Abstract
Background:
Carbapenem resistant Acinetobacter baumannii is an important nosocomial pathogen associated with a variety of infections.
Objectives:
The current study aimed to characterize the antimicrobial susceptibility, analyze the prevalence of oxacillinase and metallo-β-lactamase (MBL) genes and molecular typing of clinical isolates of A. baumannii.
Materials and Methods:
A total of 124 non-repetitive isolates of A. baumannii were collected from various clinical specimens in two teaching hospitals in Ahvaz, south-west of Iran. Antimicrobial susceptibility test was carried out by disk diffusion method. The minimum inhibitory concentrations (MICs) of imipenem, meropenem, colistin and tigecycline were determined using E-test. To screen for MBL production, double disk synergy (DDs) test and MBL E-test were performed. The presence of blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, blaVIM, blaIMP and blaSPM genes was assessed by polymerase chain reaction (PCR). To identify clonal relatedness, all isolates were subjected to repetitive sequence-based PCR (rep-PCR)
Results:
Based on disk diffusion results, the highest rate of resistance was observed in rifampin (96.8%). Colistin and polymyxin-B were the most effective agents in vitro. According to the MIC results, the rate of resistance to imipenem, meropenem, colistin and tigecycline were 78.2%, 73.4%, 0.8% and 0, respectively. Metallo-β-lactamase production was positive in 42.3% and 79.4% of the isolates by DDs test and E-test, respectively. All isolates (100%) carried blaOXA-51-like gene. According to the results of multiplex PCR, blaOXA-23-like and blaOXA-24-like genes were detected in 85.6% and 6.2% of carbapenem resistant isolates, respectively. No blaOXA-58- like, blaVIM, blaIMP and blaSPM were detected. By rep-PCR, carbapenem resistant isolates were separated into six genotypes (A to F). Genotype A (30.9%) was the most prevalent (P value < 0.001). Genotypes B and C were found in 28.9% and 26.8% of the isolates, respectively.
Conclusions:
The rate of carbapenem resistant A. baumannii isolates were high in this study. Since, blaOXA-58-like or MBL genes were not detected, it seems that resistance to carbapenems is related to blaOXA-23-like and blaOXA-24-like. Moreover, blaOXA-23-like was the most prevalent oxacillinase (OXA) gene. Most of the isolates belonged to one of the four dominant genotypes indicating clonal dissemination in the hospitals under study. In order to control the spread of carbapenem-resistant A. baumannii, infection- control strategies are needed.
Keywords: Acinetobacter baumannii, Carbapenems, Oxacillinase, Typing
1. Background
In recent years, Acinetobacter baumannii has emerged as an important pathogen in nosocomial infections and especially infects critically ill patients admitted to the intensive care units (ICUs) (1, 2). Septicemia, pneumonia, urinary tract infection, wound infection and meningitis are among the infections caused by this pathogen (3). In the hospital environment, resistance of A. baumannii to antimicrobial agents raises concerns (4). Carbapenem resistant A. baumannii are great concerns for physicians because carbapenems are common choice to treat infections caused by this pathogen (4, 5). In addition, therapeutic efficacy of carbapenems is limited due to spread of carbapenem resistant A. baumannii (4, 6). Carbapenem resistance is now observed worldwide in A. baumannii and these isolates are usually resistant to all classes of antimicrobial agents. A plenty of outbreak due to carbapenem resistant A. baumannii are reported from different countries and this situation had a worrying trend (4). Carbapenem resistance in A. baumannii is mediated by combined different mechanisms including: reduced permeability, changes in penicillin binding protein, AmpC stable derepression, efflux pumps and mostly by production of oxacillinases (OXAs) and less common by metallo-β-lactamase (MBLs) genes (7-9). Clonal transmission of drug resistant A. baumannii is reported globally (10). Inter hospital transmission of carbapenem resistant A. baumannii is demonstrated (4). It is well documented that in the nosocomial outbreaks, in most of the cases, one or two epidemic clones are involved in a given hospital (4). For epidemiological purposes and to control the spread of resistant iaolates, rapid diferrentiation of epidemic strains from the numerous incidental strains is necessary (11, 12). Molecular typing such as repetitive sequence-based polymerase chain reaction (rep-PCR) is required to determine the clonal relatedness of A. baumannii (12). The rep-PCR is beneficial for the molecular typing of A. baumannii (11).
2. Objectives
The current study aimed to determine the antimicrobial susceptibility pattern, prevalence and types of oxacillinase and metallo-β-lactamase genes and molecular typing by rep-PCR in the clinical isolates of A. baumannii.
3. Materials and Methods
3.1. Collection and Identification of Acinetobacter baumannii Isolates
From July 2011 to January 2013, a total of 124 non-duplicated A. baumannii isolates were collected from various clinical specimens in two teaching hospitals in Ahvaz, south-est of Iran. Bacterial isolates were initially identified as A. baumannii by biochemical tests (13). Suspected isolates were confirmed by PCR to identify blaOXA-51-like gene with specific primers (listed in Table 1) to amplify a 353 base pair sequence (14). DNA template for PCR was obtained by boiling method (15). Each reaction was carried out in a final volume of 25 µL containing 1x PCR buffer, 1 U Taq polymerase, 1.5 mM MgCl2, 200 µM of dNTP (SinaClon, Iran), 10 pmol of each primer (Eurofins MWG Operon, Germany) and 1 µL of the extracted DNA. PCR conditions were programmed in Mastercycler Eppendorf (Eppendorf, Germany) as follows: Initial denaturation at 94°C for 3 minutes; 35 cycles of 94°C for 45 seconds, annealing 57°C for 45 seconds, extension 72°C for 1 minute and final extension 72°C for 5 minutes. PCR products were separated on 1.5% agarose gel (SinaClon, Iran) by electrophoresis, stained with ethidium bromide (SinaClon, Iran) and then visualized under UV illumination (Syngene GeneGenius gel documentation system). Acinetobacter baumannii ATCC 19606 was used as positive control (14).
Table 1. Sequence of Primers Used in the Study.
| Primer | Forward Sequence 5’ - 3’ | Reverse Sequence 5’ - 3’ | Reference |
|---|---|---|---|
| bla OXA-51-like | TAATGCTTTGATCGGCCTTG | TGGATTGCACTTCATCTTGG | (14) |
| bla OXA-23-like | GATCGGATTGGAGAACCAGA | ATTTCTGACCGCATTTCCAT | (16) |
| bla OXA-24-like | GGTTAGTTGGCCCCCTTAAA | AGTTGAGCGAAAAGGGGATT | (16) |
| bla OXA-58-like | AAGTATTGGGGCTTGTGCTG | CCCCTCTGCGCTCTACATAC | (16) |
| bla IMP | TCGTTTGAAGAAGTTAACGG | ATGTAAGTTTCAAGAGTGATGC | (17) |
| bla VIM | GGTGTTTGGTCGCATATCGCAA | ATTCAGCCAGATCGGCATCGGC | (17) |
| bla SPM | AAAATCTGGGTACGCAAACG | ACATTATCCGCTGGAACAGG | (18) |
| REP | REP-I, III: GCGCCGICATCAGGC | REP-II: ACGTCTTATCAGGCCTAC | (19) |
3.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing of all isolates was performed using Kirby-Bauer method according to the clinical and laboratory standard institute (CLSI, 2011) guidelines. The following antimicrobial agents were tested: imipenem 10 µg, meropenem 10 µg, polymyxin-B 300 U, gentamicin 10 µg, ceftriaxone 30 µg, colistin 10 µg, piperacillin 100 µg, piperacillin-tazobactam 100/10 µg, cefepime 30 µg, tobramycin 10 µg, amikacin 30 µg, tetracycline 30 µg, ciprofloxacin 5 µg, trimethoprim-sulfamethoxazole 1.25/23.75 µg, ceftazidime 30 µg, rifampin 5 µg, tigecycline 15 µg, aztreonam 30 µg and ampicillin-sulbactam (10/10 µg), (MAST, Group Ltd, Merseyside, UK). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as control strains (20). Minimum inhibitory concentration (MIC) of imipenem, meropenem, colistin and tigecycline were determined by E-test strips (Liofilchem, Italy). Measures were obtained according to the CLSI guidelines. The US food and rug drug administration-approved criteria and Jones criteria were used for Enterobacteriacea and tigecycline breakpoint, respectively (21, 22).
3.3. Screening the metallo-β-Lactamase Producing Isolates
All isolates were screened for MBL production by an imipenem-EDTA (ethylene diamine tetra-acetic acid) double disk synergy and E-test MBL. Briefly, an overnight culture suspension of each sample was adjusted to a turbidity equivalent to 0.5 McFarland and inoculated on the surface of a Mueller-Hinton agar plate. Two 10 µg of imipenem disk (MAST, Group Ltd, Merseyside, UK) were placed on the plate 10 mm apart from edge to edge. Then 10 µL of 0.5 M EDTA solution (SinaClon, Iran) was directly added to one of them to obtain the desired concentration (750 µg). The plates were incubated at 35°C for 18 hours. After incubation, inhibition zones of the imipenem and imipenem-EDTA disks were measured and compared. If enlarged zone with imipenem-EDTA was 7 mm greater than the imipenem disk alone, it was considered as MBL positive revealing the inactivation of metallo-β-lactamase (class B) activity by EDTA (23, 24). Also an E-test MBL strip containing a double sided seven dilution range of imipenem (4 to 256 µg/mL) and imipenem (1 to 64 µg/mL) in combination with a fixed concentration of EDTA (Liofilchem, Italy) was used. The results were interpreted according to the manufacturer’s instruction.
3.4. PCR Amplification of OXA and Metallo-β-Lactamase Genes
Multiplex PCR was performed to detect blaOXA-23-like, blaOXA-24-like and blaOXA-58-like using specific primers as previously described (16). DNA template was obtained by boiling method (15). Each PCR reaction was performed in a final volume of 25 µL with 1x PCR buffer, 1 U Taq polymerase, 2 mM MgCl2, 200 µM of dNTP (SinaClon, Iran), 0.2 µM of each primer (TAG, Copenhagen A/S Denmark) and 1 µL of template DNA. PCR conditions were programmed in Mastercycler Eppendorf (Eppendorf, Germany) as follows: Initial denaturation at 94°C for 5 minutes; followed by 30 cycles at 94°C for 30 seconds, 53°C for 40 seconds and 72°C for 50 seconds and final extension at 72°C for 6 minutes. PCR products were separated by electrophoresis on 1.5% agarose gel (SinaClon, Iran) and after staining with ethidium bromide, visualized under UV gel documentation system Acinetobacter baumannii reference strains including: NCTC 13304, NCTC 13302, NCTC 13305 were used as positive control for blaOXA-23-like, blaOXA-24-like and blaOXA-58-like, respectively (16). For each gene, one amplicon was sequenced (Bioneer, South Korea) blaVIM, blaIMP and blaSPM were sought by singleplex PCR and primers previously described (17, 18). Two clinical isolates of P. aeruginosa harbored blaIMP and blaVIM were sequenced using automated sequence analyzer (Bioneer, South Korea) and used as positive control to identify the genes. The DNA of blaSPM-positive P. aeruginosa was purchased from Pasteur Institute of Iran and used as positive control in PCR reactions.
3.5. The rep-PCR
To investigate genotyping and identification of various clones, all isolates were subjected to rep-PCR with emphasis on carbapenem resistant isolates. Specific primers were used according to the previously described Bou et al. protocol (19). Template for PCR was extracted by phenol-chloroform method. Each reaction mixture was done in the total volume of 25 µL with 1x PCR buffer, 3.5 mM of MgCl2, 300 µM of dNTP, 3% dimethyl sulfoxide (DMSO) (SinaClon, Iran), 0.5 µM of each primer (TAG, Copenhagen A/S, Denmark), and 1U of Taq polymerase and 1 µL of genomic DNA. Amplification conditions were as follows: 94°C for 10 minutes; 30 cycles of 94°C for 1 minute, annealing temperatures 45°C for 1 minute, 72°C for 2 minutes and 72°C for 16 minutes. Products were separated by electrophoresis on 1.2% agarose gel (SinaClon, Iran); after staining with ethidium bromide, they were visualized under UV gel documentation system; then they were photographed and compared together by visual inspection (19). All fingerprints were observed by one observer. Snelling et al. protocol was used for classified various clones (11).
3.6. Nucleotide Sequence Accession Number
The nucleotide sequences obtained in this study were submitted to the GenBank nucleotide sequence database under the accession numbers: HG937619 for blaOXA-23-like, HG937620 for blaOXA-24-like and HG937621 for blaOXA-51-like.
3.7. Statistical Analysis
The results were analyzed using the SPSS version 16 to obtain frequencies and comparison among clones. Non-parametric chi-square test was used. A P value < 0.05 was considered statistically significant.
4. Results
4.1. Bacterial Isolates
Totally, 124 single-patient isolates were recovered. All isolates were positive for blaOXA-51-like and identified as A. baumannii. The rate of isolates from each ward and specimen are shown in Table 2.
Table 2. The Rate of Acinetobacter baumannii Species Isolated From Each Ward and Specimen.
| Ward | Rate of Isolates | Specimen | Rate of Isolates |
|---|---|---|---|
| ICU | 74.2 | Tracheal aspirate | 57.3 |
| Out patients | 8.1 | Cerebrospinal fluid | 11.3 |
| Neurosurgery | 4 | wound | 10.5 |
| Dermatology | 4 | Urine | 8.1 |
| Nephrology | 3.2 | Discharge | 5.6 |
| Orthopedic | 2.4 | Blood | 3.2 |
| Woman | 2.4 | Pleura | 1.6 |
| Surgery | 0.8 | Catheter | 1.6 |
| Neonatal | 0.8 | Eye infection | 0.8 |
Abbreviation: ICU, intensive care unit.
4.2. Antimicrobial Susceptibility
The results of antimicrobial susceptibility test by disk diffusion method are shown in Table 3. Colistin and polymyxin-B were the most active agents against the tested isolates. According to the results of MICs by E-test, among 124 isolates, 97 (78.2%) were resistant to imipenem. However, meropenem resistance was observed in 91 (73.4%) of the studied isolates. Only one isolate (0.8%) was resistant to colistin and 123 isolates (98.2%) were susceptible to this antibiotic. No tigecycline resistant isolate was observed and 99 (79.8%) and 25 (20.2%) isolates were sensitive and intermediate to this antibiotic, respectively.
Table 3. The Results of Antibiogram Analysis of Acinetobacter baumannii Isolatesa.
| Antibiotic | Sensitive | Intermediate | Resistant |
|---|---|---|---|
| Imipenem | 24.2 | 1.6 | 74.2 |
| Meropenem | 19.4 | 0.8 | 79.8 |
| Ceftazidime | 15.3 | 2.4 | 82.3 |
| Cefepime | 16.1 | 4 | 79.8 |
| Ceftriaxone | 1.6 | 12.1 | 86.3 |
| Colistin | 98.2 | NA | 0.8 |
| Piperacillin | 12.1 | 3.2 | 84.7 |
| Piperacillin-tazobactam | 16.9 | 1.6 | 81.5 |
| Polymyxin-B | 100 | NA | NA |
| Gentamicin | 28.2 | 4.8 | 66.9 |
| Tobramycin | 33.9 | 1.6 | 64.5 |
| Amikacin | 21 | 12.1 | 66.9 |
| Tetracycline | 21.8 | 12.1 | 66.1 |
| Ampicillin-sulbactam | 32.3 | 21.8 | 46 |
| Ciprofloxacin | 13.7 | 1.6 | 84.7 |
| Trimethoprim-sulfamethoxazole | 19.4 | 4.8 | 75.8 |
| Rifampin | NA | 3.2 | 96.8 |
| Aztreonam | NA | 4.8 | 95.2 |
| Tigecycline (FDA) | 6.5 | 58.1 | 35.5 |
| Tigecycline (Jones) | 45.2 | 50.8 | 4 |
Abbreviation: NA, not available.
aValues are expressed as %.
4.3. PCR Amplification of OXA Genes
Multiplex PCR analysis identified blaOXA-23-like in 83 (85.6%) of carbapenem resistant isolates. Six isolates (6.2%) contained blaOXA-24-like (Figure 1). All isolates were negative for blaOXA-58-like. Eight isolates were carbapenem resistant but had only blaOXA-51-like and other studied isolates were negative in the other studied genes. No amplification products were obtained in carbapenem susceptible isolates.
Figure 1. Agarose gel Showing OXAs Genes Obtained by Multiplex PCR.
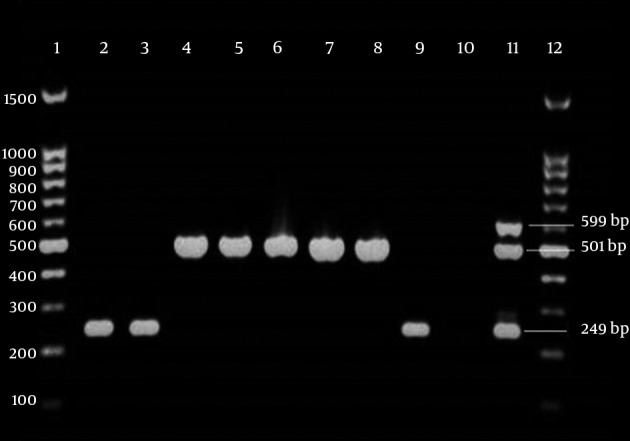
Lanes 1 and 12, 100 bp DNA ladder; lanes 2, 3 and 9, isolates with blaOXA-24-like in 249 bp; lanes 4 - 8, isolates with blaOXA-23-like in 501bp; lane 10, negative control (distilled water). lane 11, positive control Acinetobacter baumannii NCTC 13304, NCTC 13302 and NCTC 13305 were used as positive controls for blaOXA-23-like, blaOXA-24-like and blaOXA-58-like, respectively.
4.4. Metallo-β-Lactamase E-Test
To study the MBL production, carbapenem resistant isolates were evaluated by double disk synergy (DDS) test and E-test MBL strips. Of the 97 carbapenem resistant isolates, 41 (42.3%) exhibited a > 7 mm inhibitory zone and were categorized as MBL producer. However 56 isolates (57.7%) were negative for MBL production. Results of MBL E-test showed that among 97 carbapenem resistant isolates, 77 (79.4%) were positive for metallo-β-lactamase production and 20 isolates (20.6%) were negative (Figure 2). PCR did not detect metallo-β-lactamase genes, including blaVIM, blaIMP and blaSPM, among the studied isolates.
Figure 2. A MBL Producer Acinetobacter baumannii Isolate.
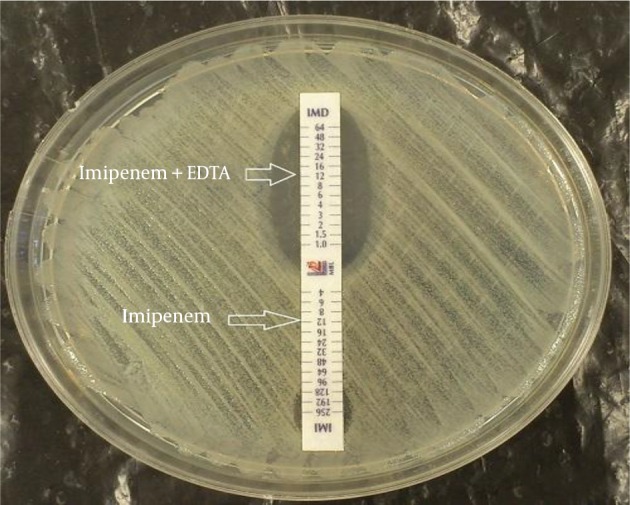
An inhibitory zone is observed in Imipenem + EDTA compared to that of imipenem only.
4.5. REP PCR
Among carbapenem resistant isolates, six clones (A - F) were observed which three of them were more prevalent (Figure 3). Genotype A was the most prevalent (P value < 0.001) and 30.9% (30 isolates) belonged to this genotype. The prevalence of other genotypes were as follows: 28 isolates (28.9%) belonged to clone B, 26 isolates (26.8%) to clone C, 10 isolates (10.3%) to clone D and 2 isolates (2.1%) to clone E. Clone F contained only one isolate (1%). Carbapenem susceptible isolates (27 isolates) had unique genotypes.
Figure 3. An Agarose gel rep-PCR Product for the Resistant Isolates.
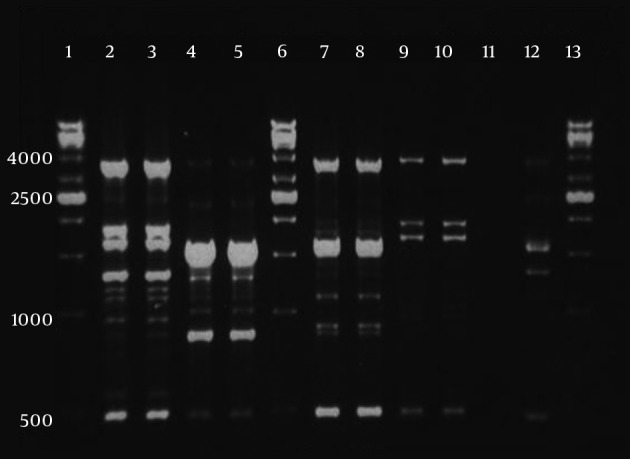
For each clone, double fingerprints are shown. Lanes 1, 6, and 13, 1 kb DNA ladder; lanes 2 and 3, clone A; lanes 4, 5, clone B; lanes 7 and 8, clone C; lane 9 and 10, clone D; lane 11, negative control; lane 12, A. baumannii NCTC 12156 (ATCC 19606).
5. Discussion
In the current study most of the isolates (74.2%) were obtained from patients in ICUs and in accordance to other researches worldwide, the rate of infection caused by A. baumannii is high in ICUs (25, 26). Previously, it was reported that A. baumannii is more prevalent among endotracheal aspirate samples. Moreover, ventilator associated pneumonia is one of the frequent nosocomial infections caused by this organism (27-31). In accordance to the mentioned studies, most of the current study samples (57.3%) were isolated from tracheal aspirates. In the present study, antimicrobial susceptibility pattern showed that colistin and polymyxin-B are the most effective agents against A. baumannii isolates, in vitro. Colistin is the last line antimicrobial agent to treat multidrug resistant A. baumannii (26, 32). Although, colistin resistant isolates are reported globally (32), in the current study only 0.8% of isolates were colistin resistant. This result indicates that colistin can be used to treat A. baumannii infections in the studied hospitals. Afterwards, among the tested antimicrobial agents, the highest rate of resistance was observed against rifampin; 96.8% and 3.2% of the isolates were resistant and intermediate to this antibiotic, respectively. No isolate was sensitive to rifampin; hence, it is suggested t that rifampin might be ineffective to treat A. baumannii infections in the studied hospitals. Carbapenems are successfully used to treat multidrug resistant A. baumannii infections; however, in recent years increase of carbapenem resistant A. baumannii isolates compromised their use (4, 26, 33). The emergence of carbapenem resistant A. baumannii is a global concern (4). In the current study the rates of resistance to imipenem by E-test and disk diffusion method were 78.2% and 74.2%, respectively. In addition, 79.8% and 73.4% of the isolates were resistant to meropenem by disk diffusion and E-test, respectively. There is a discrepancy between carbapenems E-test and disk diffusion method. Based on E-test, 78.2% of the isolates were resistant to imipenem; while disk diffusion method showed that 74.2% of the isolates were resistance to imipenem. In contrast, meropenem E-test detected 73.4% of isolates as resistant; whilst, according to disk diffusion more isolates (79.8%) were recorded as meropenem resistant. However, authors could not elucidate this difference. In A. baumannii, the most common carbapenemase genes involved in carbapenem resistance are, blaOXA-23-like, blaOXA-24-like, blaOXA-58-like and blaOXA-143-like (26, 34). In the current study, multiplex PCR detected 85.6% of carbapenem resistant isolates carrying blaOXA-23-like. The spread of OXA genes varies in different parts of the world and blaOXA-23-like is reported from 31% to 94% (7, 12, 35-39). The current study found that 6.2% of carbapenem resistant isolates harbor blaOXA-24-like. Some authors worldwide, reported the rate of blaOXA-24-like from 0 to 85.43% (7, 33, 36, 38-40). The results of the current study were consistent with those of other studies and the findings for blaOXA-23-like and blaOXA-24-like were in the reported ranges. In contrast to other studies that reported the range of blaOXA-58-like from 2% to 84.92% (12, 35-38, 40), the current study could not find any isolates positive for blaOXA-58-like. Albeit the reported co-existence of OXAs genes (29, 41, 42), coexistence between these genes was not observed in the current study and all A. baumannii isolates only had one of the blaOXA-23-like or blaOXA-24-like genes. In the current study, 42.3% and 79.4% of the isolates were MBL positive by DDs test and MBL E-test, respectively. Despite phenotypic tests, no blaIMP, blaVIM and blaSPM genes were detected by PCR and the isolates were negative for these genes. There are some possibilities about this phenomenon: 1) the MBL production may be false positive and due to bactericidal activity of EDTA, which may result in increased inhibitory zone and not associated with true MBL production (43); 2) MBL production my be true positive due to other MBL genes such as blaNDM that were not investigated in the current study (44). Similar results are reported that A. baumannii isolates were MBL producer by phenotypic tests but no MBL encoding genes were detected (45, 46). It is reported that MBL E-test has good sensitivity for MBL detection and could detected MBL both chromosomally and plasmid mediated in aerobic and anaerobic bacteria (47). According to the E-test results, it is possible that the current study isolates were true MBL producers. Interestingly the study found eight carbapenem resistant isolates that were negative for blaOXA-23-like, blaOXA-24-like, blaOXA-58-like and MBL genes and only harbored blaOXA-51-like. Similarly, Nowak et al. reported that seven isolates of carbapenem resistant A. baumannii only had blaOXA-51-like (29). Carbapenem resistance in these isolates may be associated with other mechanisms such as: modification of penicillin binding proteins, loss of porins and decreased permeability, AmpC stable derepression or over expression of efflux pump (4, 8, 9). It is noteworthy that, insertion of ISAba1 in upstream of blaOXA-51-like can lead to caebapenem resistance in A. baumannii (48). The relationship between harboring blaOXA-51-like and resistance to carbapenem in the eight isolates still need to be investigated. The current study also aimed to investigate the clonality of A. baumannii isolates by rep-PCR. Genotypic comparison by rep-PCR revealed that carbapenem resistant isolates belonged to six clones. All clones were spread in the ICUs. Clone A was dominant (30.9%) and clone F had the lowest prevalence (1%). Clonal dissemination of carbapenem resistant A. baumannii was previously reported in different studies. It has been establish that multidrug resistant of A. baumannii isolates with similar genotype can disseminate among various wards, different hospitals and even among cities (4, 10, 12, 49-51). In the current study, the vast majority of carbapenem resistant isolates (94/97, 96.9%) belonged to one of the four dominant genotypes indicating clonal dissemination of resistant isolates in the studied hospitals.
In conclusion, overall, the rate of carbapenem resistant isolates were high in the studied hospitals. Colistin and polymyxin-B were the effective antimicrobial agents, in vitro. Since in the current study blaOXA-58-like or MBL genes were not detected, it seems that carbapenem resistance is mostly related to blaOXA-23-like and blaOXA-24-like, and these genes may play an important role in carbapenem resistance in the isolates. In addition, four clones of carbapenem resistant of A. baumannii isolates are disseminated in the two studied hospitals and clone A was dominant. In accordance to other studies, in the current investigation most of the resistant isolates belonged to four clones indicating clonal dissemination of A. baumannii in the studied hospitals and that effective infection control strategies are necessary to control the spread of these resistant isolates.
Acknowledgments
This work was a part of PhD thesis, which was granted (No. 90126) and financially supported by deputy vice-chancellor for research affairs of Ahvaz Jundishapur University of Medical Sciences, and infectious and tropical diseases research center.
Footnotes
Authors’ Contribution:Study concept and design: Saeed Shoja, and Mojtaba Moosavian; acquisition of data: Saeed Shoja, and Soodabeh Rostami; analysis and interpretation of data: Saeed Shoja and Mohammad Amin Tabatabaiefar; drafting of the manuscript: Mojtaba Moosavian and Amir Peymani; critical revision of the manuscript for important intellectual content: Saeed Shoja, and Mojtaba Moosavian; statistical analysis: saeed shojia and Amir Peymani; administrative, technical, and material support: Saeed Shoja, Mojtaba Moosavian and Fariba Abbasi; study supervision: Saeed Shoja, and Mojtaba Moosavian.
Funding/Support:This work was a part of PhD thesis, which was granted (No. 90126) and financially supported by deputy vice-chancellor for research affairs of Ahvaz Jundishapur University of Medical Sciences, and Infectious and tropical disease research center.
References
- 1.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. 2013;41(1):11–9. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Karah N, Haldorsen B, Hermansen NO, Tveten Y, Ragnhildstveit E, Skutlaberg DH, et al. Emergence of OXA-carbapenemase- and 16S rRNA methylase-producing international clones of Acinetobacter baumannii in Norway. J Med Microbiol. 2011;60(Pt 4):515–21. doi: 10.1099/jmm.0.028340-0. [DOI] [PubMed] [Google Scholar]
- 3.Moosavian M, Shoja S, Nashibi R, Ebrahimi N, Tabatabaiefar MA, Rostami S, et al. Post Neurosurgical Meningitis due to Colistin Heteroresistant Acinetobacter baumannii. Jundishapur J Microbiol. 2014;7(10):e32388. doi: 10.5812/jjm.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarrilli R, Giannouli M, Tomasone F, Triassi M, Tsakris A. Carbapenem resistance in Acinetobacter baumannii: the molecular epidemic features of an emerging problem in health care facilities. J Infect Dev Ctries. 2009;3(5):335–41. doi: 10.3855/jidc.240. [DOI] [PubMed] [Google Scholar]
- 5.Dai W, Huang S, Sun S, Cao J, Zhang L. Nosocomial spread of carbapenem-resistant Acinetobacter baumannii (types ST75 and ST137) carrying blaOXA-23-like gene with an upstream ISAba1 in a Chinese hospital. Infect Genet Evol. 2013;14:98–101. doi: 10.1016/j.meegid.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Bogaerts P, Cuzon G, Naas T, Bauraing C, Deplano A, Lissoir B, et al. Carbapenem-resistant Acinetobacter baumannii isolates expressing the blaOXA-23 gene associated with ISAba4 in Belgium. Antimicrob Agents Chemother. 2008;52(11):4205–6. doi: 10.1128/AAC.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irfan S, Turton JF, Mehraj J, Siddiqui SZ, Haider S, Zafar A, et al. Molecular and epidemiological characterisation of clinical isolates of carbapenem-resistant Acinetobacter baumannii from public and private sector intensive care units in Karachi, Pakistan. J Hosp Infect. 2011;78(2):143–8. doi: 10.1016/j.jhin.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 8.Kulah C, Mooij MJ, Comert F, Aktas E, Celebi G, Ozlu N, et al. Characterisation of carbapenem-resistant Acinetobacter baumannii outbreak strains producing OXA-58 in Turkey. Int J Antimicrob Agents. 2010;36(2):114–8. doi: 10.1016/j.ijantimicag.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12(9):826–36. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 10.van Dessel H, Dijkshoorn L, van der Reijden T, Bakker N, Paauw A, van den Broek P, et al. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res Microbiol. 2004;155(2):105–12. doi: 10.1016/j.resmic.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Snelling AM, Gerner-Smidt P, Hawkey PM, Heritage J, Parnell P, Porter C, et al. Validation of use of whole-cell repetitive extragenic palindromic sequence-based PCR (REP-PCR) for typing strains belonging to the Acinetobacter calcoaceticus-Acinetobacter baumannii complex and application of the method to the investigation of a hospital outbreak. J Clin Microbiol. 1996;34(5):1193–202. doi: 10.1128/jcm.34.5.1193-1202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan ZQ, Shen DX, Cao JR, Chen R, Wei X, Liu LP, et al. Susceptibility patterns and molecular epidemiology of multidrug-resistant Acinetobacter baumannii strains from three military hospitals in China. Int J Antimicrob Agents. 2010;35(3):269–73. doi: 10.1016/j.ijantimicag.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Koneman EW. Color atlas and textbook of diagnostic microbiology. Lippincott-Raven Publishers; 1997. [Google Scholar]
- 14.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974–6. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andriamanantena TS, Ratsima E, Rakotonirina HC, Randrianirina F, Ramparany L, Carod JF, et al. Dissemination of multidrug resistant Acinetobacter baumannii in various hospitals of Antananarivo Madagascar. Ann Clin Microbiol Antimicrob. 2010;9:17. doi: 10.1186/1476-0711-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–3. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Zong Z, Lu X, Valenzuela JK, Partridge SR, Iredell J. An outbreak of carbapenem-resistant Acinetobacter baumannii producing OXA-23 carbapenemase in western China. Int J Antimicrob Agents. 2008;31(1):50–4. doi: 10.1016/j.ijantimicag.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–23. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Bou G, Cervero G, Dominguez MA, Quereda C, Martinez-Beltran J. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed-field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem- and meropenem-resistant Acinetobacter baumannii. Clin Microbiol Infect. 2000;6(12):635–43. doi: 10.1046/j.1469-0691.2000.00181.x. [DOI] [PubMed] [Google Scholar]
- 20.Wayen P. Clinical and Laboratory Standards Institute.Performance standards for antimicrobial susceptibility testing. 21st informational supplement M100-S21. Wayne: CLSI; 2011. [Google Scholar]
- 21.Jones RN, Ferraro MJ, Reller LB, Schreckenberger PC, Swenson JM, Sader HS. Multicenter studies of tigecycline disk diffusion susceptibility results for Acinetobacter spp. J Clin Microbiol. 2007;45(1):227–30. doi: 10.1128/JCM.01588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pharmaceutics W. Tygacil (tigecycline) for injection [package insert]. Philadelphia: Wyeth Pharmaceuticals Inc; 2005. [Google Scholar]
- 23.Sung JY, Kwon KC, Park JW, Kim YS, Kim JM, Shin KS, et al. [Dissemination of IMP-1 and OXA type beta-lactamase in carbapenem-resistant Acinetobacter baumannii]. Korean J Lab Med. 2008;28(1):16–23. doi: 10.3343/kjlm.2008.28.1.16. [DOI] [PubMed] [Google Scholar]
- 24.Jeon BC, Jeong SH, Bae IK, Kwon SB, Lee K, Young D, et al. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 beta-lactamase in korea. J Clin Microbiol. 2005;43(5):2241–5. doi: 10.1128/JCM.43.5.2241-2245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fontana C, Favaro M, Minelli S, Bossa MC, Testore GP, Leonardis F, et al. Acinetobacter baumannii in intensive care unit: a novel system to study clonal relationship among the isolates. BMC Infect Dis. 2008;8:79. doi: 10.1186/1471-2334-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kempf M, Rolain JM. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int J Antimicrob Agents. 2012;39(2):105–14. doi: 10.1016/j.ijantimicag.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5(12):939–51. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 28.Peymani A, Farajnia S, Nahaei MR, Sohrabi N, Abbasi L, Ansarin K, et al. Prevalence of class 1 integron among multidrug-resistant Acinetobacter baumannii in Tabriz, northwest of Iran. Pol J Microbiol. 2012;61(1):57–60. [PubMed] [Google Scholar]
- 29.Nowak P, Paluchowska P, Budak A. Distribution of blaOXA genes among carbapenem-resistant Acinetobacter baumannii nosocomial strains in Poland. New Microbiol. 2012;35(3):317–25. [PubMed] [Google Scholar]
- 30.Shoja S, Moosavian M, Peymani A, Tabatabaiefar MA, Rostami S, Ebrahimi N. Genotyping of carbapenem resistant Acinetobacter baumannii isolated from tracheal tube discharge of hospitalized patients in intensive care units, Ahvaz, Iran. Iran J Microbiol. 2013;5(4):315–22. [PMC free article] [PubMed] [Google Scholar]
- 31.Altun HU, Yagci S, Bulut C, Sahin H, Kinikli S, Adiloglu AK, et al. Antimicrobial Susceptibilities of Clinical Acinetobacter baumannii Isolates With Different Genotypes. Jundishapur J Microbiol. 2014;7(12):e32388. doi: 10.5812/jjm.13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai Y, Chai D, Wang R, Liang B, Bai N. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother. 2012;67(7):1607–15. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 33.Manageiro V, Jones-Dias D, Ferreira E, Louro D, Antimicrobial Resistance Surveillance Program in P, Canica M. Genetic diversity and clonal evolution of carbapenem-resistant Acinetobacter baumannii isolates from Portugal and the dissemination of ST118. Int J Antimicrob Agents. 2012;40(5):398–403. doi: 10.1016/j.ijantimicag.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 34.D'Arezzo S, Principe L, Capone A, Petrosillo N, Petrucca A, Visca P. Changing carbapenemase gene pattern in an epidemic multidrug-resistant Acinetobacter baumannii lineage causing multiple outbreaks in central Italy. J Antimicrob Chemother. 2011;66(1):54–61. doi: 10.1093/jac/dkq407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkier AK, Catalano M, Ramirez MS, Quiroga C, Orman B, Ratier L, et al. Polyclonal spread of bla(OXA-23) and bla(OXA-58) in Acinetobacter baumannii isolates from Argentina. J Infect Dev Ctries. 2008;2(3):235–40. doi: 10.3855/jidc.269. [DOI] [PubMed] [Google Scholar]
- 36.Sohrabi N, Farajnia S, Akhi MT, Nahaei MR, Naghili B, Peymani A, et al. Prevalence of OXA-type beta-lactamases among Acinetobacter baumannii isolates from Northwest of Iran. Microb Drug Resist. 2012;18(4):385–9. doi: 10.1089/mdr.2011.0077. [DOI] [PubMed] [Google Scholar]
- 37.Touati M, Diene SM, Racherache A, Dekhil M, Djahoudi A, Rolain JM. Emergence of blaOXA-23 and blaOXA-58 carbapenemase-encoding genes in multidrug-resistant Acinetobacter baumannii isolates from University Hospital of Annaba, Algeria. Int J Antimicrob Agents. 2012;40(1):89–91. doi: 10.1016/j.ijantimicag.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 38.Ergin A, Hascelik G, Eser OK. Molecular characterization of oxacillinases and genotyping of invasive Acinetobacter baumannii isolates using repetitive extragenic palindromic sequence-based polymerase chain reaction in Ankara between 2004 and 2010. Scand J Infect Dis. 2013;45(1):26–31. doi: 10.3109/00365548.2012.708782. [DOI] [PubMed] [Google Scholar]
- 39.Carvalho KR, Carvalho-Assef AP, Peirano G, Santos LC, Pereira MJ, Asensi MD. Dissemination of multidrug-resistant Acinetobacter baumannii genotypes carrying bla(OXA-23) collected from hospitals in Rio de Janeiro, Brazil. Int J Antimicrob Agents. 2009;34(1):25–8. doi: 10.1016/j.ijantimicag.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Ben RJ, Yang MC, Hsueh JC, Shiang JC, Chien ST. Molecular characterisation of multiple drug-resistant Acinetobacter baumannii isolates in southern Taiwan. Int J Antimicrob Agents. 2011;38(5):403–8. doi: 10.1016/j.ijantimicag.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 41.Feizabadi MM, Fathollahzadeh B, Taherikalani M, Rasoolinejad M, Sadeghifard N, Aligholi M, et al. Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. Isolated from patients at Tehran hospitals. Jpn J Infect Dis. 2008;61(4):274–8. [PubMed] [Google Scholar]
- 42.Asadollahi K, Alizadeh E, Akbari M, Taherikalani M, Niakan M, Maleki A, et al. The role of bla(OXA-like carbapenemase) and their insertion sequences (ISS) in the induction of resistance against carbapenem antibiotics among Acinetobacter baumannii isolates in Tehran hospitals. Roum Arch Microbiol Immunol. 2011;70(4):153–8. [PubMed] [Google Scholar]
- 43.Chu YW, Cheung TK, Ngan JY, Kam KM. EDTA susceptibility leading to false detection of metallo-beta-lactamase in Pseudomonas aeruginosa by Etest and an imipenem-EDTA disk method. Int J Antimicrob Agents. 2005;26(4):340–1. doi: 10.1016/j.ijantimicag.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Hrabak J, Stolbova M, Studentova V, Fridrichova M, Chudackova E, Zemlickova H. NDM-1 producing Acinetobacter baumannii isolated from a patient repatriated to the Czech Republic from Egypt, July 2011. Euro Surveill. 2012;17(7) [PubMed] [Google Scholar]
- 45.Purohit M, Mendiratta DK, Deotale VS, Madhan M, Manoharan A, Narang P. Detection of metallo-beta-lactamases producing Acinetobacter baumannii using microbiological assay, disc synergy test and PCR. Indian J Med Microbiol. 2012;30(4):456–61. doi: 10.4103/0255-0857.103770. [DOI] [PubMed] [Google Scholar]
- 46.Hu Q, Hu Z, Li J, Tian B, Xu H, Li J. Detection of OXA-type carbapenemases and integrons among carbapenem-resistant Acinetobactor baumannii in a teaching hospital in China. J Basic Microbiol. 2011;51(5):467–72. doi: 10.1002/jobm.201000402. [DOI] [PubMed] [Google Scholar]
- 47.Walsh TR, Bolmstrom A, Qwarnstrom A, Gales A. Evaluation of a new Etest for detecting metallo-beta-lactamases in routine clinical testing. J Clin Microbiol. 2002;40(8):2755–9. doi: 10.1128/JCM.40.8.2755-2759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006;258(1):72–7. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 49.Peymani A, Farajnia S, Nahaei MR, Sohrabi N, Abbasi L. Clonal Characterization of Multidrug-Resistant A. baumannii Isolated from Imam Reza Hospital in Tabriz, Iran. Iran J Infect Dis Trop Med. 2012;17(58):7–12. [Google Scholar]
- 50.Kraniotaki E, Manganelli R, Platsouka E, Grossato A, Paniara O, Palu G. Molecular investigation of an outbreak of multidrug-resistant Acinetobacter baumannii, with characterisation of class 1 integrons. Int J Antimicrob Agents. 2006;28(3):193–9. doi: 10.1016/j.ijantimicag.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 51.Ece G, Erac B, Yurday Cetin H, Ece C, Baysak A. Antimicrobial Susceptibility and Clonal Relation Between Acinetobacter baumannii Strains at a Tertiary Care Center in Turkey. Jundishapur J Microbiol. 2015;8(2):e32388. doi: 10.5812/jjm.15612. [DOI] [PMC free article] [PubMed] [Google Scholar]


