amino acid sequences within and between subgroups 1A and 1B
| Protein | % Nucleotide sequence identity within subgroup
|
% Nucleotide sequence identity between subgroups 1A and 1Ba | % Amino acid sequence identity within subgroup
|
% Amino acid sequence identity between subgroups 1A and 1Ba | ||
|---|---|---|---|---|---|---|
| Subgroup 1A | Subgroup 1Ba | Subgroup 1A | Subgroup 1Ba | |||
| N | 99 | 97-100 (99) | 93-94 (94) | 99 | 99-100 (99) | 99-100 (99) |
| P | 97 | 96-100 (98) | 92-93 (92) | 98 | 98-100 (99) | 95-96 (95) |
| M | 99 | 97-100 (99) | 94-95 (94) | 100 | 99-100 (99) | 99 (99) |
| F | 96 | 96-100 (99) | 84-86 (85) | 99 | 98-99 (99) | 98 (98) |
| M2-1 | 99 | 98-100 (99) | 93-94 (94) | 99 | 98-100 (99) | 96-97 (97) |
| M2-2 | 99 | 97-100 (99) | 95-96 (95) | 100 | 97-100 (99) | 95-97 (96) |
| SH | 98 | 95-100 (98) | 90-91 (91) | 97 | 94-100 (97) | 83-97 (86) |
| G | 92 | 92-100 (96) | 75-78 (76) | 88 | 86-100 (93) | 60-65 (62) |
Values are ranges (average) of identity obtained for comparison of strains within and between subgroups 1A and 1B.
TABLE 4.
Levels of identity of nucleotide and amino acid sequences within and between subgroups 2A and 2B
| Protein | % Nucleotide sequence identity within subgroup
|
% Nucleotide sequence identity between subgroups 2A and 2Ba | % Amino acid sequence identity within subgroup
|
% Amino acid sequence identity between subgroups 2A and 2Ba | ||
|---|---|---|---|---|---|---|
| Subgroup 2A | Subgroup 2B | Subgroup 2A | Subgroup 2B | |||
| N | NAb | 99 | 95 (95) | NA | 100 | 98 (98) |
| P | NA | 99 | 93-94 (94) | NA | 99 | 95-96 (95) |
| M | NA | 99 | 95 (95) | NA | 100 | 99 (99) |
| F | NA | 98 | 81-82 (82) | NA | 99 | 98 (98) |
| M2-1 | NA | 100 | 94 (94) | NA | 100 | 97 (97) |
| M2-2 | NA | 100 | 95 (95) | NA | 100 | 97 (97) |
| SH | NA | 98 | 88 (88) | NA | 98 | 82-83 (82) |
| G | NA | 97 | 78-79 (79) | NA | 96 | 64-66 (65) |
Values are ranges (average) of identity obtained for comparison of strains within and between subgroups 2A and 2B.
NA, not applicable because only a single isolate was tested.
SH.
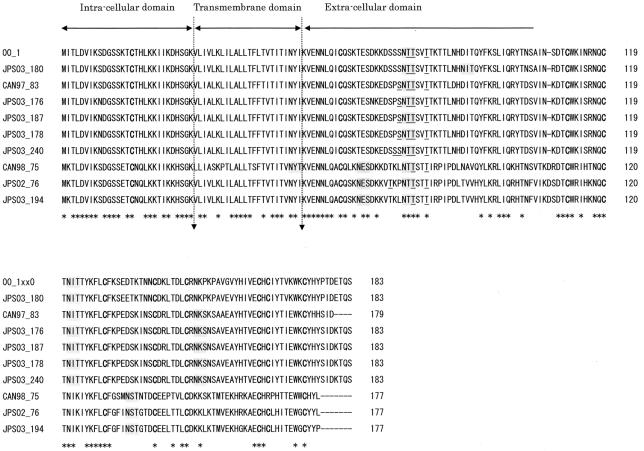
SH is predicted to be a type II glycoprotein that is inserted in the plasma membrane by a hydrophobic signal-anchor sequence located near its amino terminus (Fig. 2). The predicted length of SH in group 1 isolates except for strain CAN97-83 was 183 amino acids, as stated above (Table 1). The predicted length of SH in group 2 isolates was 177 amino acids. The predicted extracellular domain had two to four potential motifs for N-linked glycosylation and two to five potential sites for O-linked glycosylation (14) (Fig. 2). Although the NYT sequence in the CAN98-75 isolate was a consensus N-linked glycosylation site, it is unlikely to be glycosylated within the membrane. The transmembrane domain might be shifted in this sequence, or this site might not be glycosylated. The predicted SH sequences contained 9 or 10 cysteine residues, which were mostly in the extracellular domain, and 9 of these were conserved among all strains.
FIG. 2.
Comparison of the predicted amino acid sequences of SH of hMPV isolates. The predicted amino acid sequence of SH of hMPV with cysteine residues is shown in boldface type, potential N-linked glycosylation sites are shaded in gray, and potential O-glycosylation sites are underlined. Dashes indicate gaps introduced to maximize the alignment or to denote the absence of corresponding amino acids. The asterisks underneath each alignment denote amino acid identity among all sequences. Proposed intracellular, transmembrane, and extracellular domains are indicated above the sequences.
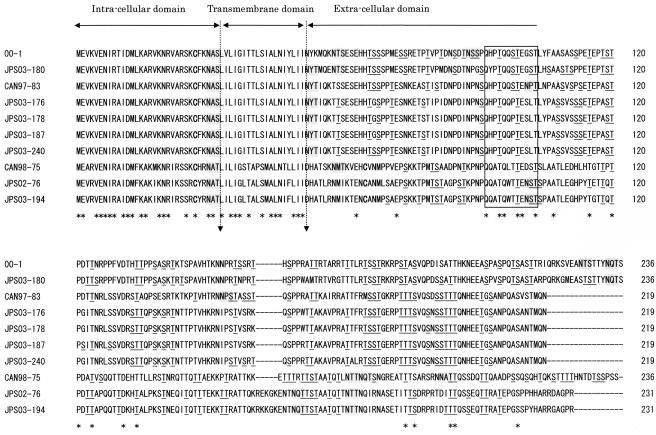
G.
G is also predicted to be a type II glycoprotein (Fig. 3). The predicted lengths of G in subgroup 1A, 1B, 2A, and 2B isolates were 236, 219, 236, and 231 amino acids, respectively. The predicted extracellular domain had one to four potential motifs for N-linked glycosylation and more than 40 potential sites for O-linked glycosylation (Fig. 3). The NAS and NAT N-linked glycosylation sequences within the intracellular domain were potential sites but are unlikely to be used. G contained one or two cysteine residues, one of which was conserved in the intracellular domain. In group 2 isolates, G had two cysteine residues, one in the intracellular domain and the other in the extracellular domain; but those in group 1 isolates had only one residue.
FIG. 3.
Comparison of the predicted amino acid sequences of G of hMPV isolates. The predicted amino acid sequence of G of hMPV with cysteine residues is shown in boldface type, potential N-linked glycosylation sites are shaded in gray, and potential O-glycosylation sites are underlined. Dashes indicate gaps introduced to maximize the alignment or to denote the absence of corresponding amino acids. The asterisks underneath each alignment denote amino acid identity among all sequences. Proposed intracellular, transmembrane, and extracellular domains are indicated above the sequences. The square indicates the positions of conserved amino acid residues.
Noncoding sequences.
hMPV contains a 3′ leader region, followed by nine putative genes and a 5′ trailer region (4, 22, 36). The transcriptional control sequences are conserved at the beginning (gene-start) and end (gene-end) of each gene. Intergenic sequences are located between the gene boundaries. Figure 4 shows the putative gene-start, gene-end, and intergenic sequences of the seven new hMPV isolates and three strains described previously. The overall gene-start signals (GGGAC/UAAA/GU) for the genes for P, M, F M2, SH, and G identified in the three strains described previously were well conserved in the seven new isolates (4, 36). As described before (4, 36), there was an additional ATG codon upstream of the gene-start motif of the gene for SH (Fig. 4).
FIG. 4.
Predicted gene boundaries, cis-acting gene-start and gene-end signals, and intergenic regions of hMPV isolates. The alignments show the N-P, P-M, M-F, F-M2, M2-SH, SH-G, and G boundaries. Conserved sequence motifs at the end and beginning of each gene are indicated by bold uppercase letters, and a consensus sequence is given below. Translational stop and start codons are underlined.
The putative gene-end signals (AGUU/AA/UnnA/UA4-7 where n is any nucleotide) of the genes for N, P, M, F, M2, SH, and G were also conserved in the seven new isolates as well as the three published strains of hMPV (Fig. 4). Strains JPS03-194 (group 2) and CAN97-83 (group 1) had one A-to-U nucleotide substitution in the poly(A) tail.
The lengths of the putative intergenic sequences of the hMPV gene varied (Fig. 4). The intergenic regions of the N-P, P-M, M-F, M2-SH, and SH-G boundaries were 2 to 3, 8, 30 to 34, 10 to 13, and 115 to 126 bp, respectively. The length of the F-M2 boundary differed in both the genetic groups and the subgroups. The F-M2 boundaries in subgroup 1A isolates (isolates 00-1 and JPS03-180) were 41 and 67 bp, respectively, and those in subgroup 1B isolates (isolates CAN97-83, JPS03-176, JPS03-178, JPS03-187, and JPS03-240) and group 2 isolates (isolates CAN98-75, JPS02-76, and JPS03-194) were 13 to 20 and 1 bp, respectively.
DISCUSSION
In the present study, the complete predicted gene sequences of N, P, M, F, M2-1, M2-2, SH, and G of seven newly isolated hMPV isolates were determined and compared with the sequences of three hMPV genomes published previously. Phylogenetic analyses indicated that hMPV isolates could be divided into two major genetic groups, tentatively named groups 1 and 2 (Fig. 1), which is consistent with the partial or complete sequences of the genes for N, P, M, F, and L reported previously (2, 5, 9, 25, 31, 37, 38). Each group could be further divided into two subgroups, tentatively named subgroups 1A and 1B and subgroups 2A and 2B (Fig. 1). The other published sequences of the genes for N, P, M, and F (GenBank accession numbers AY145242 to AY145301, AY256863 to AY256867, AY321506, AY321507, AY355328, AY355335) were classified into four subgroups (data not shown). In a previous phylogenetic analysis based on partial F sequences (80 bp) (9), 57 hMPV isolates were classified into two distinct genetic groups (groups 1 and 2) and also, potentially, into four subgroups. However, due to the small number of hMPV isolates, we cannot conclude that all hMPV isolates can be clustered into one of the four subgroups. The findings for additional sequences from around the world may require modification of the classification, but so far, the classification seems to be appropriate.
The most interesting observation in this study was that putative ORFs for G in hMPVs were classified into four subgroups by phylogenetic analysis, as were other putative ORFs for N, P, M, F, M2, and SH, and that the predicted G amino acid sequences within each subgroup were highly conserved (Tables 3 and 4). The numbers of amino acids in the ORFs within the subgroups were also identical. In hRSV, the numbers of amino acids for G ranged from 289 to 299, but the numbers for G in groups A and B are not the same (8, 12, 26, 27, 34). The nine partial sequences of hMPV G available in GenBank (accession numbers AY327802 to AY327810) belonged to subgroup 1A, and 81% amino acid identity was detected among the G sequences from subgroup 1A (data not shown). Considering that the attachment proteins of paramyxoviruses are one of the major protective antigens (8, 22), immunological pressure might function to cause the genetic diversity of hMPV G in the extracellular domain (Fig. 3). The subgroups of hMPV isolates might be useful not only for investigation of the antigenic variability of hMPV but also for the study of the neutralizing antibodies against homologous or heterologous subgroups of hMPV. In hRSV, although the extracellular domain of G is variable between groups A and B, a strictly conserved 13-amino-acid region in the central extracellular domain is noted (19). This conserved domain contains four closely spaced, exactly conserved cysteine residues and coincides with a tight turn in the predicted structure (23). In contrast, the predicted sequence of the G of hMPV lacked both the conserved 13-amino-acid region and the four cysteine residues (Fig. 3). Instead, hMPV G had a relatively well conserved region from residues 92 to 103, in which 50% (6 of 12) of the amino acids were identical (Fig. 3, square). This part might be involved in the attachment of G to cellular receptors. Cysteine residues of the predicted sequence of the SH of hMPV were well conserved, and this might be helpful in stabilizing the structure of SH.
The sequences of the ORFs of three putative surface proteins (F, SH, and G) were divergent between the two groups (Table 2). The predicted amino acid sequences of F were well conserved between the two groups, but the sequences of SH and G were less well conserved (Table 2). Nucleotide substitutions in the SH extracellular domain and the G extracellular domain were closely associated with amino acid substitutions (SH, 89.1%; G, 91.6%); the rate of amino acid substitutions was low (10.6%) for F (data not shown). This observation is consistent with previously reported findings for the G of hMPV (4) and the G of hRSV (16).
The putative gene-start signals (GGGAC/UAAA/GU) for the genes for P, M, F M2, SH, and G and the putative gene-end signals (AGUU/AA/UnnA/UA4-7) for the genes for N, P, M, F, M2, SH, and G identified in the three published strains were well conserved in the seven newly isolated hMPVs (Fig. 4) (4, 36). Those motifs were similar to those of hRSV (gene-start, GGGGCAAAUU/A; gene-end, AGUU/AAnU/AU/AA/UAAAA) (8). Further study is needed to determine whether those motifs function as gene-start and gene-end signals.
In conclusion, genes encoding the putative N, P, M, F, M2-1, M2-2, SH, and G of seven newly isolated hMPVs were analyzed. Phylogenetic analysis of the nucleotide sequences indicated that there were two genetic groups (groups 1 and 2), which were further subdivided into two subgroups, tentatively named subgroups 1A and 1B and subgroups 2A and 2B. The predicted amino acid sequences of G within members of each subgroup were highly conserved. The G of hMPV is thought to be the major antigenic determinant and to play an important role in the production of neutralizing antibodies. Clarification of the antigenic diversity of G is important for epidemiological analysis and for the establishment of strategies to prevent hMPV infections.
Acknowledgments
This study was partially supported by a grant-in-aid for exploratory research (grant 14657179 [2002]) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a grant-in-aid for the 21st century Center of Excellence for Zoonosis Control from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
We thank Stewart Chisholm for proofreading the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastien, N., S. Normand, T. Taylor, D. Ward, T. C. Peret, G. Boivin, L. J. Anderson, and Y. Li. 2003. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 93:51-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastien, N., D. Ward, P. Van Caeseele, K. Brandt, S. H. Lee, G. McNabb, B. Klisko, E. Chan, and Y. Li. 2003. Human metapneumovirus infection in the Canadian population. J. Clin. Microbiol. 41:4642-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biacchesi, S., M. H. Skiadopoulos, G. Boivin, C. T. Hanson, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315:1-9. [DOI] [PubMed] [Google Scholar]
- 5.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. [DOI] [PubMed] [Google Scholar]
- 6.Boivin, G., G. De Serres, S. Cote, R. Gilca, Y. Abed, L. Rochette, M. G. Bergeron, and P. Dery. 2003. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 9:634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cane, P. A., B. G. van den Hoogen, S. Chakrabarti, C. D. Fegan, and A. D. Osterhaus. 2003. Human metapneumovirus in a haematopoietic stem cell transplant recipient with fatal lower respiratory tract disease. Bone Marrow Transplant. 31:309-310. [DOI] [PubMed] [Google Scholar]
- 8.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, vol. 2, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 9.Ebihara, T., R. Endo, H. Kikuta, N. Ishiguro, H. Ishiko, M. Hara, Y. Takahashi, and K. Kobayashi. 2004. Human metapneumovirus infection in Japanese children. J. Clin. Microbiol. 42:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebihara, T., R. Endo, H. Kikuta, N. Ishiguro, M. Yoshioka, X. Ma, and K. Kobayashi. 2003. Seroprevalence of human metapneumovirus in Japan. J. Med. Virol. 70:281-283. [DOI] [PubMed] [Google Scholar]
- 11.Freymouth, F., A. Vabret, L. Legrand, N. Eterradossi, F. Lafay-Delaire, J. Brouard, and B. Guillois. 2003. Presence of the new human metapneumovirus in French children with bronchiolitis. Pediatr. Infect. Dis. J. 22:92-94. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, O., M. Martin, J. Dopazo, J. Arbiza, S. Frabasile, J. Russi, M. Hortal, P. Perez-Brena, I. Martinez, B. Garcia-Barreno, et al. 1994. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J. Virol. 68:5448-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta, R., and S. Brunak. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function, p. 310-322. In R. B. Altman, A. K. Dunker, L. Hunter, K. Lauderdale, and T. E. Klein (ed.), Proceedings of the Pacific Symposium on Biocomputing 2002. World Scientific Publishing Co., Singapore, Republic of Singapore. [PubMed]
- 14.Hansen, J. E., O. Lund, N. Tolstrup, A. A. Gooley, K. L. Williams, and S. Brunak. 1998. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj. J. 15:115-130. [DOI] [PubMed] [Google Scholar]
- 15.Jartti, T., B. van den Hoogen, R. P. Garofalo, A. D. Osterhaus, and O. Ruuskanen. 2002. Metapneumovirus and acute wheezing in children. Lancet 360:1393-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, P. R., and P. L. Collins. 1989. The 1B (NS2), 1C (NS1) and N proteins of human respiratory syncytial virus (RSV) of antigenic subgroups A and B: sequence conservation and divergence within RSV genomic RNA. J. Gen. Virol. 70:1539-1547. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, P. R., and P. L. Collins. 1988. The fusion glycoproteins of human respiratory syncytial virus of subgroups A and B: sequence conservation provides a structural basis for antigenic relatedness. J. Gen. Virol. 69:2623-2628. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, P. R., Jr., R. A. Olmsted, G. A. Prince, B. R. Murphy, D. W. Alling, E. E. Walsh, and P. L. Collins. 1987. Antigenic relatedness between glycoproteins of human respiratory syncytial virus subgroups A and B: evaluation of the contributions of F and G glycoproteins to immunity. J. Virol. 61:3163-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. USA 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juhasz, K., and A. J. Easton. 1994. Extensive sequence variation in the attachment (G) protein gene of avian pneumovirus: evidence for two distinct subgroups. J. Gen. Virol. 75:2873-2880. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 22.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Field’s virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Langedijk, J. P., W. M. Schaaper, R. H. Meloen, and J. T. van Oirschot. 1996. Proposed three-dimensional model for the attachment of protein G of respiratory syncytial virus. J. Gen. Virol. 77:1249-1257. [DOI] [PubMed] [Google Scholar]
- 24.Levine, S., R. Klaiber-Franco, and P. R. Paradiso. 1987. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J. Gen. Virol. 68:2521-2524. [DOI] [PubMed] [Google Scholar]
- 25.Maggi, F., M. Pifferi, M. Vatteroni, C. Fornai, E. Tempestini, S. Anzilotti, L. Lanini, E. Andreoli, V. Ragazzo, M. Pistello, S. Specter, and M. Bendinelli. 2003. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J. Clin. Microbiol. 41:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez, I., O. Valdes, A. Delfraro, J. Arbiza, J. Russi, and J. A. Melero. 1999. Evolutionary pattern of the G glycoprotein of human respiratory syncytial viruses from antigenic group B: the use of alternative termination codons and lineage diversification. J. Gen. Virol. 80:125-130. [DOI] [PubMed] [Google Scholar]
- 27.Melero, J. A., B. Garcia-Barreno, I. Martinez, C. R. Pringle, and P. A. Cane. 1997. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J. Gen. Virol. 78:2411-2418. [DOI] [PubMed] [Google Scholar]
- 28.Olmsted, R. A., N. Elango, G. A. Prince, B. R. Murphy, P. R. Johnson, B. Moss, R. M. Chanock, and P. L. Collins. 1986. Expression of the F glycoprotein of respiratory syncytial virus by a recombinant vaccinia virus: comparison of the individual contributions of the F and G glycoproteins to host immunity. Proc. Natl. Acad. Sci. USA 83:7462-7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peiris, J. S., W. H. Tang, K. H. Chan, P. L. Khong, Y. Guan, Y. L. Lau, and S. S. Chiu. 2003. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg. Infect. Dis. 9:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelletier, G., P. Dery, Y. Abed, and G. Boivin. 2002. Respiratory tract reinfections by the new human metapneumovirus in an immunocompromised child. Emerg. Infect. Dis. 8:976-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peret, T. C., G. Boivin, Y. Li, M. Couillard, C. Humphrey, A. D. Osterhaus, D. D. Erdman, and L. J. Anderson. 2002. Characterization of human metapneumoviruses isolated from patients in North America. J. Infect. Dis. 185:1660-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stockton, J., I. Stephenson, D. Fleming, and M. Zambon. 2002. Human metapneumovirus as a cause of community-acquired respiratory illness. Emerg. Infect. Dis. 8:897-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoffel, W., M. Duker, and K. Hofmann. 1993. Molecular cloning and gene organization of the mouse mitochondrial 3,2-trans-enoyl-CoA isomerase. FEBS Lett. 333:119-122. [DOI] [PubMed] [Google Scholar]
- 34.Sullender, W. M. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 13:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Hoogen, B. G., T. M. Bestebroer, A. D. Osterhaus, and R. A. Fouchier. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119-132. [DOI] [PubMed] [Google Scholar]
- 37.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viazov, S., F. Ratjen, R. Scheidhauer, M. Fiedler, and M. Roggendorf. 2003. High prevalence of human metapneumovirus infection in young children and genetic heterogeneity of the viral isolates. J. Clin. Microbiol. 41:3043-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walsh, E. E., and J. Hruska. 1983. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J. Virol. 47:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wertz, G. W., P. L. Collins, Y. Huang, C. Gruber, S. Levine, and L. A. Ball. 1985. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. USA 82:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]