Abstract
Human metapneumovirus (hMPV) has been associated with respiratory illnesses like those caused by human respiratory syncytial virus (HRSV) infection. Similar to other pneumoviruses, genetic diversity has been reported for hMPV. Little information is currently available on the genetic variability of the G glycoprotein (G), which is the most variable gene in RSV and avian pneumovirus. The complete nucleotide sequences of the G open reading frame (ORF) of 24 Canadian hMPV isolates were determined. Phylogenetic analysis showed the existence of two major groups or clusters (1 and 2). All but one of the hMPV isolates that we examined belonged to cluster 1. Additional genetic variability was observed in cluster 1, which separated into two genetic subclusters. Within cluster 1 the nucleotide sequence identity for the G ORF was 74.2 to 100%, and the identity for the predicted amino acid sequence was 61.4 to 100%. The G genes of cluster 1 isolates were more divergent from the cluster 2 isolates, with 45.6 to 50.5% and 34.2 to 37.2% identity levels for the nucleotide and amino acid sequences, respectively. Sequence analysis also revealed changes in stop codon usage, resulting in G proteins of different lengths (217, 219, 228, and 236 residues). Western blot analysis with the use of hMPV-specific polyclonal antisera to each hMPV cluster showed significant antigenic divergence between the G proteins of clusters 1 and 2. These results suggest that the G protein of hMPV is continuously evolving and that the genetic diversity observed for the hMPV genes is reflected in the antigenic variability, similar to HRSV.
Human metapneumovirus (HMPV), a newly discovered paramyxovirus, has been associated with respiratory illnesses ranging from upper respiratory tract disease to severe bronchiolitis and pneumonia similar to those caused by human respiratory syncytial virus (HRSV) infection (4, 17, 19). Since its initial identification in The Netherlands, hMPV has been isolated from patients with respiratory disease in several countries, indicating that hMPV is present throughout the world (4, 12, 14, 17, 19). As with other pneumoviruses, genetic diversity has been reported for hMPV. Phylogenetic analysis of the M, N, P, F, and L genes of hMPV showed that hMPV isolates were divided into two genetic clusters tentatively named 1 and 2 (1, 14, 19). Recently the complete consensus nucleotide sequences of isolates representative of both genotypes revealed an overall amino acid sequence identity of 90%, and the greatest diversity was observed for the G protein (37% identity) (3). Furthermore, analysis of several hMPV G open reading frames (ORFs) revealed amino acid sequence identities ranging from 31 to 35% between the two clusters (13). HRSV isolates are divided into two antigenic subgroups, A and B, on the basis of their reaction with monoclonal antibodies (18). Although HRSV A and B strains differ in all 10 viral proteins, the G glycoprotein shows the greatest antigenic and sequence divergence, with only 47% amino acid homology between prototype HRSV A and B viruses (18). It has been suggested that changes in amino acid sequences benefit the virus, possibly by modifying epitopes and allowing it to escape preexisting immunity. Similarly, the genetic diversity observed for the hMPV genes may lead to antigenic variations, and these two hMPV genetic clusters may also represent two different antigenic groups. In this study, to evaluate the genetic and antigenic diversity of the G proteins of hMPV, we sequenced the G ORFs of two Canadian isolates representative of hMPV clusters 1 and 2 and expressed the corresponding protein in recombinant baculoviruses. Furthermore, to estimate the extent of genetic diversity within a cluster, we also analyzed the sequences of the G protein of 22 Canadian isolates belonging to hMPV genetic cluster 1.
MATERIALS AND METHODS
Specimens.
The sequence of the G ORF was determined for 22 hMPV specimens collected from January to April 2002. The specimens originated from four provincial public health laboratories across Canada: Nova Scotia (1 specimen), Manitoba (15 specimens), Saskatchewan (5 specimens), and British Columbia (1 specimen).
Primer sequences.
Primers specific to hMPV G and L gene sequences, hMPVG1F (5′-ATG GAG GTG AAA GTG GAG AAC AT-3′) and hMPVL1R (5′-7189-GTG GAT TCA TTG AGA GGA TCC AT-7167-3′) were used to amplify the G ORF.
RT-PCR and sequence analysis.
Viral RNA was extracted from 100 μl of original sample or tissue culture fluid with the RNeasy Mini kit (Qiagen). Viral RNA was amplified in a one-step reverse transcription-PCR (RT-PCR; Qiagen) according to the manufacturer's recommendations. Briefly, 5 μl of RNA was added to the RT-PCR mixture containing 2 μl of Qiagen OneStep RT-PCR enzyme mix, 10 μl of 5× Qiagen OneStep RT-PCR buffer, 400 μM deoxynucleoside triphosphates, 0.6 μM (each) primer, and 10 μl of Q solution in a final volume of 50 μl. The thermocycler conditions used were 50°C for 30 min for reverse transcription; 95°C for 15 min for the activation of the HotStart DNA polymerase; and then 35 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min, followed by an extension of 10 min at 72°C. The PCR products were purified using the QIAquick PCR purification kit (Qiagen) and sequenced on an ABI 377 Sequencer with the use of a fluorescent dye-terminator kit (Applied Biosystems). Adequate positive and negative controls were used to ensure the accuracy of the results. The DNA sequences were assembled and analyzed with SEQMAN, EDITSEQ, and MEGALIGN programs in Lasergene (DNAStar, Madison, Wis.). Phylogenetic trees were generated by the neighbor-joining method with the MEGA program (7).
Production of anti-hMPV polyclonal antisera.
Guinea pigs were immunized intranasally with 1 ml of cell culture supernatant containing 104 50% tissue culture infective doses of the CAN99-80 or CAN99-81 hMPV isolate. Twenty-eight days postinfection, the animals were exsanguinated and the serum was retrieved.
Generation of recombinant baculoviruses, Western blot analysis, and immunoprecipitation.
hMPV G ORFs from CAN99-81 (cluster 1) and CAN99-80 (cluster 2) were amplified by RT-PCR with the following primer sets: G-CAN99-81-forward (5′-GCGCAGATCTATGGAAGTAAGAGTG-3′) and G-CAN99-80-forward (5′-GCGCAGATCTATGGAGGTGAAAGTG-3′), which introduce the BglII site (underlined) before the start of the coding region, and G-CAN99-81-reverse (5′-GCGCGAATTCTTAACTAGTTTGGTT-3′) and G-CAN99-80-reverse (5′-GCGCGAATTCTTAACTACTTGGAGA-3′), which add an EcoRI site (underlined) downstream of the ORF. The amplicons were inserted into the baculovirus transfer vector pBacPAK8 (Clontech) digested by BglII and EcoRI restriction enzymes. Recombinant plasmids carrying the hMPV G gene were further confirmed by DNA sequencing. Recombinant baculoviruses were generated by cotransfecting Spodoptera frugiperda 21 (SF21) cells with BacPAK6 viral DNA and hMPV G recombinant plasmid DNA according to the standard procedure (Clontech).
Recombinant antigen for Western blot analysis was generated by infecting six-well tissue culture plates of SF21 cells (106 cells/well) with recombinant baculovirus carrying the hMPV G gene of CAN99-80 or CAN99-81. Three days postinfection, cells were harvested by centrifugation at 1,620 × g for 5 min. Cell pellets were washed once with cold (4°C) phosphate-buffered saline and pelleted under the same condition. Cells were resuspended in distilled water, and an equal volume of 2× sample buffer (100 mM Tris-HCl [pH 7.5], 10% β-mercaptoethanol, 0.04% bromophenol blue, 8% sodium dodecyl sulfate [SDS], 25% glycerol) was added. The cell lysate was then heated at 95°C for 5 min, and proteins were separated by electrophoresis on an SDS-12% polyacrylamide gel and transferred by electroblotting onto a nitrocellulose membrane (Bio-Rad Laboratories; catalog no. 162-0215). The membrane was blocked with 5% skim milk powder in Tris-buffered saline for 1 h at room temperature, incubated with polyclonal antibodies derived from guinea pigs immunized with hMPV (CAN99-80 or CAN99-81) (1:2,000 diluted in 5% skim milk) for 1 h at room temperature, washed with Tris-buffered saline containing 0.2% Triton X-100 and 0.05% Tween 20, and incubated with goat anti-guinea pig antibody conjugated with horseradish peroxidase (1:50,000 diluted in 10% skim milk) for 1 h at room temperature. Afterwards, the membrane was subjected to the chemifluorescent substrate (Pierce; catalog no. 33250) for 1 min. The detected proteins were visualized by autoradiography.
For the metabolic radiolabeling and immunoprecipitation, virus-infected cells (48 h postinfection) were placed in methionine-free Grace's medium (Gibco/BRL) for 30 min, after which they were pulse-labeled for 60 min with 300 μCi of [35S]methionine (Amersham)/ml. After pulse-labeling, cells were lysed on ice for 15 min with extraction buffer containing 1% Nonidet P-40, 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), and 1 mM phenylmethylsulfonyl fluoride. After centrifugation for 5 min at 16,000 × g, the cell extract was incubated for 2 h at 4°C with appropriate serum, after which 5% protein A-Sepharose (Amersham) was added for 1 h. The precipitated immune complex was washed three times with radioimmunoprecipitation assay buffer consisting of 50 mM Tris-HCl (pH 7.5), 1% Nonidet P-40, 0.1% SDS, 1% sodium deoxycholate, 150 mM NaCl, and 1 mM EDTA (pH 8.0). Samples were solubilized by being boiled for 5 min in 1× sample buffer containing 50 mM Tris-HCl (pH 7.5), 5% β-mercaptoethanol, 0.02% bromophenol blue, 4% SDS, and 12.5% glycerol and analyzed on an SDS-10% polyacrylamide gel followed by fluorography.
Nucleotide sequence database accession number.
The hMPV sequences described in this paper have been deposited in GenBank under accession numbers AY574224 to AY574247. The nucleotide sequence database accession numbers for hMPV Netherlands isolate 00-1 (NLD00-1) and Canadian isolates CAN97-83 and CAN98-75 are AF371367, AY297749, and AY297748, respectively.
RESULTS
Phylogenetic analysis of the G gene.
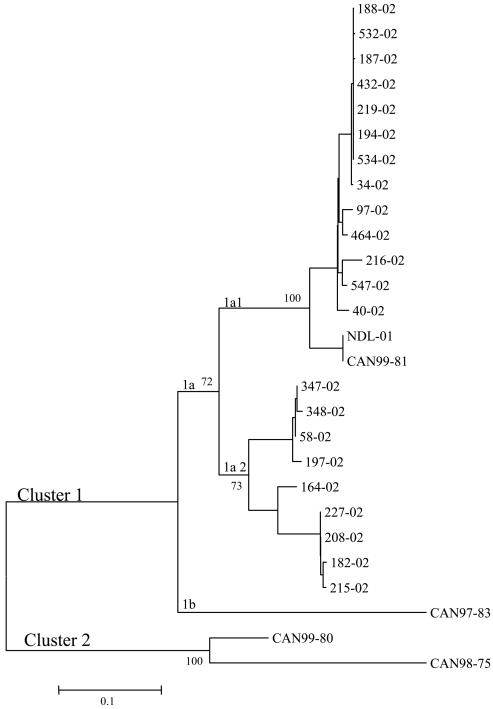
Nucleotide sequences of the G ORF were determined for two Canadian hMPV isolates collected in 1999 (CAN99-81 and CAN99-80) as well as for 22 Canadian isolates collected in 2002. Initial molecular characterization of the F gene of these hMPV specimens determined that CAN99-81 and the 2002 specimens belonged to hMPV genetic cluster 1 and that CAN99-80 belonged to cluster 2 (1, 2, 14). Phylogenetic analysis based on the G ORF sequences of the hMPV Canadian isolates and the published full-length hMPV sequence (NDL-01, CAN97-83, and CAN98-75) showed similar results (3, 19). All 2002 isolates clustered with NDL-01 and CAN99-81 in genetic subcluster 1a, which was further divided into two genetic lineages (1a1 and 1a2), and CAN97-83 was found in subcluster 1b (Fig. 1). Isolate CAN99-80 was found in cluster 2 with CAN98-75 (Fig. 1).
FIG. 1.
Phylogenetic analysis of hMPV isolates. Nucleotide sequences were determined for the G ORF. The corresponding ORF sequences from previously reported hMPV Canadian (CAN97-83 and CAN98-75) and Dutch (NDL-01) isolates were also analyzed. Phylogenetic analysis was performed using the neighbor-joining method of the MEGA program. Bootstrap proportions were plotted at the main internal branches of the phylogram to show support values. The year in which the isolates was collected is indicated by the first (for example, CAN99-81) or last (for example, 40-02) two numerals in the isolate name.
Sequence analysis of the G gene.
Results of sequence analysis of the G ORF of the 2002 Canadian isolates, CAN99-81, CAN99-80, and the published full-length hMPV sequences (NDL-01, CAN98-75, and CAN97-83) are summarized in Table 1. Comparison between the G nucleotide sequences of cluster 1 (CAN99-81, CAN97-83, and 2002 isolates) and cluster 2 (CAN99-80 and CAN98-75) isolates showed low levels of identity (45.6 to 50.5%), similar to what is observed between HRSV subgroups A and B. Identity levels increased to 74.2 to 100% for isolates within the same cluster. Similar results were reported for comparison of 25 hMPV field isolates obtained during five epidemic seasons (13). Predicted amino acid identity between the different 2002 isolates was 61.4 to 100%, and they showed identity levels of 65.0 to 99.6% and 65.0 to 88.2% with CAN99-81 and NDL-01, respectively (Table 1). Levels of identity were lower, 34.2 to 37.2%, when 2002 isolates were compared to the cluster 2 CAN99-80 isolate.
TABLE 1.
Nucleotide and predicted amino acid sequence identity levels among the G glycoproteins of hMPV 2002 Canadian isolates, CAN99-81, CAN99-80, and the indicated hMPV isolates
| hMPV isolates | % of nucleotide (amino acid) identity
|
||
|---|---|---|---|
| 2002 Canadian isolatesa | CAN99-81 | CAN99-80 | |
| Cluster 1 | |||
| 2002 Canadian isolates | 74.2-100 (61.4-100) | ||
| NDL-01 | 76.2-93.2 (65.0-88.2) | 100 (100) | 47.0 (35.4) |
| CAN97-83 | 78.9-97.3 (65.0-95.0) | 80.6 (69.5) | 49.7 (36.8) |
| CAN99-81 | 75.9-93.2 (65.0-99.6) | 47.0 (35.4) | |
| Cluster 2 | |||
| CAN98-75 | 45.6-47.1 (32.5-35.8) | 46.0 (33.8) | 97.9 (95.4) |
| CAN99-80 | 46.5-50.5 (34.2-37.2) | 47.0 (35.4) | |
The results represent the range of identity (percent) obtained for the comparison of the most divergent to the most conserved sequences.
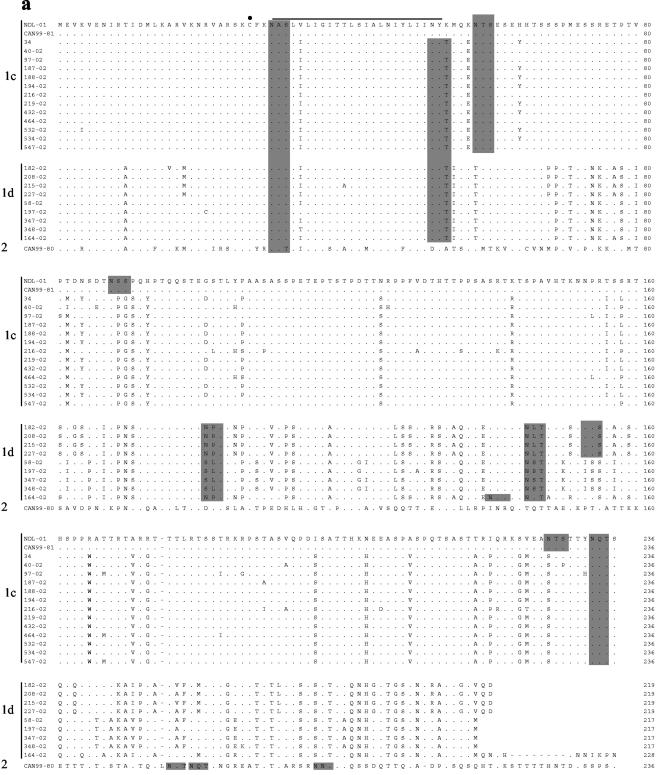
The alignment of the deduced amino acid sequences of the G protein of the 2002 isolates with those of NDL-01, CAN99-80, and CAN99-81 is given in Fig. 2. All changes were base substitutions, and no deletions, insertions, or frameshift mutations were observed. Amino acid substitutions were distributed along the entire protein but were more frequent in the extracellular region. The predicted amino acid sequence identity between the cluster 1 (CAN99-81) and 2 (CAN99-80) isolates was 35.4% (Table 1). The predicted hydrophilicity profiles of the two proteins were similar for the cytoplasmic and transmembrane domains, but minor differences were observed for the rest of the protein (Fig. 2b). In contrast to the high degree of amino acid divergence observed for the overall G protein sequence, the cytoplasmic and transmembrane domains exhibit considerable amino acid homology, both within and between hMPV clusters of the 27 hMPV isolates analyzed (89.6 to 100% and 60.4 to 75.0%, respectively).
FIG. 2.
(a) Alignment of the predicted amino acid sequences of the hMPV G protein of the hMPV Canadian isolates with the Dutch isolate NDL-01. Only residues that differ from isolate NDL-01 are shown; identical amino acids are represented by periods; gaps are represented by dashes. Potential N-linked glycosylation sites are shaded, and the conserved cysteine residue at position 27 is marked by a dot. (b) Predicted hydrophilicity profile of CAN99-81 (cluster 1) and CAN99-80 (cluster 2) isolates.
The G ORF contained 7.3 to 8.4% and 5.4 to 5.9% proline residues for cluster 1 and 2 isolates, respectively. The high content of proline residues found in the G glycoprotein is consistent with glycoproteins of mucinous origin. The cysteine residue at position 27 is strictly conserved among all isolates, and one additional cysteine residue at position 21 is found in isolate 197-02. The conserved cysteine (amino acid [aa] 27) was also present in all isolates analyzed by Peret et al. (13). The sequence comparison also showed a threonine and serine content of 30 to 34%, threonine and serine residues being potential acceptor sites for O-linked sugars. Nine threonine and five serine residues were conserved among all the sequences analyzed. Cluster 1 isolates also possessed an additional 7 conserved threonine and 10 serine residues. The number of N-linked glycosylation sites present in the G protein of hMPV isolates varied from three to six, and only one was strictly conserved (aa 30, Fig. 2). The N-linked glycosylation site at aa 30 is conserved in all hMPV G sequences analyzed to date (3, 13). However, since it is located at the junction of the cytoplasmic tail and the transmembrane domain, it is unlikely that this site is used for N-linked glycosylation. As mentioned previously, this region of the protein is highly conserved among isolates, and the conservation of these amino acids might be important for the integrity of the cytoplasmic tail and the transmembrane domain.
Similar to what was observed by Peret et al. (13), nucleotide changes were found at the C-terminal end of the protein, leading to changes in stop codon usage, resulting in G proteins of different length. G proteins of 217, 219, 228, and 236 residues were observed, corresponding to usage of four in-frame termination codons located at nucleotide (nt) 652 (UAA), 658 (UAG), 685 (UAA), and 709 (UAA), respectively (13). Four isolates used the stop codon at nt 652, four used the one at nt 658, one used the codon at nt 685, and 13 used that at nt 709. All isolates using the stop codon at nt 709 were found in subcluster 1c, whereas isolates using the other stop codons were found in subcluster 1d.
Antigenic analysis of the G protein.
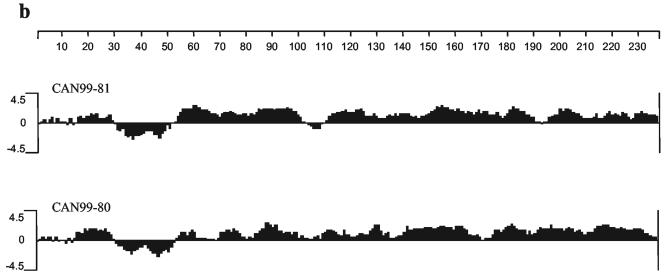
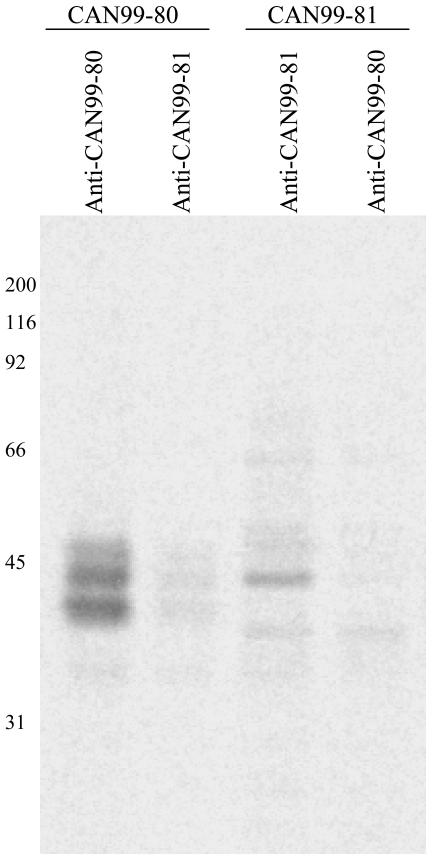
In order to evaluate the antigenic diversity, the G proteins of two representative hMPV isolates, CAN99-81 (cluster 1) and CAN99-80 (cluster 2), were expressed from recombinant baculoviruses. The G protein synthesized in insect cells was examined by Western blotting with hMPV-specific guinea pig polyclonal antisera raised against both hMPV isolates (CAN99-80 and CAN99-81) (Fig. 3). The results show differences in G protein mobility between the two hMPV clusters. CAN99-81 G protein expressed from infected cells migrated as a heterogeneous smear with an estimated molecular mass of 38 to 80 kDa, with a major band at 50 kDa (Fig. 3a), and CAN99-80 G protein migrated as a smear of 45 to 75 kDa, with the dominant species migrating as a broad band of 45 and 58 kDa (Fig. 3b). Similar results were obtained with several independent virus stocks. The G ORF sequence predicts a protein of 25.9 kDa, which contrasts with the higher molecular mass observed for the mature protein expressed in baculovirus. This observation suggests that, like that of HRSV, the G glycoprotein of hMPV is N linked and O linked glycosylated. The smear pattern is also indicative of glycosylation. The lower-molecular-mass species observed for CAN99-81 (38 kDa) corresponds to the predicted molecular mass for the G protein plus the N-linked carbohydrates for this isolate (37 kDa; four sites in the extracellular domain). In contrast, the lower-molecular-mass species observed for CAN99-80 (45 kDa) is larger than the predicted molecular mass for the G protein plus the N-linked carbohydrates for this isolates (34 kDa; three sites in the extracellular domain), suggesting the presence of additional O-linked carbohydrates. CAN99-80 has 24 serine and 53 threonine residues, whereas CAN99-81 has 37 serine and 43 threonine residues. Therefore, different usage of potential O-linked glycosylation sites could explain the differences observed in the migration patterns.
FIG. 3.
Antigenic analysis of CAN99-80 (cluster 2) and CAN99-81 (cluster 1) G proteins by Western blotting with hMPV-specific polyclonal antisera raised against CAN99-81 (a) and CAN99-80 (b). Whole-cell lysates of recombinant baculovirus-infected SF21 cells expressing the G protein were analyzed. Mock-infected SF21 cells were used as a negative control. Numbers at left of each panel are molecular masses in kilodaltons.
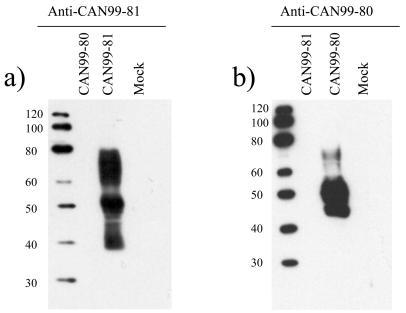
The Western blotting results also show that the detection of the G protein was cluster specific. The G protein of CAN99-81 was detected only with the antiserum raised against CAN99-81 and not with the one raised against CAN99-80. Similarly, the G protein of CAN99-80 was detected with the CAN99-80 antiserum and not the CAN99-81 antiserum. To ensure that the specificity observed was not due to experimental conditions promoting the detection of linear epitopes versus discontinuous epitopes, we performed an immunoprecipitation experiment. The CAN99-80 antiserum immunoprecipitated lower-molecular-mass proteins migrating as a heterogeneous smear with estimated molecular masses of 40 to 50 kDa, and the CAN99-81 antiserum immunoprecipitated proteins with molecular masses ranging from 38 to 66 kDa (Fig. 4). High-molecular-mass species of the G protein were not immunoprecipitated by both antisera, suggesting that antibodies recognizing the heavily glycosylated forms of the native G protein are poorly induced in guinea pigs. This also indicates that the heavy glycosylation may contribute to the interference with the induction of antibodies to the G protein. Although both antisera could immunoprecipitate the CAN99-80 and CAN99-81 G glycoproteins, the intensity of the signal was much stronger for the homologous protein, suggesting some level of cluster specificity. However, these results also suggest that CAN99-80 and CAN99-81 share a limited number of conformational epitopes that could provide some cross-reactivity.
FIG. 4.
Immunoprecipitation of the G glycoprotein of CAN99-80 and CAN99-81 with hMPV-specific polyclonal antisera raised against CAN99-80 and CAN99-81. The immunoprecipitated G proteins were analyzed on an SDS-10% polyacrylamide gel followed by fluorography. Numbers at left are molecular masses in kilodaltons.
DISCUSSION
In this study, we examined the genetic diversity of 24 hMPV isolates collected in Canada. Phylogenetic analysis based on the nucleotide sequences of the G ORF of hMPV isolates revealed the presence of two genetic clusters (1 and 2) and that cluster 1 evolved with multiple lineages that cocirculated during epidemics. The results also showed a low level of amino acid identity between the G proteins of the two hMPV clusters (32.5 to 37.2%). Furthermore, sequence analysis of 22 hMPV cluster 1 isolates, all collected in 2002, revealed extensive variability of the G protein within cluster 1, with identity levels as low as 61.4%, which clearly indicates that two different genetic lineages of cluster 1 isolates were cocirculating in Canada during 2002. These results confirm data from recent studies showing levels of amino acid identity ranging from 31 to 37% between the G proteins of the two hMPV clusters and the presence of additional diversity within clusters (3, 13). As with HRSV, the G protein of which is involved in neutralizing and protective immunity, the high percentage of nucleotide changes that resulted in amino acid changes suggests that there may be a selective advantage to G protein changes (5, 10, 16). Another similarity with HRSV is the use of different stop codons, resulting in G proteins of different lengths and the correlation observed between the G protein length and the position of the isolate in the phylogenetic tree (6, 8, 15). Changes in stop codon usage have been found in HRSV escape mutants selected with monoclonal antibodies that recognize strain-specific epitopes (9). Furthermore, reports show that, for HRSV, carbohydrates in the C-terminal third of the G protein influence the expression of certain epitopes by either masking or contributing to antibody recognition (11). Although the N-linked and O-linked glycosylation sites were conserved in some positions for the hMPV G protein, sequence analysis also revealed lineage-specific patterns. Therefore, as with HRSV, the extensive glycosylation of the hMPV G protein may help the virus to evade the immune system. Although strain-specific monoclonal antibodies are not available to determine the precise antigenic and genetic relatedness of hMPV isolates, these results taken together suggest that hMPV may evade the immune system through epitope modification like HRSV. The highly specific reactivity of antisera raised against viruses from the two clusters of hMPV supports this hypothesis.
REFERENCES
- 1.Bastien, N., S. Normand, T. Taylor, D. Ward, T. C. Peret, G. Boivin, L. J. Anderson, and Y. Li. 2003. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 93:51-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastien, N., D. Ward, P. Van Caeseele, K. Brandt, S. H. Lee, G. McNabb, B. Klisko, E. Chan, and Y. Li. 2003. Human metapneumovirus infection in the Canadian population. J. Clin. Microbiol. 41:4642-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biacchesi, S., M. H. Skiadopoulos, G. Boivin, C. T. Hanson, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Barreno, B., A. Portela, T. Delgado, J. A. Lopez, and J. A. Melero. 1990. Frame shift mutations as a novel mechanism for the generation of neutralization resistant mutants of human respiratory syncytial virus. EMBO J. 9:4181-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamasaki, H., H. Tsutsumi, K. Seki, and S. Chiba. 2001. Genetic variability of respiratory syncytial virus subgroup B strain isolated during the last 20 years from the same region in Japan: existence of time-dependent linear genetic drifts. Arch. Virol. 146:457-466. [DOI] [PubMed] [Google Scholar]
- 7.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 8.Martinez, I., O. Valdes, A. Delfraro, J. Arbiza, J. Russi, and J. A. Melero. 1999. Evolutionary pattern of the G glycoprotein of human respiratory syncytial viruses from antigenic group B: the use of alternative termination codons and lineage diversification. J. Gen. Virol. 80:125-130. [DOI] [PubMed] [Google Scholar]
- 9.Melero, J. A., B. Garcia-Barreno, I. Martinez, C. R. Pringle, and P. A. Cane. 1997. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J. Gen. Virol. 78:2411-2418. [DOI] [PubMed] [Google Scholar]
- 10.Norrby, E., M. A. Mufson, H. Alexander, R. A. Houghten, and R. A. Lerner. 1987. Site-directed serology with synthetic peptides representing the large glycoprotein G of respiratory syncytial virus. Proc. Natl. Acad. Sci. USA 84:6572-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palomo, C., P. A. Cane, and J. A. Melero. 2000. Evaluation of the antibody specificities of human convalescent-phase sera against the attachment (G) protein of human respiratory syncytial virus: influence of strain variation and carbohydrate side chains. J. Med. Virol. 60:468-474. [PubMed] [Google Scholar]
- 12.Pelletier, G., P. Dery, Y. Abed, and G. Boivin. 2002. Respiratory tract reinfections by the new human metapneumovirus in an immunocompromised child. Emerg. Infect. Dis. 8:976-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peret, T. C., Y. Abed, L. J. Anderson, D. D. Erdman, and G. Boivin. 2004. Sequence polymorphism of the predicted human metapneumovirus G glycoprotein. J. Gen. Virol. 85:679-686. [DOI] [PubMed] [Google Scholar]
- 14.Peret, T. C., G. Boivin, Y. Li, M. Couillard, C. Humphrey, A. D. Osterhaus, D. D. Erdman, and L. J. Anderson. 2002. Characterization of human metapneumoviruses isolated from patients in North America. J. Infect. Dis. 185:1660-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peret, T. C., C. B. Hall, K. C. Schnabel, J. A. Golub, and L. J. Anderson. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 79:2221-2229. [DOI] [PubMed] [Google Scholar]
- 16.Rueda, P., T. Delgado, A. Portela, J. A. Melero, and B. Garcia-Barreno. 1991. Premature stop codons in the G glycoprotein of human respiratory syncytial viruses resistant to neutralization by monoclonal antibodies. J. Virol. 65:3374-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stockton, J., I. Stephenson, D. Fleming, and M. Zambon. 2002. Human metapneumovirus as a cause of community-acquired respiratory illness. Emerg. Infect. Dis. 8:897-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullender, W. M. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 13:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]