Abstract
An evaluation of the sensitivities of three DNA extraction methods, i.e., FTA filter paper, a QIAamp stool mini kit, and a conventional phenol-chloroform method, by using specimens with known concentrations of Enterocytozoon bieneusi spores was performed. FTA filter paper and the QIAamp stool mini kit were the most sensitive methods, which could detect E. bieneusi in specimens with a concentration of 800 spores/ml. We also compared five previously described PCR methods that use five different primer pairs for the detection of E. bieneusi and showed that MSP3-MSP4B and EBIEF1-EBIER1 were the most sensitive primers. Although both sets of primers showed the same sensitivity, using the MSP3-MSP4B primers can directly provide genotypic information by sequencing. A blinded diagnostic test to compare PCR and light microscopy methods for the detection of E. bieneusi in stool specimens was also conducted. The use of FTA filter paper for DNA extraction together with the PCR method using the primer pair MSP3-MSP4B showed 100% sensitivity and 100% specificity for the detection of E. bieneusi in stool specimens, while the light microscopy method gave a sensitivity of 86.7% and a specificity of 100%.
Enterocytozoon bieneusi is an emerging pathogen causing diarrhea in patients with human immunodeficiency virus infection and other immunosuppressive conditions (9, 20, 33). Self-limited diarrhea as well as chronic diarrhea in immunocompetent patients has also been reported (25, 32). The prevalence of E. bieneusi in human immunodeficiency virus-infected patients with diarrhea was 2 to 50%, depending on the study population and methods of diagnosis (2, 8, 16, 20). Several staining methods such as Gram-chromotrope (17), modified trichrome (31), and chemofluorescence stains such as Calcofluor White M2R (29) have been developed for the detection of E. bieneusi. However, the accuracy of microscopic diagnosis of this organism depends on the experience of the microscopist. Moreover, staining methods cannot differentiate down to the species level. Thus, electron microscopy is still necessary for confirmation of the diagnosis and species identification (6). Molecular techniques that rely on PCR-based methods to amplify different regions of the small subunit rRNA (SSU rRNA) gene for the identification of E. bieneusi have been successfully developed (4, 7, 26, 30, 35). More recently, a real-time PCR method was used to quantify E. bieneusi DNA in stool specimens for monitoring treatment in immunocompromised patients (15). A multicenter study has shown that PCR can detect E. bieneusi in concentrations as low as 102 spores/g of stool, while a detection limit of 104 spores/g of stool was apparent for light microscopy (22). Thus, epidemiological studies based on only light microscopy may give prevalence data which do not reflect the true prevalence of E. bieneusi.
PCR amplification using stool specimens for the detection of E. bieneusi could be insensitive because of PCR inhibitors and the difficulty of spore disruption. To raise the sensitivity of PCR to diagnose E. bieneusi infection, an efficient DNA extraction method is needed. Commercial DNA extraction kits such as the QIAamp stool mini kit (QIAGEN, Hilden, Germany), Instagene Matrix (Bio-Rad, Hercules, Calif.), and RapidPrep Micro Genomic DNA isolation kit (Pharmacia Biotech Inc., Piscataway, N.J.) have shown their usefulness for DNA extraction from stool specimens. Recently, the extraction-free FTA filter method (Whatman Bioscience, Cambridge, United Kingdom) has been demonstrated to have high sensitivity for DNA detection by PCR (19). These DNA extraction methods, however, have never been compared. We aimed to evaluate the sensitivities of three DNA extraction methods, i.e., a QIAamp stool mini kit, FTA filter paper, and a conventional phenol-chloroform method.
In recent years, researchers have developed PCR methods for the detection of E. bieneusi in stool specimens. These methods have also never been compared. We chose to evaluate five previously described single-step PCR methods (4, 7, 14, 26, 30) with species-specific primer sets for the detection of E. bieneusi in stool specimens. Moreover, the sensitivities and specificities of the most sensitive PCR method using the most sensitive DNA extraction method were compared with results obtained from light microscopy by using electron microscopy as the gold standard.
MATERIALS AND METHODS
Stool samples.
Stool specimens were collected from 290 children who lived in an orphanage situated in Bangkok, Thailand, during a routine stool examination that was performed every 6 months by the Department of Parasitology, Phramongkutklao College of Medicine, Bangkok, Thailand. Stool specimens were stained with Gram-chromotrope as previously described (17) and examined under a 100× objective by light microscopy for E. bieneusi. Confirmation of the presence of E. bieneusi was performed by electron microscopy or PCR. From these samples, a single positive specimen was used to evaluate the sensitivities of three DNA extraction and five species-specific PCR methods. All positive specimens confirmed by electron microscopy were used for the evaluation of sensitivities and specificities of light microscopy and the PCR method. These specimens were stored at 4°C for less than 3 months. Stool specimens with no E. bieneusi spores were from healthy persons who lived in a rural community outside Bangkok. These samples were ruled out for E. bieneusi by negative Calcofluor staining.
Evaluation of DNA extraction methods.
Three DNA extraction methods, i.e., FTA filter paper, a QIAamp stool mini kit, and a conventional phenol-chloroform method were compared. For extraction with the FTA filter paper, 6-mm disks were punched out from FTA filter paper (Whatman Bioscience) by using a modified hole punch and placed in a 1.5-ml microcentrifuge tube. Fifteen microliters of each diluted sample was spotted onto the FTA disks and dried on a heating block at 56°C. One quarter of each disk was used for one test since it could fit in a 0.2-ml PCR tube. The FTA disk was washed twice with 200 μl of FTA purification buffer (Life Technologies, Gaithersburg, Md.) for 15 min and then washed twice with 200 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA [pH 8.0]) for 5 min and again dried on a heating block at 56°C. The washed FTA disks were used as the DNA template in PCR amplification. It is critical that the TE buffer used to wash the FTA disks have a pH of 8.0; otherwise, there may be interference with the DNA amplification. Moreover, FTA disks have to be completely dried before the PCR is performed. For the QIAamp stool mini kit (QIAGEN), 200 μl of stool specimen was used for DNA extraction, following the manufacturer's instructions. The extracted DNA of each sample was kept frozen at −20°C until used. The phenol-chloroform extraction method was performed as described by Katzwinkel-Wladarsch et al. (7). Two hundred microliters of diluted stool was added with 33.3 μl of 1 M KOH and 9.3 μl of 1 M dithiothreitol and mixed thoroughly in a microcentrifuge tube. After incubation at 65°C for 15 min, the samples were neutralized with 4.3 μl of 25% HCl, buffered with 80 μl of 2 M Tris-HCl (pH 8.3), and the suspension was mixed again. DNA was extracted by shaking with 250 μl of phenol-chloroform-isoamyl alcohol (25:24:1) saturated with 10 mM Tris (pH 8.0) and 1 mM EDTA. To precipitate nucleic acids from the aqueous phase, 0.1 volume of 3 M sodium acetate, pH 5.0, and 2 volumes of cold absolute ethanol were used. DNA was collected immediately by centrifugation at 10,000 × g for 20 min; then ethanol was removed, and the pellet was collected after washing with 0.5 ml of cold 70% ethanol. To remove the residual ethanol, 1 ml of acetone was added, and the pellet was then dried at 37 to 50°C with the lid open. The DNA pellet was resuspended in 200 μl of TE buffer and kept at −20°C until used.
To determine the sensitivities of these three DNA extraction methods, a stool specimen with a known concentration of microsporidial spores was used. A spore count of E. bieneusi was performed by using a positive stool sample dissolved in 200 μl of phosphate-buffered saline, pH 7.5. This specimen was stained by Gram-chromotrope and counted by light microscopy. Dilutions of homogenized stools were made to give different spore concentrations at 100,000, 20,000, 4,000, 800, and 160 spores/ml. For the QIAamp stool mini kit and phenol-chloroform methods, 200 μl of diluted stool sample was used for each test. A total of 10 μl of each extracted DNA specimen was used in the PCR amplification to obtain the equivalent of 1,000, 200, 40, 8, and 1.6 spores per PCR mixture. The sensitivity of the FTA filter was assessed by using the same samples as used for the QIAamp stool mini kit and conventional phenol-chloroform method. Since the amount of specimen placed on the FTA disk was limited to 15 μl and one-fourth of the FTA disk was used for each test, the number of spores was equivalent to 375, 75, 15, 3, and 0.6 spores per PCR mixture. The most sensitive extraction method is defined as the method that can extract DNA from the specimen with the lowest spore concentration and give a positive band of E. bieneusi by PCR amplification with the primer pair MSP3-MSP4B with PCR conditions as described by Katzwinkel-Wladarsch et al. (7). The evaluation of each method was performed three times.
Evaluation of PCR amplification methods.
Five single-step PCR methods for the detection of E. bieneusi were compared for their sensitivities. Stool specimens with five different concentrations of E. bieneusi spores, i.e., 100,000, 20,000, 8,000, 400, and 160 spores/ml, were extracted for DNA by FTA filter paper. The most sensitive PCR method is defined as the method that can amplify the specimen containing the lowest spore concentration. These five sets of primer pairs included MSP3-MSP4B (7), EBIEF1-EBIER1 (4), Primer set 2 (26), Eb.gc-Eb.gt (30), and V1-Mic3 (14). Genomic DNA and the primer pairs were used with PCR conditions as previously described (4, 7, 14, 26, 30). PCR amplification was performed by using a Perkin Elmer 480 thermal cycler. A 10-μl PCR product from each reaction mixture was run on a 2% agarose gel (FMC Bioproducts, Rockland, Maine) with 1% Tris-borate-EDTA buffer. Gels were stained with ethidium bromide and visualized under UV light and documented on high-density printing paper by using a UV-save gel documentation system I (UVItech, Cambridge, United Kingdom). The evaluation of the sensitivities of each PCR method was performed three times.
Comparison of PCR and light microscopy for the detection of E. bieneusi in stool specimens.
The most sensitive DNA extraction and PCR method was chosen for the comparison of the light microscopy and PCR methods for the detection of E. bieneusi in stool samples. The blinded evaluation of both techniques was performed by using 30 positive and 30 negative stool samples. Stool specimens were examined microscopically by an experienced microscopist by using Gram-chromotrope staining. The sources of all positive and negative stool samples were described above. Sensitivities and specificities were calculated by using two-by-two tables and Epi Info version 6.01 software. A chi-square test was used to determine the significance of the difference between two proportions for sensitivities and specificities of the two diagnostic methods.
RESULTS
Sensitivities of DNA extraction methods.
To evaluate the sensitivities of these three extraction methods, specimens with known spore concentrations were used for the experiment. As shown in Table 1, FTA filter paper and the QIAamp stool mini kit could detect E. bieneusi at concentrations as low as 800 spores/ml. The sensitivity of the phenol-chloroform extraction method was equivalent to 4,000 spores/ml. The comparison shows that FTA filter paper and the QIAamp stool mini kit are more sensitive compared to the conventional phenol-chloroform method for DNA extraction of E. bieneusi in stool specimens.
TABLE 1.
Comparison of the sensitivities of three DNA extraction methods for detection of E. bieneusi in stool specimens
| Extraction method | Results by spore concn (spores/ml)
|
||||
|---|---|---|---|---|---|
| 100,000 | 20,000 | 4,000 | 800 | 160 | |
| Phenol-chloroform | + | + | + | − | − |
| QIAamp stool mini kit | + | + | + | + | − |
| FTA filter paper | + | + | + | + | − |
Sensitivities of PCR methods.
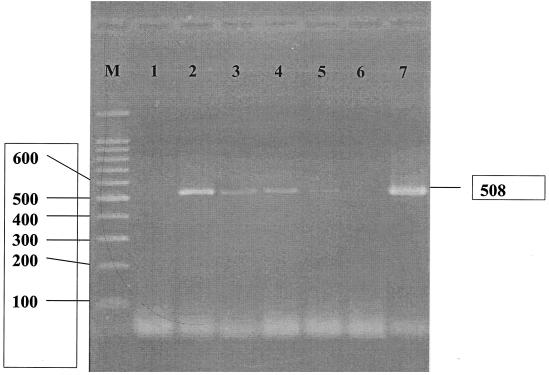
A comparison of the sensitivities of five PCR methods is shown in Table 2. Our results demonstrated that the two protocols that use the MSP3-MSP4B and EBIEF1-EBIER1 primer pairs were the most sensitive methods for detecting E. bieneusi at concentrations as low as 800 spores/ml. Figure 1 shows the sensitivity testing of the PCR method using the primer pair MSP3-MSP4B to amplify E. bieneusi in stool specimens. The protocols which used the Eb.gc-Eb.gt and V1-Mic3 primers could detect E. bieneusi in specimens with 20,000 spores/ml. Following the technique of Schuitema et al. (26) and using Primer set 2, we could not amplify the DNA of E. bieneusi at a concentration of 100,000 spores/ml. Positive results were found only in specimens with spore concentrations higher than 100,000 spores/ml.
TABLE 2.
Comparison of the sensitivities of five PCR methods for the detection of E. bieneusi in stool specimens
| Primer pair | Expected amplicon (bp) | Reference | Results by spore concn (spores/ml)
|
||||
|---|---|---|---|---|---|---|---|
| 100,000 | 20,000 | 4,000 | 800 | 160 | |||
| MSP3-MSP4B | 508 | 7 | + | + | + | + | − |
| EBIEF1-EBIER1 | 607 | 4 | + | + | + | + | − |
| Primer set 2 | 1,294 | 24 | − | − | − | − | − |
| Eb.gc-Eb.gt | 210 | 29 | + | + | − | − | − |
| V1-Mic3 | 446 | 14 | + | + | − | − | − |
FIG. 1.
PCR analysis using the MSP3-MSP4B primer pair specific to the SSU rRNA gene of E. bieneusi. Lane M, molecular markers of 100-bp DNA ladder; lane 1, negative control; lanes 2 to 6, different concentrations of spores (100,000, 20,000, 4,000, 800, and 160 spores/ml, respectively); lane 7, positive control.
Sensitivities and specificities of PCR and light microscopy for the detection of E. bieneusi.
Based on the above results, FTA filter paper was chosen for DNA extraction together with the PCR method using the primer pair MSP3-MSP4B to test their sensitivities and specificities for the detection of E. bieneusi in stool specimens. Although the PCR method using the EBIEF1-EBIER1 primer pair gave the same sensitivity as the primer pair MSP3-MSP4B, we chose the MSP3-MSP4B primer pair because of its usefulness in terms of genotypic characterization. Table 3 shows the sensitivities and specificities of the PCR method and light microscopy with Gram-chromotrope staining. The sensitivities of PCR and light microscopy with Gram-chromotrope staining were 100% (95% confidence interval [CI], 90.5 to 100) and 86.7% (95% CI, 70.9 to 95.6), respectively. The specificities of both PCR and light microscopy with Gram-chromotrope staining were 100% (95% CI, 90.5 to 100). The difference between the sensitivities of both methods was 13.3% (95% CI, 4.4 to 29.1). There were no significant differences between the results from PCR and light microscopy with Gram-chromotrope staining in terms of sensitivity (P = 0.9) and specificity (P = 0.36).
TABLE 3.
Comparison of PCR and light microscopy for the detection of E. bieneusi
| Method and result |
E. bieneusi specimens
|
|
|---|---|---|
| No. positive (%) | No. negative (%) | |
| PCR | ||
| Positive | 30 (100) | 0 (0) |
| Negative | 0 (0) | 30 (100) |
| Light microscopy | ||
| Positive | 26 (86.7) | 0 (0) |
| Negative | 4 (13.3) | 30 (100) |
DISCUSSION
The detection of protozoan parasites in stool specimens by PCR usually requires a highly sensitive DNA extraction method because of the presence of inhibitors in stools. To choose a sensitive method for the PCR detection of E. bieneusi DNA in stool specimens, we evaluated three DNA extraction methods: a QIAamp stool mini kit, FTA filter paper, and a conventional phenol-chloroform method. Our study showed that the detection limit varied between 800 to 4,000 spores/ml, depending on the extraction method. Although the phenol-chloroform method has been widely used as a standard method for DNA extraction, our data showed that this less sensitive method could detect spores at a concentration of 4,000 spores/ml. Since DNA was extracted directly from stool samples, poor DNA extraction efficiency or incomplete removal of inhibitors could have occurred. The extraction protocol involved several steps; thus, limited numbers of specimens could be tested each time. To raise the sensitivity of DNA extraction, spore concentration or purification and/or DNA purification afterward may be required (12, 18, 27, 28).
Our experiments showed that both the QIAamp stool mini kit and FTA filter paper could detect E. bieneusi in stool specimens with the same concentration of 800 spores/ml. The QIAamp stool mini kit procedure involves digestion of proteins, elimination of inhibitors, binding DNA to silica gel membrane, and elution of DNA by spin column. In this study, stool homogenate was incubated at 70°C for 5 min with lysis buffer for efficient spore lysis before the extraction. Since spores of E. bieneusi are resistant to disruption and lysis, increasing the incubation temperature of the stool homogenates in the lysis buffer up to 95°C may be helpful to improve sensitivity. The QIAamp stool mini kit involves several steps and takes approximately 1 h to be completed, thus limiting the numbers of specimens that can be handled at one time.
FTA filter paper is an extraction-free, filter-based template which is impregnated with denaturants, chelating agents, and a free-radical trap. Lysis of organisms occurs upon contact with the FTA filter paper, and then DNA is trapped on the matrix. Inhibitors or other debris are effectively removed by washing reagent. Since the application of samples onto the FTA filters is very simple, a large number of samples for field epidemiological studies can be prepared simultaneously by untrained personnel with less technical equipment. The whole procedure, including drying and washing to get the DNA template ready for PCR amplification, takes less than 3 h after spotting the specimens. The disks can be stored at room temperature so that they are easy to handle and transport for further analysis. FTA filter paper and the QIAamp stool mini kit are commercially available with approximately the same price on a sample basis. Modification of the FTA filters by using an individual hole punch to get an FTA disk with a 6-mm diameter for tested samples can reduce the cost. Thus, for long-term use, FTA filters are a low-cost method with high sensitivity, which makes them a good choice for use with large numbers of samples. In addition, the amount of sample required for FTA filter paper is less than that for the QIAamp stool mini kit.
A number of E. bieneusi-specific PCR protocols, which are usually based on the amplification of the intergenic transcribed spacer region of the SSU rRNA gene, have been developed. However, there has been no evaluation of these PCR methods. We evaluated single-step PCR rather than nested PCR methods for the detection of E. bieneusi in stool specimens because the single-step PCR method is convenient and less time consuming. Based on the above data, we used the FTA filter method for DNA extraction to evaluate five PCR protocols, i.e., PCR with the primer pairs MSP3-MSP4B, EBIEF1-EBIER1, Primer set 2, Eb.gc-Eb.gt, and V1-Mic3. Our study showed that the MSP3-MSP4B and EB1EF1-EB1ER primer pairs were the most sensitive primers to detect E. bieneusi in stool specimens. Both MSP3-MSP4B and EB1EF1-EB1ER have been shown to be E. bieneusi specific and do not cross-amplify with other human microsporidia (4, 7). These two PCR protocols have been used for the identification of E. bieneusi both in humans and animals (1, 12, 13, 21). The MSP3-MSP4B primer set has shown satisfactory results for species-specific identification (5, 23, 24). For the purpose of genotypic characterization, comparisons of the intergenic transcribed spacer sequences amplified by MSP3-MSP4B can differentiate their genotype.
The protocol of the Eb.gc-Eb.gt primer set was also tested and was shown to have less sensitivity in the present study. In contrast to the protocols using the MSP3-MSP4B and EB1EF1-EB1ER primer pairs, the Eb.gc-Eb.gt primer pair requires PCR-restriction fragment length polymorphism for genotypic characterization of E. bieneusi (11). However, the PCR-restriction fragment length polymorphism technique has the disadvantage that any mutations or deletions between the restriction enzyme recognition sites are not accessible. The usefulness of genotypic differentiation might be limited.
The use of forward primer V1 (35) and reverse primer Mic3 was developed for E. bieneusi identification (14). The PCR protocol with the V1-Mic3 primer pair and in situ hybridization procedures was a sensitive diagnostic tool and enabled the differentiation of E. bieneusi from other microsporidia (3). Our study demonstrated that the PCR protocol using the V1-Mic3 primer pair was not as sensitive as the protocols with the MSP3-MSP4B and EB1EF1-EB1ER primer pairs. Primer set 2 was designed by Schuitema et al. (26) and was shown to be useful for the detection of E. bieneusi in stool specimens (10, 14). However, we found that Primer set 2 was the least sensitive protocol in this study.
Rinder et al. (22) conducted a blinded, externally controlled multicenter evaluation of light microscopy and PCR for the detection of E. bieneusi in stool specimens. The sensitivities reported from six different laboratories were between 71 to 100%. The differences in the sensitivities might be dependent on DNA extraction methods and PCR protocols. Recently, a real-time PCR method for the detection of Encephalitozoon intestinalis from stool specimens has been developed. It has been shown that real-time PCR is more sensitive than light microscopy (34). In the present study, we evaluated the sensitivities and specificities of PCR and light microscopy for the detection of E. bieneusi in stool specimens. We used the most sensitive techniques to detect E. bieneusi, i.e., FTA filter paper for DNA extraction and the MSP3-MSP4B primer pair for PCR amplification. Although light microscopy seemed to be less sensitive than the PCR method, there is no statistically significant difference between these two methods. The nonsignificant difference might be due to the specimens used in this study, which contained high concentrations of spores and were examined by a highly experienced microscopist. Although light microscopy is an inexpensive method, it cannot differentiate species of microsporidia. Light microscopy, thus, is suitable for a routine screening test. The PCR method using the MSP3-MSP4B primer pair together with FTA filter paper for DNA extraction gave 100% sensitivity and 100% specificity for the detection of E. bieneusi in stool specimens. Hence, this method is a powerful diagnostic method for evaluating clinical specimens and also a meaningful tool for epidemiological study of this infection. Moreover, genotype can be characterized by sequence analysis of the PCR product. Understanding the genetic variations of E. bieneusi strains among the population will be useful for exploring the source, transmission, and pathogenesis of this organism.
In conclusion, we demonstrate that both FTA filter paper and the QIAamp stool mini kit were sensitive methods for the DNA extraction of E. bieneusi in stool specimens. We chose FTA filter paper for further investigation since it is easy to use, is a rapid test, does not require experienced persons to handle specimens, and requires smaller amounts of stool specimens. Comparison of five PCR protocols showed that the PCR protocols using the primer pairs MSP3-MSP4B and EB1EF1-EB1ER are the most sensitive methods for the detection of E. bieneusi in the present study. Using these sensitive DNA extraction and PCR methods together gave high sensitivity and specificity for the detection of E. bieneusi.
Acknowledgments
This work was supported by Thailand-Tropical Diseases Research Programme (T-2), ID 02-2-ARI-24-007.
REFERENCES
- 1.Breitenmoser, A. C., A. Mathis, E. Bürgi, R. Weber, and P. Deplazes. 1999. High prevalence of Enterocytozoon bieneusi in swine with four genotypes that differ from those identified in humans. Parasitology 118:447-453. [DOI] [PubMed] [Google Scholar]
- 2.Canning, E. U., and W. S. Hollister. 1990. Enterocytozoon bieneusi (Microspora): prevalence and pathogenicity in AIDS patients. Trans. R. Soc. Trop. Med. Hyg. 84:181-186. [DOI] [PubMed] [Google Scholar]
- 3.Carville, A., K. Manfield, G. Widmer, A. Lackner, D. Kotler, P. Wiest, T. Gumbo, S. Sarbah, and S. Tzipori. 1997. Development and application of genetic probes for detection of Enterocytozoon bieneusi in formalin-fixed stools and in intestinal biopsy specimens from infected patients. Clin. Diagn. Lab. Immunol. 4:405-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.da Silva, A. J., D. A. Schwartz, G. S. Visvesvara, H. de Moura, S. B. Slemenda, and N. J. Pieniazek. 1996. Sensitive PCR diagnosis of infections by Enterocytozoon bieneusi (microsporidia) using primers based on the region coding for small-subunit rRNA. J. Clin. Microbiol. 34:986-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dengjel, B., M. Zahler. W. Hermanns, K. Heinritzi, T. Spillmann, A. Thomschke, T. Loscher, R. Gothe, and H. Rinder. 2001. Zoonotic potential of Enterocytozoon bieneusi. J. Clin. Microbiol. 39:4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franzen, C., and A. Müller. 1999. Molecular techniques for detection, species differentiation, and phylogenetic analysis of microsporidia. Clin. Microbiol. Rev. 12:243-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katzwinkel-Wladarsch, S., M. Lieb, W. Helse, T. Löscher, and H. Rinder. 1996. Direct amplification and species determination of microsporidian DNA from stool specimen. Trop. Med. Int. Health 1:373-378. [DOI] [PubMed] [Google Scholar]
- 8.Kotler, D. P., and J. M. Orenstein. 1994. Prevalence of enteric pathogens in HIV-infected individuals referred for gastrointestinal evaluation. Am. J. Gastroenterol. 89:1998-2002. [PubMed] [Google Scholar]
- 9.Kotler, D. P., and J. M. Orenstein. 1998. Clinical syndromes associated with microsporidiosis. Adv. Parasitol. 40:321-349. [DOI] [PubMed] [Google Scholar]
- 10.Ligoury, O., F. David, C. Sarfati, A. R. J. Schuitema, R. A. Hartskeerl, F. Derouin, J. Modaï, and J. M. Molina. 1997. Diagnosis of infections caused by Enterocytozoon bieneusi and Encephalitozoon intestinalis using polymerase chain reaction in stool specimens. AIDS 11:723-726. [DOI] [PubMed] [Google Scholar]
- 11.Ligoury, O., F. David, C. Sarfati, F. Derouin, and J. M. Molina. 1998. Determination of types of Enterocytozoon bieneusi strains isolated from patients with intestinal microsporidiosis. J. Clin. Microbiol. 36:1882-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lores, B., C. del Aguila, and C. Arias. 2002. Enterocytozoon bieneusi (microsporidia) in faecal samples from domestic animal from Galicia, Spain. Mem. Inst. Oswaldo Cruz 97:941-945. [DOI] [PubMed] [Google Scholar]
- 13.Lores, B., I. Lopez-Miragaya, C. Arias, S. Fenoy, J. Torres, and C. del Aguila. 2002. Intestinal microsporidiosis due to Enterocytozoon bieneusi in elderly human immunodeficiency virus-negative patients from Vigo, Spain. Clin. Infect. Dis. 34:918-921. [DOI] [PubMed] [Google Scholar]
- 14.Manfield, K. G., A. Carville, D. Shvetz, J. Mackey, S. Tzipori, and A. A. Lackner. 1997. Identification of an Enterocytozoon bieneusi-like microsporidian parasite in simian-immunodeficiency-virus-inoculated macaques with hepatobiliary disease. Am. J. Pathol. 150:1395-1405. [PMC free article] [PubMed] [Google Scholar]
- 15.Menotti, J., B. Cassinat, R. Porcher, C. Sarfati, F. Derouin, and J. M. Molina. 2003. Development of a real-time polymerase-chain-reaction assay for quantitative detection of Enterocytozoon bieneusi DNA in stool specimens from immunocompromised patients with intestinal microsporidiosis. J. Infect. Dis. 187:1469-1474. [DOI] [PubMed] [Google Scholar]
- 16.Molina, J. M., C. Sarfati, B. Beauvais, M. Lemann, A. Lesourd, F. Ferchal, I. Casin, P. Lagrange, R. Modigliani, F. Derouni, and J. Modai. 1993. Intestinal microsporidiosis in human immunodeficiency virus-infected patients with chronic unexplained diarrhea: prevalence and clinical and biologic features. J. Infect. Dis. 167:217-221. [DOI] [PubMed] [Google Scholar]
- 17.Moura, H., J. L. da Silva, F. C. Sodre, P. Brasil, D. Walmo, S. Wahlquist, S. Wallace, G. P. Croppo, and G. S. Visvesvara. 1996. Gram-chromotrope: a new technique that enhances detection of microsporidia spores in clinical specimens. J. Eukaryot. Microbiol. 43:94S-95S. [DOI] [PubMed] [Google Scholar]
- 18.Muller, A., K. Stellermann, P. Hartmann, M. Schrappe, G. Fatkenheuer, B. Salzberger, V. Diehl, and C. Franzen. 1999. A powerful DNA extraction method and PCR for detection of microsporidia in clinical stool specimens. Clin. Diag. Lab. Immunol. 6:243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlandi, P. A., and K. A. Lampel. 2000. Extraction-free, filter-based template preparation for rapid and sensitive PCR detection of pathogenic parasitic protozoa. J. Clin. Microbiol. 38:2271-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabenek, L., F. Gyorkey, R. M. Genta, P. Gyorkey, L. W. Foote, and J. M. Risser. 1993. The role of Microsporidia in the pathogenesis of HIV-related chronic diarrhea. Ann. Intern. Med. 119:895-899. [DOI] [PubMed] [Google Scholar]
- 21.Reetz, J., H. Rinder, A. Thomschke, H. Manke, M. Schwebs, and A. Bruderek. 2002. First detection of the microsporidium Enterocytozoon bieneusi in non-mammalian hosts (chickens). Int. J. Parasitol. 32:785-787. [DOI] [PubMed] [Google Scholar]
- 22.Rinder, H., K. Janitschke, H. Aspöck, A. J. Da Silva, P. Deplazes, D. P. Fedorko, C. Franzen, U. Futh, F. Hünger, A. Lehmacher, C. G. Meyer, J. M. Molina, J. Sandfort, R. Weber, T. Löscher, and the Diagnostic Multicenter Study Group on Microsporidia. 1998. Blinded, externally controlled multicenter evaluation of light microscopy and PCR for detection of microsporidia in stool specimens. J. Clin. Microbiol. 36:1814-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rinder, H., A. Thomschke, B. Dengjel, R. Gothe, T. Luscher, and M. Zahler. 2000. Close genotypic relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J. Parasitol. 86:185-188. [DOI] [PubMed] [Google Scholar]
- 24.Sadler, F., N. Peake, R. Borrow, P. L. Rowl, E. G. L. Wilkin, and A. Curry. 2002. Genotyping of Enterocytozoon bieneusi in AIDS patients from the north west of England. J. Infect. 44:39-42. [DOI] [PubMed] [Google Scholar]
- 25.Sandfort, J., A. Hannemann, H. Gelderblom, K. Stark, R. L. Owen, and B. Ruf. 1994. Enterocytozoon bieneusi infection in an immunocompetent patient who had acute diarrhea and who was not infected with the human immunodeficiency virus. Clin. Infect. Dis. 19:514-516. [DOI] [PubMed] [Google Scholar]
- 26.Schuitema, A. R. J., R. A. Hartskeerl, T. van Gool, R. Laxminarayan, and W. J. Terpstra. 1993. Application of the polymerase chain reaction for the diagnosis of microsporidiosis. AIDS 7(Suppl. 3):S62-S63. [Google Scholar]
- 27.Sulaiman, I. M., R. Fayer, A. A. Lal, J. M. Trout, F. W. Schaefer III, and L. Xiao. 2003. Molecular characterization of microsporidia indicated that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 69:4495-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Gool, T., E. U. Canning, and J. Dankert. 1994. An improved practical and sensitive technique for the detection of microsporidian spores in stool samples. Trans. R. Soc. Trop. Med. Hyg. 88:189-190. [DOI] [PubMed] [Google Scholar]
- 29.Vávra, J., R. Dahbiová, W. S. Hollister, and E. U. Canning. 1993. Staining of microsporidian spores by optical brighteners with remarks on the use of brighteners for the diagnosis of AIDS associated human microsporidioses. Folia Parasitol. 40:267-272. [PubMed] [Google Scholar]
- 30.Velasquez, J. N., S. Carnevale, E. A. Guarnera, J. H. Labbe, A. Chertcoff, M. G. Cabrera, and M. I. Rodriguez. 1996. Detection of the microsporidian parasite Enterocytozoon bieneusi in specimens from patients with AIDS by PCR. J. Clin. Microbiol. 34:3230-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber, R., R. T. Bryan, R. L. Owen, C. M. Wilcox, L. Gorelkin, and G. S. Visvesvara. 1992. Improved light-microscopical detection of microsporidia spores in stool and duodenal aspirates. N. Engl. J. Med. 326:161-166. [DOI] [PubMed] [Google Scholar]
- 32.Weber, R., and R. T. Bryan. 1994. Microsporidial infections in immunodeficient and immunocompetent patients. Clin. Infect. Dis. 19:517-521. [DOI] [PubMed] [Google Scholar]
- 33.Weber, R., R. T. Bryan, D. A. Schwartz, and R. L. Owen. 1994. Human microsporidial infection. Clin. Microbiol. Rev. 7:426-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolk, D. M., S. K. Schneider, N. L. Wengenack, L. M. Sloan, and J. E. Rosenblatt. 2002. Real-time PCR method for detection of Encephalitozoon intestinalis from stool specimens. J. Clin. Microbiol. 40:3922-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu, X., M. Wittner, H. B. Tanowitz, D. Kolter, A. Cali, and L. M. Weiss. 1993. Small subunit rRNA sequence of Enterocytozoon bieneusi and its potential diagnostic role with use of the polymerase chain reaction. J. Clin. Microbiol. 168:1570-1575. [DOI] [PubMed] [Google Scholar]