Abstract
Epithelial ovarian cancer (EOC) is one of the deadliest common cancers. The five most common types of disease are high-grade and low-grade serous, endometrioid, mucinous and clear cell carcinoma. Each of these subtypes present distinct molecular pathogeneses and sensitivities to treatments. Recent studies show that certain genetic variants confer susceptibility to all subtypes while other variants are subtype-specific. Here, we perform an extensive analysis of the genetic architecture of EOC subtypes. To this end, we used data of 10,014 invasive EOC patients and 21,233 controls from the Ovarian Cancer Association Consortium genotyped in the iCOGS array (211,155 SNPs). We estimate the array heritability (attributable to variants tagged on arrays) of each subtype and their genetic correlations. We also look for genetic overlaps with factors such as obesity, smoking behaviors, diabetes, age at menarche and height. We estimated the array heritabilities of high-grade serous disease ( ), endometrioid ( ), clear cell ( ) and all EOC ( ). Known associated loci contributed approximately 40 % of the total array heritability for each subtype. The contribution of each chromosome to the total heritability was not proportional to chromosome size. Through bivariate and cross-trait LD score regression, we found evidence of shared genetic backgrounds between the three high-grade subtypes: serous, endometrioid and undifferentiated. Finally, we found significant genetic correlations of all EOC with diabetes and obesity using a polygenic prediction approach.
Introduction
In developed countries, epithelial ovarian cancer (EOC) is the leading gynecological malignancy with an estimated annual incidence rate of 12 per 100,000 and a poor 5 year survival between 20 and 50 % (Chornokur et al. 2015; Sopik et al. 2015; Sung et al. 2014). About 90 % of invasive tumors in the ovary are of epithelial origin (Kurman et al. 2014). These tumors are divided into various histological subtypes that include: serous, mucinous, endometrioid, clear cell, Brenner, other minor types, as well as undifferentiated, mixed and unclassified carcinomas (Prat 2012; Sung et al. 2014). Serous carcinomas can be subdivided into high-grade (90 %) and low-grade disease (10 %) (Kurman and Shih Ie 2008; Malpica et al. 2004; Shih Ie and Kurman 2004).
Each epithelial ovarian cancer histologic subtype exhibits a distinct etiologic and molecular pathogenesis and sensitivity to treatment (e.g., chemotherapeutic agents) (Anglesio et al. 2013; Della Pepa et al. 2015; Risch et al. 1996; Shih Ie and Kurman 2004; Soslow 2008). It has been suggested that serous carcinomas arise from the epithelial mucosal lining of the fallopian tube fimbriae or from endosalpingiotic deposits on the ovarian or peritoneal surfaces. Clear cell and endometrioid subtypes may arise from endometriotic lesions (Kurman et al. 2014; Wiegand et al. 2010), while mucinous tumors do not yet have a clear origin, though metaplastic transformation of the epithelial lining of ovarian inclusion cysts has been suggested. Serous carcinoma is by far the most deadly type of EOC, with 5-year survival of less than 20% for patients suffering from high-grade disease and 50 % for those with low-grade disease (Malpica et al. 2004). In contrast, women with mucinous, endometrioid or clear cell carcinomas tend to have better prognosis, with estimated 5-year survivals of 50–60 % (Malpica et al. 2004; Simons et al. 2015). These differences in survival are due at least in part to the fact that high-grade serous carcinomas are usually detected at advanced stages of disease but the other subtypes at earlier stages (Devouassoux-Shisheboran and Genestie 2015; Malpica et al. 2004; Simons et al. 2015).
Genetic studies have shown that around 20 % of patients with high-grade serous cancers carry germ-line and somatic mutations in BRCA1 or BRCA2 (Alsop et al. 2012; Berchuck et al. 1998) along with somatic mutations in TP53 that are present in most tumors (Cancer Genome Atlas Research Network 2011). Alterations in KRAS and BRAF but not TP53 have been associated with low-grade serous carcinomas (Della Pepa et al. 2015; Grisham et al. 2013; Jones et al. 2012). Mucinous carcinomas also frequently have somatic mutations in KRAS (Cuatrecasas et al. 1997) in addition to mutations in HER2 (Anglesio et al. 2013). Endometrioid and clear cell carcinomas often carry somatic mutations in AR1D1A and PIK3CA (Jones et al. 2010). In addition, genome-wide association studies (GWAS) have found 20 common polymorphisms associated with risk of EOC (Bojesen et al. 2013; Bolton et al. 2010; Goode et al. 2010; Permuth-Wey et al. 2013; Pharoah et al. 2013; Song et al. 2009).
Specific germ-line SNPs are commonly found in the different EOC subtypes. However, these variants explain only a fraction of the cases, thus, it is not known whether or not other genetic components are shared among the subtypes. One of our previous studies (Lu et al. 2015) estimated the array heritability (i.e., heritability explained by about 200,000 genotyped SNPs but not all the genome) of all EOC to be 5.6 %, and 8.8 % for the most common EOC subtype, high-grade serous.
Beside genetic factors predisposing to these diseases, some environmental factors such as smoking (Collaborative Group on Epidemiological Studies of Ovarian Cancer et al. 2012; Faber et al. 2013) and obesity (Aune et al. 2015; Collaborative Group on Epidemiological Studies of Ovarian Cancer 2012; Olsen et al. 2013) may be associated with increases in risk of some subtypes of EOC. In addition, traits including achieved height (Aune et al. 2015; Wiren et al. 2014) and diabetes mellitus (Gapstur et al. 2012; Lee et al. 2013) have been positively associated to EOC. In contrast, some studies have shown that age at menarche (Gong et al. 2013) is inversely associated with risk of EOC. Evidence suggests that all these traits have heritable components. Genetic variation may explain as much as 80 % of the total variance of height (Yang et al. 2010) or even 40 % for smoking behavior (Vink and Boomsma 2011; Vink et al. 2005). It is possible that part of the heritability of EOC may be explained by the heritability of these traits, if they are associated with EOC risk.
In this work, we investigate three aspects of the genetic architecture of EOC and its subtypes: (1) the total genetic contribution of all array genotyped SNPs (genome-wide, per chromosome and after accounting for known EOC associated loci); (2) the genetic correlations between EOC subtypes; and (3) the genetic correlations between EOC subtypes and risk factors such as obesity and smoking. To this end, we use genotype and risk-factor data from studies participating in the Ovarian Cancer Association Consortium (OCAC). We quantify genetic contributions to disease using genome-wide complex trait analysis (GCTA) (Lee et al. 2011; Yang et al. 2010, 2011a). Then, we evaluate shared genetic backgrounds between EOC subtypes and candidate risk factors using complementary approaches: bivariate linear mixed models (Lee et al. 2012), cross-trait LD score regression (Bulik-Sullivan et al. 2015a) and polygenic risk prediction (International Schizophrenia Consortium et al. 2009).
Methods
Data
We used data from the Ovarian Cancer Association Consortium (OCAC). This dataset consists of custom Illumina iCOGS array genotyping of 47,630 cases and controls in 43 OCAC studies. Detailed description of the content of the array can be found elsewhere (Pharoah et al. 2013). In brief, the array consists of 211,155 variants within breast, ovarian and prostate cancer susceptibility loci as well as candidate SNPs, SNPs associated with other cancers and SNPs associated with relevant quantitative traits such as body mass index (BMI) and the onset of menarche.
We applied standard quality control (QC) for the genotype data. First, we selected only samples from European ancestry studies and that were within 6 s.d. from the genotype-derived PC1 and PC2 from the 1000 Genomes European population (Supplementary figure 1). We excluded individuals with missing genotypes in 5 % or more of the SNPs. Likewise, we removed SNPs with call rates below 99 %, minor allele frequencies (MAF) below 1 % and SNPs that deviated from Hardy–Weinberg equilibrium at P < 0.0001 (Lu et al. 2014). Further, given that our analytic methods are sensitive to relatedness (e.g., results may be biased by common environmental factors in relatives) we removed individuals such that no sample pairs had identity by descent (IBD) >10 % (i.e., less than second cousins), giving more priority to keeping cases than controls. In concordance with one of our previous work (Lu et al. 2015), we focused only on those with invasive EOC tumors. In total, 10,014 EOC cases and 21,233 controls met these criteria and were genotyped for 195,183 SNPs. The number of cases according to histologic subtype is displayed in Table 1. The numbers of initial cases and controls per study are summarized in Supplementary Table 1.
Table 1.
Array heritabilities ( ) and standard errors (s.e.) for invasive EOC according to histological subtype
| Subtype | Cases | Controls | Life-time risk | All SNPs
|
Removing known locia
|
||||
|---|---|---|---|---|---|---|---|---|---|
|
|
s.e. | P value |
|
s.e. | P value | ||||
| High-grade Serous | 4098 | 21,233 | 0.0055 | 0.088 | 0.010 | 2.2E–16 | 0.047 | 0.009 | 1.83E–09 |
| Clear cell | 620 | 21,233 | 0.0005 | 0.067 | 0.033 | 0.017 | 0.046 | 0.029 | 0.058 |
| Endometrioid (all) | 1342 | 21,233 | 0.001 | 0.032 | 0.016 | 0.016 | 0.020 | 0.014 | 0.077 |
| Endometrioid G1/G2 | 906 | 21,233 | 0.001 | 0.044 | 0.024 | 0.025 | 0.037 | 0.021 | 0.037 |
| Endometrioid G3 | 436 | 21,233 | 0.001 | 0.049 | 0.046 | 0.127 | 0.009 | 0.041 | 0.417 |
| Mucinous | 658 | 21,233 | 0.0005 | 0.000 | 0.028 | 0.5 | 0.000 | 0.025 | 0.5 |
| Unknown | 2934 | 21,233 | 0.009 | 0.070 | 0.015 | 1.1E–10 | 0.041 | 0.012 | 1.1E–04 |
| All | 10,014 | 21,233 | 0.009 | 0.056 | 0.006 | 2.2E–16 | 0.036 | 0.005 | 2.2E–16 |
Results for all iCOGS SNPs, and after removing known associated loci. Disease prevalence of EOC subtypes is calculated as the lifetime risk of ovarian cancer multiplied by the relative proportion of the corresponding EOC subtype. See “Methods” section. Bolded estimates are statistically significantly different from 0
Loci removed: WNT4, RSPO1, SYNPO2, GPX6, ABO, ATAD5, C19orf62, CMYC, TIPARP, BNC2, ARHGAP27, TERT, RAD51B/C/D, BRIP1, BARD1, PALB2, NDN, CHMP4C, MLLT10, HNF1B, BRCA1, BRCA2, KRAS, TP53, HER2, AR1D1A and PIK3CA
Analysis
We estimated the variance explained by all SNPs in the array ( ) (Lee et al. 2011), the variance after removing known loci, and the variance explained by each chromosome for each of the EOC subtypes. We used GCTA to calculate one genetic relationship matrix (GRM) for all autosomes.
The estimated variance explained was transformed from the observed scale to an unobserved continuous “liability” scale using a probit transformation (Lee et al. 2011) taking into account the disease prevalence. The lifetime risk of the various EOC subtypes were calculated as the lifetime risk of ovarian cancer (~1 % according to the Surveillance, Epidemiology and End Results (SEER), http://seer.cancer.gov/statfacts) multiplied by the relative proportion of each subtype according to SEER program DevCan database (http://surveillance.cancer.gov/devcan/canques.html) in all ovarian cancer. Given that around 90 % of ovarian cancers are of epithelial origin, we used 0.9 % as the prevalence for all EOC. As , is derived solely from the SNPs tagged on the genotyping array instead of the whole genome, it provides a lower bound on heritability estimates (Lu et al. 2014). Phenotypes were modeled as a linear function of the sum of the additive effects due to all SNPs associated with trait-associated variants and residual effects. Variance components were estimated using residual maximum likelihood (REML) (Yang et al. 2010). For tests of whether a variance component is zero or not, the test is one-sided and under the null hypothesis that the test statistic follows a 50:50 mixture of a point mass at zero and the χ1 distribution (Yang et al. 2010, 2011a). One-sided p values were calculated to estimate the statistical significance. Likewise, to estimate the proportion of that is explained by the known loci [WNT4, RSPO1, SYNPO2, GPX6, ABO, ATAD5, C19orf62, CMYC, TIPARP, BNC2, ARHGAP27, TERT, RAD51B/C/D, BRIP1, BARD1, PALB2, NDN, CHMP4C, MLLT10, HNF1B, BRCA1, BRCA2, KRAS, TP53, HER2, AR1D1A and PIK3CA (Bojesen et al. 2013; Bolton et al. 2010; Goode et al. 2010; Permuth-Wey et al. 2013; Pharoah et al. 2013; Song et al. 2009)], we recomputed the GRM with the SNPs (6391 SNPs) close to the known loci SNPs (±1 megabase either side) removed.
Similarly, to investigate the genetic contributions within each of the chromosomes, we computed one GRM per chromosome and performed analyses using REML fitting the 22 genetic variance components in the model as implemented in GCTA with the flag –mgrm (multiple GRMs) (Yang et al. 2011b). Given that loading 22 GRMs with the 21,051 controls and the cases of the various histotypes was computationally intractable, we assigned to each case just one control of the same study, yielding smaller GRMs (e.g., for high-grade Serous cancer there were 3705 cases and 3705 controls). We then normalized the contribution of each chromosome by the number of independent SNPs (percentage) in the iCOGs array per chromosome. This number of independent SNPs was estimated through LD pruning using the PLINK command –indep 50 5 1.2, where 50 is the window size (#SNPs), 5 is the number of SNPs the window can shift, and 1.2 is 1/(1 − R2), where R2 is the multiple correlation coefficient for a SNP regressed on all other SNPs simultaneously (Chang et al. 2015). To approximate the s.e. of the variance explained by each chromosome, we performed a jackknifing procedure up to 1000 times, taking 80 % of the cases and 80 % of the controls each time. Given the complexity of the sample, around 20 % of the jackknifing repetitions did not converge within 1000 iterations so the standard errors were computed from just the 800 successful jackknifings.
To investigate the genetic correlations between the subtypes, to remove potential biases from overlapping control samples from the different studies, we matched each case to 1 control of the same study, and distributed controls in such a way that each EOC subtype had separate sets of controls. For example, all of the controls for mucinous EOC were different from the endometrioid EOC controls.
Genetic correlation (rg) represents the proportion of the total genetic variance that two traits share. To investigate the rg between EOC subtypes, we used two distinct approaches that can be applied to population-based samples. We first used the GRM in a bivariate mixed-effects linear model implemented in GCTA (Cross-Disorder Group of the Psychiatric Genomics Consortium et al. 2013) to compute the genetic correlations between the various EOC subtypes. The estimated genetic correlation is the additive genetic covariance between traits, normalized by the geometric mean of the individual trait genetic variances (producing values from −1 to +1). The additive genetic covariance was estimated by relating trait covariances between unrelated individuals to genetic relationship estimates from marker data. Increased covariance between traits with high genetic relationship values implies a positive genetic correlation between traits. To control for any potential effects of population stratification, all the analyses were performed using the first ten principal components (PCs) of the genotypes as covariates. Estimates are reported as genetic correlation ± standard error.
We also used cross-trait LD score regression (Bulik-Sullivan et al. 2015a), a recently developed approach that is able to estimate the genetic correlations using solely GWAS summary statistics and is not affected by sample overlap. We first ran genome-wide association analyses using the same samples as when computing per each EOC subtype (i.e., we repeatedly made use of all of the controls for analysis of each subtype) and with the ten first PCs and study site as covariates. Genomic inflation factors for these GWAS analyses ranged from 0.99 for mucinous cancer to 1.07 for all EOC. We used the LD-scores estimated by Bulik-Sullivan et al. (2015a, b) available at http://www.broadinstitute.org/~bulik/eur_ldscores/which are based on the 1000 Genomes European population and estimated within 1-cM windows. We then estimated the genetic correlation using software available at https://github.com/bulik/ldsc with the default parameters.
Genetic correlations between EOC subtypes and risk factors
Using cross-trait LD score regression, we estimated genetic correlations between risk factors and EOC histotypes. To this end, we used publicly available GWAS summary results from the latest GWAS meta-analyses of BMI and height from the Genetic Investigation of Anthropometric Traits (GIANT) Consortium. These analyses included 339,225 (Locke et al. 2015) and 253,288 (Wood et al. 2014) individuals, respectively. We also estimated genetic correlations using the GIANT extreme anthropometric traits GWAS which used obesity class 1 (BMI > 30), class 2 (BMI > 35) and class 3 (BMI > 40) groups as cases, and individuals with BMI ≤25 as controls, in a sample of 263,407 individuals (Berndt et al. 2013). Genetic overlaps with age at menarche were carried out based on the GWAS of the Reproductive Genetics Consortium which involved 182,416 women (Perry et al. 2014). Smoking behavior genetic predisposition was approximated based on the Tobacco and Genetics Consortium GWAS which involved 74,053 participants (Tobacco and Genetics Consortium 2010). Finally, for diabetes, we used the summary results for type 2 diabetes GWAS of the DIAGRAM (diabetes genetics replication and meta-analysis) consortium, which involved 34,840 cases and 114,981 controls (Morris et al. 2012).
We also carried out a polygenic risk prediction approach. This method involves the computation of polygenic risk scores (PGRS) of each of the risk factors and uses these scores to predict disease status (International Schizophrenia Consortium et al. 2009). The PGRS describes a predicted phenotypic value based on the genetic component and is computed by aggregating the magnitude of associations of many variants. These associations are estimated using a discovery set of subjects (e.g., for height or BMI) to identify the relevant SNPs and estimate the magnitude of association of each, and these magnitudes or the number of “high-risk” alleles in each SNP are then summed to create a score. Subsequently, we examine the association of this score within a target subject set (e.g., EOC cases and controls). If the score association is significant, it implies a genetic correlation between the two traits. In this study, we selected variants to compute the PGRS based on 11 p value thresholds (<0.00001, 0.001, 0.01, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.75, 1). Given the nature of the iCOGS array in which many loci have high densities of tagged SNPs, we performed linkage disequilibrium (LD) clumping to remove the correlated variants (r2 > 0.2) within 500 kb windows for each component of the PGRS. The computations for PGRS and LD clumping were performed with PLINK (Chang et al. 2015). Finally, we standardized each of the PGRS to have mean 0 and variance 1 and examined their associations with the various EOC subtypes through logistic regression, adjusted for the first ten PCs.
Multiple testing corrections
The polygenic risk prediction approach carries a high multiple testing burden, as does consideration of the various histologic groups and risk factors. However, given that we computed 11 PGRS for each trait based on sequential p-value thresholds, our statistics are not independent. To estimate the real number of independent hypotheses, we computed the correlation matrix of all the PGRS used in this study and fed this into a Matrix Spectral Decomposition (matSpD) algorithm (Nyholt 2004), to estimate the number of independent variables. This algorithm provides an equivalent number of independent variables in a correlation matrix, by examining the ratio of the observed eigenvalue variance to its theoretical maximum. We estimated the number of independent PGRS to be 35 out of the 88 PGRS. As we examined these 35 independent PGRS in five separate EOC subtypes (high-grade serous, endometrioid, clear cell, mucinous and unknown), our significance threshold for the polygenic risk prediction analyses was 0.05/(35 × 5) = .00029.
Results
Genetic contribution of each chromosome and known loci
Fitting a GRM computed after removing known EOC-associated loci in univariate mixed-effect linear models implemented in GCTA (Yang et al. 2010, 2011a), we found that the known loci contributed about 40 % of the total heritability of EOC and each of the subtypes (Table 1). The estimated heritability of all EOC dropped from 5.6 to 3.6 % once we removed known EOC-associated loci from the GRM. We observed a similar reduction of variance explained by the polygenic component for the EOC subtypes high-grade serous (8.8–4.7 %), endometrioid (3.2–2.0 %) and clear cell (6.7–4.6 %) (Table 1). Interestingly, in contrast to grade 1 and grade 2 (G1/G2) endometrioid where the heritability did not drop substantially (4.4–3.7 %), grade 3 (G3) endometrioid dropped from 4.9 to 0.9 %. As shown previously (Lu et al. 2015), the heritability of mucinous cancer was not detectably different from 0. We were unable to perform any analyses for low-grade serous cancer given the small sample size (Ncases = 350). We also had a set of cases with unknown EOC subtype classification; we expect that a high portion of these are individuals with undifferentiated high-grade serous, endometrioid or mixed serous EOC subtypes. For these, the heritability dropped from 7.0 to 4.1 % after removing known loci.
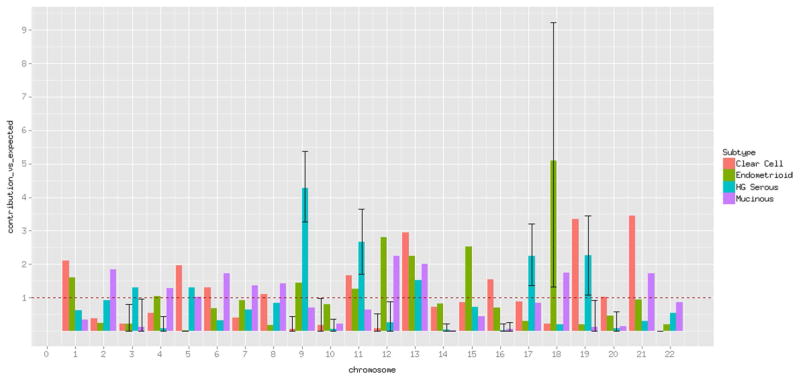
To inspect the contributions of heritability per chromosome, we computed one GRM per chromosome, and fitted the multiple genetic variance components into linear mixed models as above. We found that the chromosomal contributions were not proportional to the number of independent SNPs in each of the chromosomes (Fig. 1). For example, the contribution of chromosomes 9, 11, 17 and 19 to high-grade serous EOC were larger than expected the 95 % confidence interval (approximated through jack-knifing 1000 times) did not overlap with 1. In contrast chromosomes 4, 10, 12, 14, 18 and 20 contributed less than expected.
Fig. 1.
Contribution to the heritability by chromosome versus expected. Black vertical lines show the 95 % confidence intervals approximated through jackknifing up to 1000 times. These are only shown for those instances that do not overlap with 1 to facilitate visualization. The same graph with all confidence intervals is included as supplementary figure 2
Genetic correlation between EOC subtypes
We used the GRM as a random effect in a bivariate mixed-effects linear model implemented in GCTA to assess genetic heterogeneity across EOC histologic subtypes. Table 2 summarizes the genetic correlations between the various EOC subtypes. We found significant genetic overlap between high-grade serous EOC and endometrioid EOC (rg = 0.63 ± 0.27; P = .0029). Given that high-grade serous disease is not infrequently misclassified as endometrioid EOC (Gilks et al. 2008), we also estimated the genetic correlations separating (G1/G2) endometrioid disease from (G3). Here we found that the genetic correlation between high-grade serous and G1/G2 endometrioid cancer was lower (rg = 0.33 ± 0.23; P = .062) than between G3 endometrioid and high-grade serous cancer (rg = 1.00 ± 0.83; P = .00078), suggesting that potential misclassification may have inflated the genetic correlation estimate when using all endometrioid EOC. Interestingly, we observed an appreciable but non-significant genetic overlap of about rg = 0.5 between low-grade endometrioid and clear cell EOC. We also found that the genetic correlations between “unknown/unclassified” EOC and high-grade serous and high-grade endometrioid disease were significant and essentially 1 (rg = 1.0 ± 0.30; P = 10−7 and rg = 1.0 ± 0.96 P = .0049, respectively). The REML bivariate analyses involving Mucinous did not converge so did not yield any meaningful estimates. Further, removing known associated loci from the analyses affected the genetic correlation between endometrioid EOC (high and low grade) in a way that this was no longer significant (Table 2).
Table 2.
Genetic correlations and (standard error) between major EOC subtypes as estimated from iCOGS array
| Subtype | High-grade serous | Endometrioid (all) | Endometrioid G1/G2 | Endometrioid G3 | Clear cell | Unknown |
|---|---|---|---|---|---|---|
| High-grade serous | – | 0.48 (0.35) P = 0.072 | 0.24 (0.30) P = 0.21 | 1.0 (2.66) P = 0.5 | 0.29 (0.42) P = 0.24 | 1.0 (0.510) P = 5.1E – 04 |
| Endometrioid (all) | 0.63 (0.27) P = 0.0029 | – | – | – | 0.73 (0.64) P = 0.088 | 0.50 (0.47) P = 0.12 |
| Endometrioid G1/G2 | 0.33 (0.23) P = 0.062 | – | – | 0.36 (1.25) P = 0.30* | 0.42 (0.53) P = 0.20 | 0.37 (0.41) P = 0.18 |
| Endometrioid G3 | 1.0 (0.83) P = 7.8E–04 | – | 0.42 (0.56) P = 0.2* | – | 1.00 (1.68) P = 0.5 | 1.0 (4.44) P = 0.5 |
| Clear cell | 0.28 (0.33) P = 0.18 | 0.69 (0.56) P = 0.074 | 0.52 (0.54) P = 0.14 | 0.99 (0.87) P = 0.073 | – | 0.09 (0.55) P = 0.43 |
| Unknown | 1.0 (0.30) P = 1.0E–07 | 0.68 (0.33) P = 0.0082 | 0.42 (0.29) P = 0.057 | 1.0 (0.96) P = 0.0049 | 0.15 (0.39) P = 3.5E–01 | – |
Lower triangular matrix shows the genetic correlation using all the SNPs in the iCOGS array, while the upper triangular matrix shows the genetic correlation after removing known associated loci. For these calculations, each case was matched to one control in a way that none of the subtypes share any controls. Analyses for mucinous and low-grade serous EOC subtypes were underpowered to yield reliable estimates Bolded estimates are significantly different from 0
Significance (P value) where the null hypothesis rG = 1
Given that splitting the controls during the bivariate analyses to avoid sample overlap could have resulted in decreased power to detect genetic correlations; we complemented the genetic correlation analysis with the cross-trait LD score regression method, which is not biased by overlapping samples. In line with our results above, we found a statistically significant genetic correlation between high-grade serous EOC and endometrioid EOC (rg = 0.67 ± 0.25; P = 7.4E–03), high-grade serous EOC and unknown EOC (rg = 0.63 ± 0.25; P = .013) and endometrioid EOC and unknown EOC (rg = 1.00 ± 0.30; P = 5.7E–04) (Table 3).
Table 3.
Cross-trait LD score regression between EOC subtypes
| HG serous | Endometrioid | Endometrioid G1/G2 | Endometrioid G3 | Clear Cell | Unknown | |
|---|---|---|---|---|---|---|
| HG serous | – | 0.82 (0.49) P = 0.095 | 0.35 (0.41) P = 0.41 | 1.0 (1.17) P = 0.20 | – | 0.46 (0.46) P = 0.31 |
| Endometrioid | 0.67 (0.25) P = 0.0074 | – | – | – | – | 1.0 (0.41) P = 0.01 |
| Endometrioid G1/G2 | 0.35 (0.25) P = 0.15 | – | – | 0.49 (0.70) P = 0.47* | – | 0.85 (0.40) P = 0.035 |
| Endometrioid G3 | 1.0 (0.79) P = 0.15 | – | 0.53 (0.67) P = 0.48* | – | – | 1.0 (0.73) P = 0.15 |
| Clear Cell | 0.53 (0.57) P = 0.35 | 0.91 (0.80) P = 0.26 | 0.71 (0.59) P = 0.23 | 1.00 (1.06) P = 0.29 | – | – |
| Unknown | 0.63 (0.25) P = 1.3E–02 | 1.0 (0.30) P = 5.7E–04 | 0.77 (0.33) P = 0.02 | 1.00 (0.79) P = 0.14 | 0.38 (0.53) P = 0.47 | – |
Estimates and (standard errors) are reported. Analyses for mucinous and low-grade serous EOC subtypes were underpowered to yield reliable estimates
Bolded estimates are significantly different from 0
Genetic overlap of EOC subtypes and associated environmental factors
To investigate the genetic overlap between all EOC and age at menarche, BMI, obesity, smoking, height and diabetes we used the cross-trait LD score regression method as well as a polygenic risk prediction approach. We did not detect any significant genetic correlations using cross-trait LD score regression (Table 4). However, through the polygenic risk prediction approach, we found significant genetic overlap (at Bonferroni P value threshold = .00029) of all EOC with obesity and with diabetes (Table 5). The genetic overlap with diabetes appeared mainly in association with mucinous EOC. Overall, the directions of association are consistent with what has been reported in observational studies (Aune et al. 2015; Collaborative Group on Epidemiological Studies of Ovarian Cancer 2012; Faber et al. 2013; Olsen et al. 2013), although most of these associations are not significant.
Table 4.
Genetic correlation between risk factors and EOC subtypes using cross-trait LD score regression
| All | HG serous | Endometrioid | Clear cell | Unknown | |
|---|---|---|---|---|---|
| BMI | 0.045 (0.07) P = 0.52 | −0.04 (0.08) P = 0.63 | 0.18 (0.11) P = 0.10 | −0.01 (0.16) P = 0.96 | 0.07 (0.08) P = 0.38 |
| Smoking | −0.34 (0.29) P = 0.23 | −0.43 (0.33) P = 0.20 | −0.37 (0.43) P = 0.39 | −0.44 (0.66) P = 0.51 | −0.17 (0.31) P = 0.58 |
| Height | 0.081 (0.062) P = 0.19 | 0.13 (0.09) P = 0.15 | 0.03 (0.09) P = 0.69 | 0.24 (0.17) P = 0.17 | 0.00 (0.08) P = 0.98 |
| Menarche | −0.07 (0.08) P = 0.38 | −0.23 (0.13) P = 0.06 | −0.04 (0.12) P = 0.75 | 0.32 (0.36) P = 0.36 | 0.05 (0.09) P = 0.59 |
| Obesity* >30 BMI | 0.05 (0.09) P = 0.58 | −0.02 (0.09) P = 0.86 | 0.26 (0.17) P = 0.13 | −0.18 (0.26) P = 0.50 | 0.12 (0.11) P = 0.27 |
| Obesity* >35 BMI | 0.019 (0.087) P = 0.83 | –0.03 (0.11) P = 0.80 | 0.02 (0.18) P = 0.90 | −0.23 (0.37) P = 0.54 | 0.17 (0.12) P = 0.17 |
| Obesity* >40 BMI | −0.02 (0.15) P = 0.88 | −0.02 (0.17) P = 0.92 | −0.06 (0.30) P = 0.84 | NA | 0.03 (0.19) P = 0.89 |
| Diabetes | 0.04 (0.12) P = 0.75 | −0.04 (0.14) P = 0.74 | 0.04 (0.19) P = 0.84 | −0.29 (0.38) P = 0.45 | 0.21 (0.14) P = 0.15 |
Estimates and (standard errors) are reported. Analyses for mucinous and low-grade serous EOC subtypes were underpowered to yield reliable estimates
Reference group was individuals with BMI ≤25
Table 5.
Odds Ratios corresponding to 1 standard deviation increase in the PGRS and significance estimates (P values) from the polygenic risk prediction approach between “environmental factors” PGRS and EOC subtypes
| HG serous | Mucinous | Clear cell | Endometrioid | Unknown | ALL | |
|---|---|---|---|---|---|---|
| Menarche | 0.99 (0.54) | 1.09 (0.036) | 1.05 (0.2) | 1.04 (0.12) | 1.04 (0.086) | 1.02 (0.17) |
| BMI | 1.04 (0.028) | 1.05 (0.26) | 1.06 (0.17) | 1.07 (0.011) | 1.04 (0.068) | 1.04 (0.003) |
| Smoking | 1.03 (0.11) | 0.93 (0.067) | 0.92 (0.049) | 1.04 (0.18) | 0.95 (0.0071) | 0.97 (0.019) |
| Height | 1.03 (0.14) | 1.1 (0.015) | 1.1 (0.025) | 1.04 (0.17) | 0.96 (0.06) | 1.03 (0.022) |
| Diabetes | 1.04 (0.021) | 1.18 (1.1e–05) | 1.08 (0.067) | 1.07 (0.011) | 1.04 (0.034) | 1.05 (4.1e–04) |
| Obesity >30 BMI | 1.05 (0.0051) | 1.06 (0.15) | 1.06 (0.14) | 1.04 (0.19) | 1.04 (0.032) | 1.05 (2.6e–04) |
| Obesity >35 BMI | 1.03 (0.08) | 1.05 (0.21) | 0.9 (0.012) | 1.02 (0.42) | 1.05 (0.028) | 1.04 (0.0053) |
| Obesity >40 BMI | 1.03 (0.15) | 1.06 (0.14) | 0.87 (0.0015) | 0.96 (0.13) | 1.03 (0.19) | 0.98 (0.21) |
The displayed numbers correspond to the best association p value out of the 11 different PGRS which were derived using different p value thresholds. In this part, we used the total set of controls with each of the EOC subtypes
Bolded estimates are statistically significant (Bonferroni P value threshold 2.9 × 10−4)
Reference group was individuals with BMI ≤25
Discussion
In this work, we have investigated the genetic architecture of EOC and its different subtypes. Our univariate analyses show an extent of hidden heritability inherent in the iCOGS array, with known associated loci accounting for about 40 % of the total array heritability for most EOC histotypes, except for high-grade endometrioid, where they account for most of . Its important to note that to reach these estimates we removed 2 Mb per locus, which was done to ensure that no effect of these loci remained; however, this could also have inflated the estimates. We also showed that the hidden heritability is not spread proportionally across the chromosomes, with some contributing very little to the array heritability and others up to five times more than expected given their iCOGS SNP compositions. A limitation in our univariate experiments was that it was underpowered to compute meaningful estimates for low-grade serous and mucinous EOC. Although we had a bigger sample size for mucinous EOC than clear cell EOC, the analyses could have been affected by how each individual study deal with mucin-producing peritoneal tumors.
Using bivariate linear mixed model and cross-trait LD score regression approaches, we investigated genetic correlations between the various EOC subtypes. The bivariate linear mixed model provides unbiased estimates of genetic correlation and it requires individual genotype data to compute the GRM. Cross-trait LD score regression only requires summary results from the discovery set, and in contrast to the bivariate mixed model approach, it allows sample overlap (in this case, overlapping controls) (Bulik-Sullivan et al. 2015a). While studies have shown shared germ-line risk mutations across the various EOC subtypes, these account for only a small fraction of general heritability (Bojesen et al. 2013; Bolton et al. 2010; Goode et al. 2010; Permuth-Wey et al. 2013; Pharoah et al. 2013; Song et al. 2009). We found a very high genetic correlation between high-grade serous EOC and poorly differentiated (G3, high-grade) endometrioid disease, and with unknown/unclassified EOC, which represents undifferentiated epithelial carcinoma. These correlations seem entirely reasonable, because high-grade endometrioid disease is sometimes misdiagnosed as high-grade serous, or may constitute a version of high-grade serous with slightly different differentiation. Undifferentiated ovarian carcinoma clinically resembles high-grade serous in response to treatment and in mortality. Low-grade serous, low-grade endometrioid and clear cell carcinoma (which is relatively low grade) are heritability-distinct from the high-grade diseases and behave that way. Mucinous ovarian cancer seems to be a largely separate disease and has its own set of risk factors (Risch et al. 1996). It does not appear to be related heritably to the other ovarian cancer histotypes.
We also considered whether the heritability of EOC and its subtypes could be explained (at least partly) via factors such as obesity, height, diabetes, smoking and age at menarche. As these factors have genetic components, it is plausible that the heritability of EOC could reflect the heritability of a causal factor. Using cross-trait LD score regression, we had insufficient power to detect genetic correlations, as this approach is greatly affected by small numbers of SNPs and by small sample sizes. However, through a polygenic risk prediction approach—which, although it does not directly quantify genetic overlap, is powerful for detecting genetic correlations between traits when the discovery and target sets are well powered (Dudbridge 2013), we found a significant positive genetic overlap between diabetes, obesity and all EOC. This genetic overlap appeared to be concentrated within mucinous disease and may not reflect other EOC histotypes. Genetic correlation in this analysis is estimated based on a large number of SNPs, so it is possible that the correlations seen between diabetes and obesity and EOC may be mediated by an upstream phenotype (e.g., hormonal changes). Genetic overlap analyses between EOC and the other risk factors did not reveal any other significant associations. Potential reasons for this include small sample sizes for some of the EOC subtypes, and incomplete mapping of relevant variants of the risk factors (i.e., variants in the iCOGS array explain only a limited amount of variance of the risk factors).
Its important to note that our results were derived from SNPs tagged in the iCOGS array. Hence, the numbers of SNPs included in the analyses (195,183 SNPs) are smaller than in a typical GWAS array. Additional analyses could be performed on imputed genotypes from the iCOGS data; however, the iCOGS array is not designed to tag the whole genome, so imputation would likely still be limited to the existing tagged regions. Nevertheless, this array, which included several SNPs associated with other cancer types as well as with relevant quantitative traits such as BMI and the onset of menarche (Pharoah et al. 2013), allowed us to establish reasonably accurate estimates where the target sample sizes were well powered (e.g., high-grade serous, endometrioid, unknown/undifferentiated, and all EOC).
In summary, our results show that the major important EOC subtypes are genetically very homogeneous, and likely arise from a combination of known risk factors plus genetic contributions (beyond the known genetic predisposition mutations). This commonality highlights that high-grade disease could be considered a single clinical entity, with perhaps only minor variation between the serous, endometrioid and undifferentiated types. Low-grade histotypes, as well as mucinous ovarian cancer, likely represent more distinct pathologic variation. We also found that a great proportion of heritability is “missing”. Our analyses will be complemented once data of individuals genotyped in the OncoArray, which integrates a GWAS backbone, becomes available.
Supplementary Material
Acknowledgments
The Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund, thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07). The Nurses’ Health Studies would like to thank the participants and staff of the Nurses’ Health Study and Nurses’ Health Study II for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. Funding of the constituent studies was provided by the California Cancer Research Program (00-01389V-20170, N01-CN25403, 2II0200); the Canadian Institutes of Health Research (MOP-86727); Cancer Australia; Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124); the Danish Cancer Society (94-222-52); the ELAN Program of the University of Erlangen-Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Helse Vest; the Norwegian Cancer Society; the Norwegian Research Council; the Ovarian Cancer Research Fund; National Kankerplan of Belgium; the L & S Milken Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); the Roswell Park Cancer Institute Alliance Foundation; the US National Cancer Institute (K07-CA095666, K07-CA80668, K07-CA143047, K22-CA138563, N01-CN55424, N01-PC67001, N01-PC067010, N01-PC035137, P01-CA017054, P01-CA087696, P30-CA072720, P30-CA15083, P30-CA008748, P50-CA159981, P50-CA105009, P50-CA136393, R01-CA149429, R01-CA014089, R01-CA016056, R01-CA017054, R01-CA049449, R01-CA050385, R01-CA054419, R01-CA058598, R01-CA058860, R01-CA061107, R01-CA061132, R01-CA063678, R01-CA063682, R01-CA067262, R01-CA071766, R01-CA074850, R01-CA080978, R01-CA083918, R01-CA087538, R01-CA092044, R01-CA095023, R01-CA122443, R01-CA112523, R01-CA114343, R01-CA126841, R01-CA136924, R03-CA113148, R03-CA115195, U01-CA069417, U01-CA071966, UM1-CA186107, UM1-CA176726 and Intramural research funds); the NIH/National Center for Research Resources/General Clinical Research Center (MO1-RR000056); the US Army Medical Research and Material Command (DAMD17-01-1-0729, DAMD17-02-1-0666, DAMD17-02-1-0669, W81XWH-07-0449, W81XWH-10-1-02802); the US Public Health Service (PSA-042205); the National Health and Medical Research Council of Australia (199600 and 400281); the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research (01 GB 9401); the State of Baden-Wurttemberg through Medical Faculty of the University of Ulm (P.685); the German Cancer Research Center; the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Lon V. Smith Foundation (LVS-39420); the Oak Foundation; Eve Appeal; the OHSU Foundation; the Mermaid I project; the Rudolf-Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital ‘Womens Health Theme’ and the Royal Marsden Hospital; and WorkSafeBC 14. Investigator-specific funding: G.C.P receives scholarship support from the University of Queensland and QIMR Berghofer. Y. L. was supported by the NHMRC Early Career Fellowship. G. C. T. is supported by the National Health and Medical Research Council. S. M. was supported by an ARC Future Fellowship.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-016-1663-9) contains supplementary material, which is available to authorized users.
Contributor Information
Gabriel Cuellar-Partida, Email: gabriel.cuellar@qimrberghofer.edu.au.
Stuart MacGregor, Email: Stuart.MacGregor@qimrberghofer.edu.au.
References
- Alsop K, Fereday S, Meldrum C, deFazio A, Emmanuel C, George J, Dobrovic A, Birrer MJ, Webb PM, Stewart C, Friedlander M, Fox S, Bowtell D, Mitchell G. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol. 2012;30:2654–2663. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglesio MS, Kommoss S, Tolcher MC, Clarke B, Galletta L, Porter H, Damaraju S, Fereday S, Winterhoff BJ, Kalloger SE, Senz J, Yang W, Steed H, Allo G, Ferguson S, Shaw P, Teoman A, Garcia JJ, Schoolmeester JK, Bakkum-Gamez J, Tinker AV, Bowtell DD, Huntsman DG, Gilks CB, McAlpine JN. Molecular characterization of mucinous ovarian tumours supports a stratified treatment approach with HER2 targeting in 19% of carcinomas. J Pathol. 2013;229:111–120. doi: 10.1002/path.4088. [DOI] [PubMed] [Google Scholar]
- Aune D, Navarro Rosenblatt DA, Chan DS, Abar L, Vingeliene S, Vieira AR, Greenwood DC, Norat T. Anthropometric factors and ovarian cancer risk: a systematic review and nonlinear dose-response meta-analysis of prospective studies. Int J Cancer. 2015;136:1888–1898. doi: 10.1002/ijc.29207. [DOI] [PubMed] [Google Scholar]
- Berchuck A, Heron KA, Carney ME, Lancaster JM, Fraser EG, Vinson VL, Deffenbaugh AM, Miron A, Marks JR, Futreal PA, Frank TS. Frequency of germline and somatic BRCA1 mutations in ovarian cancer. Clin Cancer Res. 1998;4:2433–2437. [PubMed] [Google Scholar]
- Berndt SI, Gustafsson S, Magi R, Ganna A, Wheeler E, Feitosa MF, Justice AE, Monda KL, Croteau-Chonka DC, Day FR, Esko T, Fall T, Ferreira T, Gentilini D, Jackson AU, Luan J, Randall JC, Vedantam S, Willer CJ, Winkler TW, Wood AR, Workalemahu T, Hu YJ, Lee SH, Liang L, Lin DY, Min JL, Neale BM, Thorleifsson G, Yang J, Albrecht E, Amin N, Bragg-Gresham JL, Cadby G, den Heijer M, Eklund N, Fischer K, Goel A, Hottenga JJ, Huffman JE, Jarick I, Johansson A, Johnson T, Kanoni S, Kleber ME, Konig IR, Kristiansson K, Kutalik Z, Lamina C, Lecoeur C, Li G, Mangino M, McArdle WL, Medina-Gomez C, Muller-Nurasyid M, Ngwa JS, Nolte IM, Paternoster L, Pechlivanis S, Perola M, Peters MJ, Preuss M, Rose LM, Shi J, Shungin D, Smith AV, Strawbridge RJ, Surakka I, Teumer A, Trip MD, Tyrer J, Van Vliet-Ostaptchouk JV, Vandenput L, Waite LL, Zhao JH, Absher D, Asselbergs FW, Atalay M, Attwood AP, Balmforth AJ, Basart H, Beilby J, Bonnycastle LL, Brambilla P, Bruinenberg M, Campbell H, Chasman DI, Chines PS, Collins FS, Connell JM, Cookson WO, de Faire U, de Vegt F, Dei M, Dimitriou M, Edkins S, Estrada K, Evans DM, Farrall M, Ferrario MM, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, Hillman KM, Mai PL, Lawrenson K, Stutz MD, Lu Y, Karevan R, Woods N, Johnston RL, French JD, Chen X, Weischer M, Nielsen SF, Maranian MJ, Ghoussaini M, Ahmed S, Baynes C, Bolla MK, Wang Q, Dennis J, McGuffog L, Barrowdale D, Lee A, Healey S, Lush M, Tessier DC, Vincent D, Bacot F, Vergote I, Lambrechts S, Despierre E, Risch HA, Gonzalez-Neira A, Rossing MA, Pita G, Doherty JA, Alvarez N, Larson MC, Fridley BL, Schoof N, Chang-Claude J, Cicek MS, Peto J, Kalli KR, Broeks A, Armasu SM, Schmidt MK, Braaf LM, Winterhoff B, Nevanlinna H, Konecny GE, Lambrechts D, Rogmann L, Guenel P, Teoman A, Milne RL, Garcia JJ, Cox A, Shridhar V, Burwinkel B, Marme F, Hein R, Sawyer EJ, Haiman CA, Wang-Gohrke S, Andrulis IL, Moysich KB, Hopper JL, Odunsi K, Lindblom A, Giles GG, Brenner H, Simard J, Lurie G, Fasching PA, Carney ME, Radice P, Wilkens LR, Swerdlow A, Goodman MT, et al. Australian Cancer Study, Australian Ovarian Cancer Study, Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer, Gene Environment Interaction and Breast Cancer, Swedish Breast Cancer Study, Hereditary Breast and Ovarian Cancer Research Group Netherlands, Epidemiological study of BRCA1 and BRCA2 Mutation Carriers (EMBRACE); Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers (GEMO) Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–384. 384e1–2. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, Sher T, Gentry-Maharaj A, Wozniak E, Tsai YY, Weidhaas J, Paik D, Van Den Berg DJ, Stram DO, Pearce CL, Wu AH, Brewster W, Anton-Culver H, Ziogas A, Narod SA, Levine DA, Kaye SB, Brown R, Paul J, Flanagan J, Sieh W, McGuire V, Whittemore AS, Campbell I, Gore ME, Lissowska J, Yang HP, Medrek K, Gronwald J, Lubinski J, Jakubowska A, Le ND, Cook LS, Kelemen LE, Brook-Wilson A, Massuger LF, Kiemeney LA, Aben KK, van Altena AM, Houlston R, Tomlinson I, Palmieri RT, Moorman PG, Schildkraut J, Iversen ES, Phelan C, Vierkant RA, Cunningham JM, Goode EL, Fridley BL, Kruger-Kjaer S, Blaeker J, Hogdall E, Hogdall C, Gross J, Karlan BY, Ness RB, Edwards RP, Odunsi K, Moyisch KB, Baker JA, Modugno F, Heikkinenen T, Butzow R, Nevanlinna H, Leminen A, Bogdanova N, Antonenkova N, Doerk T, Hillemanns P, Durst M, Runnebaum I, Thompson PJ, Carney ME, Goodman MT, Lurie G, Wang-Gohrke S, Hein R, Chang-Claude J, Rossing MA, Cushing-Haugen KL, Doherty J, Chen C, Rafnar T, Besenbacher S, Sulem P, Stefansson K, Birrer MJ, Terry KL, Hernandez D, Cramer DW, Vergote I, Amant F, Lambrechts D, Despierre E, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM ReproGen Consortium, Psychiatric Genomics Consortium, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015a doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015b;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chornokur G, Lin HY, Tyrer JP, Lawrenson K, Dennis J, Amankwah EK, Qu X, Tsai YY, Jim HS, Chen Z, Chen AY, Permuth-Wey J, Aben K, Anton-Culver H, Antonenkova N, Bruinsma F, Bandera EV, Bean YT, Beckmann MW, Bisogna M, Bjorge L, Bogdanova N, Brinton LA, Brooks-Wilson A, Bunker CH, Butzow R, Campbell IG, Carty K, Chang-Claude J, Cook LS, Cramer DW, Cunningham JM, Cybulski C, Dansonka-Mieszkowska A, du Bois A, Despierre E, Dicks E, Doherty JA, Dork T, Durst M, Easton DF, Eccles DM, Edwards RP, Ekici AB, Fasching PA, Fridley BL, Gao YT, Gentry-Maharaj A, Giles GG, Glasspool R, Goodman MT, Gronwald J, Harrington P, Harter P, Hein A, Heitz F, Hildebrandt MA, Hillemanns P, Hogdall CK, Hogdall E, Hosono S, Jakubowska A, Jensen A, Ji BT, Karlan BY, Kelemen LE, Kellar M, Kiemeney LA, Krakstad C, Kjaer SK, Kupryjanczyk J, Lambrechts D, Lambrechts S, Le ND, Lee AW, Lele S, Leminen A, Lester J, Levine DA, Liang D, Lim BK, Lissowska J, Lu K, Lubinski J, Lundvall L, Massuger LF, Matsuo K, McGuire V, McLaughlin JR, McNeish I, Menon U, Milne RL, Modugno F, Moysich KB, Ness RB, Nevanlinna H, Eilber U, Odunsi K, Olson SH, Orlow I, et al. Common genetic variation in cellular transport genes and epithelial ovarian cancer (EOC) risk. PLoS One. 2015;10:e0128106. doi: 10.1371/journal.pone.0128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Group on Epidemiological Studies of Ovarian Cancer. Ovarian cancer and body size: individual participant meta-analysis including 25,157 women with ovarian cancer from 47 epidemiological studies. PLoS Med. 2012;9:e1001200. doi: 10.1371/journal.pmed.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Group on Epidemiological Studies of Ovarian Cancer. Beral V, Gaitskell K, Hermon C, Moser K, Reeves G, Peto R. Ovarian cancer and smoking: individual participant meta-analysis including 28,114 women with ovarian cancer from 51 epidemiological studies. Lancet Oncol. 2012;13:946–956. doi: 10.1016/S1470-2045(12)70322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, Mowry BJ, Thapar A, Goddard ME, Witte JS, Absher D, Agartz I, Akil H, Amin F, Andreassen OA, Anjorin A, Anney R, Anttila V, Arking DE, Asherson P, Azevedo MH, Backlund L, Badner JA, Bailey AJ, Banaschewski T, Barchas JD, Barnes MR, Barrett TB, Bass N, Battaglia A, Bauer M, Bayes M, Bellivier F, Bergen SE, Berrettini W, Betancur C, Bettecken T, Biederman J, Binder EB, Black DW, Blackwood DH, Bloss CS, Boehnke M, Boomsma DI, Breen G, Breuer R, Bruggeman R, Cormican P, Buccola NG, Buitelaar JK, Bunney WE, Buxbaum JD, Byerley WF, Byrne EM, Caesar S, Cahn W, Cantor RM, Casas M, Chakravarti A, Chambert K, Choudhury K, Cichon S, Cloninger CR, Collier DA, Cook EH, Coon H, Cormand B, Corvin A, Coryell WH, Craig DW, Craig IW, Crosbie J, Cuccaro ML, Curtis D, Czamara D, Datta S, Dawson G, Day R, De Geus EJ, Degenhardt F, Djurovic S, Donohoe GJ, Doyle AE, Duan J, Dudbridge F, Duketis E, Ebstein RP, Edenberg HJ, Elia J, Ennis S, Etain B, Fanous A, Farmer AE, Ferrier IN, Flickinger M, Fombonne E, Foroud T, Frank J, Franke B, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuatrecasas M, Villanueva A, Matias-Guiu X, Prat J. K-ras mutations in mucinous ovarian tumors: a clinicopathologic and molecular study of 95 cases. Cancer. 1997;79:1581–1586. doi: 10.1002/(sici)1097-0142(19970415)79:8<1581::aid-cncr21>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Della Pepa C, Tonini G, Santini D, Losito S, Pisano C, Di Napoli M, Cecere SC, Gargiulo P, Pignata S. Low grade serous ovarian carcinoma: from the molecular characterization to the best therapeutic strategy. Cancer Treat Rev. 2015;41:136–143. doi: 10.1016/j.ctrv.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Devouassoux-Shisheboran M, Genestie C. Pathobiology of ovarian carcinomas. Chin J Cancer. 2015;34:50–55. doi: 10.5732/cjc.014.10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber MT, Kjaer SK, Dehlendorff C, Chang-Claude J, Andersen KK, Hogdall E, Webb PM, Jordan SJ, Rossing MA, Doherty JA, Lurie G, Thompson PJ, Carney ME, Goodman MT, Ness RB, Modugno F, Edwards RP, Bunker CH, Goode EL, Fridley BL, Vierkant RA, Larson MC, Schildkraut J, Cramer DW, Terry KL, Vitonis AF, Bandera EV, Olson SH, King M, Chandran U, Kiemeney LA, Massuger LF, van Altena AM, Vermeulen SH, Brinton L, Wentzensen N, Lissowska J, Yang HP, Moysich KB, Odunsi K, Kasza K, Odunsi-Akanji O, Song H, Pharaoh P, Shah M, Whittemore AS, McGuire V, Sieh W, Sutphen R, Menon U, Gayther SA, Ramus SJ, Gentry-Maharaj A, Pearce CL, Wu AH, Pike MC, Risch HA, Jensen A Australian Cancer Study, Australian Ovarian Cancer Study Group, Ovarian Cancer Association Consortium. Cigarette smoking and risk of ovarian cancer: a pooled analysis of 21 case-control studies. Cancer Causes Control. 2013;24:989–1004. doi: 10.1007/s10552-013-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapstur SM, Patel AV, Diver WR, Hildebrand JS, Gaudet MM, Jacobs EJ, Campbell PT. Type II diabetes mellitus and the incidence of epithelial ovarian cancer in the cancer prevention study-II nutrition cohort. Cancer Epidemiol Biomarkers Prev. 2012;21:2000–2005. doi: 10.1158/1055-9965.EPI-12-0867. [DOI] [PubMed] [Google Scholar]
- Gilks CB, Ionescu DN, Kalloger SE, Kobel M, Irving J, Clarke B, Santos J, Le N, Moravan V, Swenerton K Cheryl Brown Ovarian Cancer Outcomes Unit of the British Columbia Cancer Agency. Tumor cell type can be reproducibly diagnosed and is of independent prognostic significance in patients with maximally debulked ovarian carcinoma. Hum Pathol. 2008;39:1239–1251. doi: 10.1016/j.humpath.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Gong TT, Wu QJ, Vogtmann E, Lin B, Wang YL. Age at menarche and risk of ovarian cancer: a meta-analysis of epidemiological studies. Int J Cancer. 2013;132:2894–2900. doi: 10.1002/ijc.27952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, Widschwendter M, Vierkant RA, Larson MC, Kjaer SK, Birrer MJ, Berchuck A, Schildkraut J, Tomlinson I, Kiemeney LA, Cook LS, Gronwald J, Garcia-Closas M, Gore ME, Campbell I, Whittemore AS, Sutphen R, Phelan C, Anton-Culver H, Pearce CL, Lambrechts D, Rossing MA, Chang-Claude J, Moysich KB, Goodman MT, Dork T, Nevanlinna H, Ness RB, Rafnar T, Hogdall C, Hogdall E, Fridley BL, Cunningham JM, Sieh W, McGuire V, Godwin AK, Cramer DW, Hernandez D, Levine D, Lu K, Iversen ES, Palmieri RT, Houlston R, van Altena AM, Aben KK, Massuger LF, Brooks-Wilson A, Kelemen LE, Le ND, Jakubowska A, Lubinski J, Medrek K, Stafford A, Easton DF, Tyrer J, Bolton KL, Harrington P, Eccles D, Chen A, Molina AN, Davila BN, Arango H, Tsai YY, Chen Z, Risch HA, McLaughlin J, Narod SA, Ziogas A, Brewster W, Gentry-Maharaj A, Menon U, Wu AH, Stram DO, Pike MC, Beesley J, Webb PM, Chen X, Ekici AB, Thiel FC, Beckmann MW, Yang H, Wentzensen N, Lissowska J, Fasching PA, Despierre E, Amant F, Vergote I, Doherty J, Hein R, Wang-Gohrke S, Lurie G, et al. Wellcome Trust Case-Control Consortium, Australian Cancer Study, Australian Ovarian Cancer Study Group, Ovarian Cancer Association Consortium. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42:874–879. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisham RN, Iyer G, Garg K, DeLair D, Hyman DM, Zhou Q, Iasonos A, Berger MF, Dao F, Spriggs DR, Levine DA, Aghajanian C, Solit DB. BRAF mutation is associated with early stage disease and improved outcome in patients with low-grade serous ovarian cancer. Cancer. 2013;119:548–554. doi: 10.1002/cncr.27782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Kurman RJ, Nakayama K, Velculescu VE, Vogelstein B, Kinzler KW, Papadopoulos N, Shih Ie M. Low-grade serous carcinomas of the ovary contain very few point mutations. J Pathol. 2012;226:413–420. doi: 10.1002/path.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurman RJ, Shih Ie M. Pathogenesis of ovarian cancer: lessons from morphology and molecular biology and their clinical implications. Int J Gynecol Pathol. 2008;27:151–160. doi: 10.1097/PGP.0b013e318161e4f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurman RJ, Carcangiu M, Herrington C, Young R. WHO classification of tumours of female reproductive organs. IARC Press; 2014. [Google Scholar]
- Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Jeon I, Kim JW, Song YS, Yoon JM, Park SM. Diabetes mellitus and ovarian cancer risk: a systematic review and meta-analysis of observational studies. Int J Gynecol Cancer. 2013;23:402–412. doi: 10.1097/IGC.0b013e31828189b2. [DOI] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Magi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Hua Zhao J, Zhao W, Chen J, Fehrmann R, Hedman AK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Mateo Leach I, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stancakova A, Strawbridge RJ, Ju Sung Y, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Arnlov J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Bluher M, Bohringer S, Bonnycastle LL, Bottcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Ida Chen YD, Clarke R, Daw EW, de Craen AJ, Delgado G, Dimitriou M, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Cuellar-Partida G, Painter JN, Nyholt DR, Morris AP, Fasching PA, Hein A, Burghaus S, Beckmann MW, Lambrechts D, Van Nieuwenhuysen E, Vergote I, Vanderstichele A, Doherty JA, Rossing MA, Wicklund KG, Chang-Claude J, Eilber U, Rudolph A, Wang-Gohrke S, Goodman MT, Bogdanova N, Dork T, Durst M, Hillemanns P, Runnebaum IB, Antonenkova N, Butzow R, Leminen A, Nevanlinna H, Pelttari LM, Edwards RP, Kelley JL, Modugno F, Moysich KB, Ness RB, Cannioto R, Hogdall E, Jensen A, Giles GG, Bruinsma F, Kjaer SK, Hildebrandt MA, Liang D, Lu KH, Wu X, Bisogna M, Dao F, Levine DA, Cramer DW, Terry KL, Tworoger SS, Missmer S, Bjorge L, Salvesen HB, Kopperud RK, Bischof K, Aben KK, Kiemeney LA, Massuger LF, Brooks-Wilson A, Olson SH, McGuire V, Rothstein JH, Sieh W, Whittemore AS, Cook LS, Le ND, Gilks CB, Gronwald J, Jakubowska A, Lubinski J, Gawelko J, Song H, Tyrer JP, Wentzensen N, Brinton L, Trabert B, Lissowska J, McLaughlin JR, Narod SA, Phelan C, Anton-Culver H, Ziogas A, Eccles D, Gayther SA, Gentry-Maharaj A, Menon U, Ramus SJ, Wu AH, Dansonka-Mieszkowska A, Kupryjanczyk J, Timorek A, Szafron L, Cunningham JM, Fridley BL, Winham SJ, Bandera EV, et al. Australian Ovarian Cancer Study, International Endogene Consortium. Shared genetics underlying epidemiological association between endometriosis and ovarian cancer. Hum Mol Genet. 2015;24:5955–5964. doi: 10.1093/hmg/ddv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Ek WE, Whiteman D, Vaughan TL, Spurdle AB, Easton DF, Pharoah PD, Thompson DJ, Dunning AM, Hayward NK, Chenevix-Trench G, Macgregor S Q-MEGA and AMFS Investigators, ANECS-SEARCH, UKOPS-SEARCH, BEACON Consortium. Most common ‘sporadic’ cancers have a significant germline genetic component. Hum Mol Genet. 2014;23:6112–6118. doi: 10.1093/hmg/ddu312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica A, Deavers MT, Lu K, Bodurka DC, Atkinson EN, Gershenson DM, Silva EG. Grading ovarian serous carcinoma using a two-tier system. Am J Surg Pathol. 2004;28:496–504. doi: 10.1097/00000478-200404000-00009. [DOI] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Muller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJ, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WH, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JR, Platou CG, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stancakova A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney AS, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutskov K, Langford C, Leander K, Lindholm E, Lobbens S, Mannisto S, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen CM, Nagle CM, Whiteman DC, Ness R, Pearce CL, Pike MC, Rossing MA, Terry KL, Wu AH, Risch HA, Yu H, Doherty JA, Chang-Claude J, Hein R, Nickels S, Wang-Gohrke S, Goodman MT, Carney ME, Matsuno RK, Lurie G, Moysich K, Kjaer SK, Jensen A, Hogdall E, Goode EL, Fridley BL, Vierkant RA, Larson MC, Schildkraut J, Hoyo C, Moorman P, Weber RP, Cramer DW, Vitonis AF, Bandera EV, Olson SH, Rodriguez-Rodriguez L, King M, Brinton LA, Yang H, Garcia-Closas M, Lissowska J, Anton-Culver H, Ziogas A, Gayther SA, Ramus SJ, Menon U, Gentry-Maharaj A, Webb PM Australian Cancer Study, Australian Ovarian Cancer Study Group, Ovarian Cancer Association Consortium. Obesity and risk of ovarian cancer subtypes: evidence from the Ovarian Cancer Association Consortium. Endocr Relat Cancer. 2013;20:251–262. doi: 10.1530/ERC-12-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permuth-Wey J, Lawrenson K, Shen HC, Velkova A, Tyrer JP, Chen Z, Lin HY, Chen YA, Tsai YY, Qu X, Ramus SJ, Karevan R, Lee J, Lee N, Larson MC, Aben KK, Anton-Culver H, Antonenkova N, Antoniou AC, Armasu SM, Bacot F, Baglietto L, Bandera EV, Barnholtz-Sloan J, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Cai Q, Campbell I, Chang-Claude J, Chanock S, Chenevix-Trench G, Cheng JQ, Cicek MS, Coetzee GA, Cook LS, Couch FJ, Cramer DW, Cunningham JM, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dork T, du Bois A, Durst M, Easton DF, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher DA, Flanagan JM, Garcia-Closas M, Gentry-Maharaj A, Giles GG, Glasspool RM, Gonzalez-Bosquet J, Goodman MT, Gore M, Gorski B, Gronwald J, Hall P, Halle MK, Harter P, Heitz F, Hillemanns P, Hoatlin M, Hogdall CK, Hogdall E, Hosono S, Jakubowska A, Jensen A, Jim H, Kalli KR, Karlan BY, Kaye SB, Kelemen LE, Kiemeney LA, Kikkawa F, Konecny GE, Krakstad C, Kjaer SK, Kupryjanczyk J, Lambrechts D, Lambrechts S, Lancaster JM, Le ND, Leminen A, Levine DA, Liang D, Lim BK, Lin J, et al. Australian Cancer Study; Australian Ovarian Cancer Study, Consortium of Investigators of Modifiers of BRCA1/2. Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nat Commun. 2013;4:1627. doi: 10.1038/ncomms2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JR, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, He C, Chasman DI, Esko T, Thorleifsson G, Albrecht E, Ang WQ, Corre T, Cousminer DL, Feenstra B, Franceschini N, Ganna A, Johnson AD, Kjellqvist S, Lunetta KL, McMahon G, Nolte IM, Paternoster L, Porcu E, Smith AV, Stolk L, Teumer A, Tsernikova N, Tikkanen E, Ulivi S, Wagner EK, Amin N, Bierut LJ, Byrne EM, Hottenga JJ, Koller DL, Mangino M, Pers TH, Yerges-Armstrong LM, Hua Zhao J, Andrulis IL, Anton-Culver H, Atsma F, Bandinelli S, Beckmann MW, Benitez J, Blomqvist C, Bojesen SE, Bolla MK, Bonanni B, Brauch H, Brenner H, Buring JE, Chang-Claude J, Chanock S, Chen J, Chenevix-Trench G, Collee JM, Couch FJ, Couper D, Coviello AD, Cox A, Czene K, D’Adamo AP, Davey Smith G, De Vivo I, Demerath EW, Dennis J, Devilee P, Dieffenbach AK, Dunning AM, Eiriksdottir G, Eriksson JG, Fasching PA, Ferrucci L, Flesch-Janys D, Flyger H, Foroud T, Franke L, Garcia ME, Garcia-Closas M, Geller F, de Geus EE, Giles GG, Gudbjartsson DF, Gudnason V, Guenel P, Guo S, Hall P, Hamann U, Haring R, Hartman CA, Heath AC, Hofman A, Hooning MJ, Hopper JL, Hu FB, Hunter DJ, Karasik D, Kiel DP, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H, Weber R, Karevan R, Larson MC, Song H, Tessier DC, Bacot F, Vincent D, Cunningham JM, Dennis J, Dicks E, Aben KK, Anton-Culver H, Antonenkova N, Armasu SM, Baglietto L, Bandera EV, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brenton JD, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Campbell I, Carney ME, Carvalho RS, Chang-Claude J, Chen YA, Chen Z, Chow WH, Cicek MS, Coetzee G, Cook LS, Cramer DW, Cybulski C, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dork T, du Bois A, Durst M, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher D, Flanagan J, Gao YT, Garcia-Closas M, Gentry-Maharaj A, Giles G, Gjyshi A, Gore M, Gronwald J, Guo Q, Halle MK, Harter P, Hein A, Heitz F, Hillemanns P, Hoatlin M, Hogdall E, Hogdall CK, Hosono S, Jakubowska A, Jensen A, Kalli KR, Karlan BY, Kelemen LE, Kiemeney LA, Kjaer SK, Konecny GE, Krakstad C, Kupryjanczyk J, Lambrechts D, Lambrechts S, Le ND, Lee N, Lee J, Leminen A, Lim BK, Lissowska J, Lubinski J, Lundvall L, Lurie G, Massuger LF, et al. Australian Cancer Study, Australian Ovarian Cancer Study Group. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45:362–370. 370e1–2. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat J. New insights into ovarian cancer pathology. Ann Oncol. 2012;23(Suppl 10):x111–x117. doi: 10.1093/annonc/mds300. [DOI] [PubMed] [Google Scholar]
- Risch HA, Marrett LD, Jain M, Howe GR. Differences in risk factors for epithelial ovarian cancer by histologic type. Results of a case–control study. Am J Epidemiol. 1996;144:363–372. doi: 10.1093/oxfordjournals.aje.a008937. [DOI] [PubMed] [Google Scholar]
- Shih Ie M, Kurman RJ. Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am J Pathol. 2004;164:1511–1518. doi: 10.1016/s0002-9440(10)63708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Ezendam N, Bulten J, Nagtegaal I, Massuger L. Survival of patients with mucinous ovarian carcinoma and ovarian metastases: a population-based cancer registry study. Int J Gynecol Cancer. 2015 doi: 10.1097/IGC.0000000000000473. [DOI] [PubMed] [Google Scholar]
- Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, Anton-Culver H, Chang-Claude J, Cramer DW, DiCioccio R, Dork T, Goode EL, Goodman MT, Schildkraut JM, Sellers T, Baglietto L, Beckmann MW, Beesley J, Blaakaer J, Carney ME, Chanock S, Chen Z, Cunningham JM, Dicks E, Doherty JA, Durst M, Ekici AB, Fenstermacher D, Fridley BL, Giles G, Gore ME, De Vivo I, Hillemanns P, Hogdall C, Hogdall E, Iversen ES, Jacobs IJ, Jakubowska A, Li D, Lissowska J, Lubinski J, Lurie G, McGuire V, McLaughlin J, Medrek K, Moorman PG, Moysich K, Narod S, Phelan C, Pye C, Risch H, Runnebaum IB, Severi G, Southey M, Stram DO, Thiel FC, Terry KL, Tsai YY, Tworoger SS, Van Den Berg DJ, Vierkant RA, Wang-Gohrke S, Webb PM, Wilkens LR, Wu AH, Yang H, Brewster W, Ziogas A, Houlston R, Tomlinson I, Whittemore AS, Rossing MA, Ponder BA, Pearce CL, Ness RB, Menon U, Kjaer SK, Gronwald J, Garcia-Closas M, Fasching PA, Easton DF, Chenevix-Trench G, Berchuck A, Pharoah PD, Gayther SA Australian Cancer Study, Australian Ovarian Cancer Study Group, Ovarian Cancer Association Consortium. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopik V, Iqbal J, Rosen B, Narod SA. Why have ovarian cancer mortality rates declined? Incidence. Gynecol Oncol, Part I. 2015 doi: 10.1016/j.ygyno.2015.06.017. [DOI] [PubMed] [Google Scholar]
- Soslow RA. Histologic subtypes of ovarian carcinoma: an overview. Int J Gynecol Pathol. 2008;27:161–174. doi: 10.1097/PGP.0b013e31815ea812. [DOI] [PubMed] [Google Scholar]
- Sung PL, Chang YH, Chao KC, Chuang CM Task Force on Systematic Review and Meta-analysis of Ovarian Cancer. Global distribution pattern of histological subtypes of epithelial ovarian cancer: a database analysis and systematic review. Gynecol Oncol. 2014;133:147–154. doi: 10.1016/j.ygyno.2014.02.016. [DOI] [PubMed] [Google Scholar]
- Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Boomsma DI. Interplay between heritability of smoking and environmental conditions? A comparison of two birth cohorts. BMC Public Health. 2011;11:316. doi: 10.1186/1471-2458-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35:397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih Ie M, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiren S, Haggstrom C, Ulmer H, Manjer J, Bjorge T, Nagel G, Johansen D, Hallmans G, Engeland A, Concin H, Jonsson H, Selmer R, Tretli S, Stocks T, Stattin P. Pooled cohort study on height and risk of cancer and cancer death. Cancer Causes Control. 2014;25:151–159. doi: 10.1007/s10552-013-0317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, Amin N, Buchkovich ML, Croteau-Chonka DC, Day FR, Duan Y, Fall T, Fehrmann R, Ferreira T, Jackson AU, Karjalainen J, Lo KS, Locke AE, Magi R, Mihailov E, Porcu E, Randall JC, Scherag A, Vinkhuyzen AA, Westra HJ, Winkler TW, Workalemahu T, Zhao JH, Absher D, Albrecht E, Anderson D, Baron J, Beekman M, Demirkan A, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Fraser RM, Goel A, Gong J, Justice AE, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Lui JC, Mangino M, Mateo Leach I, Medina-Gomez C, Nalls MA, Nyholt DR, Palmer CD, Pasko D, Pechlivanis S, Prokopenko I, Ried JS, Ripke S, Shungin D, Stancakova A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Afzal U, Arnlov J, Arscott GM, Bandinelli S, Barrett A, Bellis C, Bennett AJ, Berne C, Bluher M, Bolton JL, Bottcher Y, Boyd HA, Bruinenberg M, Buckley BM, Buyske S, Caspersen IH, Chines PS, Clarke R, Claudi-Boehm S, Cooper M, Daw EW, De Jong PA, Deelen J, Delgado G, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011a;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011b;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.