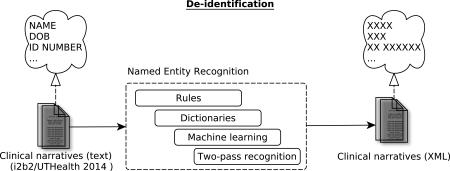
Abstract
A recent promise to access unstructured clinical data from electronic health records on large-scale has revitalized the interest in automated de-identification of clinical notes, which includes the identification of mentions of Protected Health Information (PHI). We describe the methods developed and evaluated as part of the i2b2/UTHealth 2014 challenge to identify PHI defined by 25 entity types in longitudinal clinical narratives. Our approach combines knowledge-driven (dictionaries and rules) and data-driven (machine learning) methods with a large range of features to address de-identification of specific named entities. In addition, we have devised a two-pass recognition approach that creates a patient-specific run-time dictionary from the PHI entities identified in the first step with high confidence, which is then used in the second pass to identify mentions that lack specific clues. The proposed method achieved the overall micro F1-measures of 91% on strict and 95% on token-level evaluation on the test dataset (514 narratives). Whilst most PHI entites can be reliably identified, particularly challenging were mentions of Organisations and Professions. Still, the overall results suggest that automated text mining methods can be used to reliably process clinical notes to identify personal information and thus providing a crucial step in large-scale de-identification of unstructured data for further clinical and epidemiological studies.
Keywords: De-identification, Named Entity Recognition, Information Extraction, Clinical Text Mining, Electronic Health Record
Graphical abstract
1. Introduction
A recent promise and the potential of wider availability of data from Electronic Health Records (EHRs) to support clinical research are often hindered by personal health information that is present in EHRs, raising a number of ethical and legal issues. De-identification of such data is therefore one of the main pre-requisites for using EHRs in clinical research. As a result, there is a growing interest for automated de-identification methods to ultimately aid accessibility to data by removing Protected Health Information (PHI) from clinical records. De-identification of unstructured data in particular is challenging, as PHI can appear virtually anywhere in a clinical narrative or letter. This task is often considered as Named Entity Recognition (NER), where mentions of specific PHI data types (e.g. patient names, their age and address) need to be identified in the text of clinical narratives.
Automated de-identification of unstructured documents has been a research topic for more than twenty years. As early as 1996, Sweeney et al. proposed a rule-based approach to recognize twenty five overlapping entity types they identified as PHI in EHRs1. Since then, a large number of systems have been introduced, including knowledge-based2,3,4,5 and data-driven6,7,8,9,10,11, as well as hybrid12,13,14 methods that combine various approaches. In terms of types of clinical narrative, previous de-identification research has explored varied clinical documents such as discharge summaries11,15, pathology reports9, nursing progress notes2 and mental health records4.
The 2006 i2b2 de-identification challenge15 was the first effort to provide a common test-bed for eight PHI entity types (mentions of Patients, Doctors, Hospitals, IDs, Dates, Locations, Phone numbers and Age) in clinical discharge summaries. The submitted systems ranged from rule-based5 and machine-learning (ML) methods (e.g. using Conditional Random Fields13, Hidden Markov Models13, and Decision Trees8) with a wide range of features, to hybrid approaches (e.g. combining rules and Support Vector Machines12). A notable observation across methods was the use of knowledge-driven techniques (in particular rules) both for the direct recognition of PHI and in support of data-driven and hybrid methods. For example, rules were used as features in ML models (e.g. indicating whether a particular rule was triggered)12, as a post-processing correction module13 or combined with data-driven results at the final step (e.g. integration of ML and rule-based annotations)14. This trend was often motivated by the presence of a number of categories that are characterized by regularized expressions (e.g., date, phone, zip/postcode, and identification numbers), which make rules an efficient modelling technique. In general, the 2006 shared task showed that data-driven methods with features generated by rules for regularized expressions performed best8,13. They were followed by hybrid methods12, while the pure rule-based systems proved to perform less well5.
The 2014 i2b2/UTHealth16 Shared Task in de-identification17 of longitudinal clinical narratives focused on 25 entity types, inclusive of twelve types as defined by the Health Insurance Portability and Accountability Act (HIPAA). The entity types were grouped into seven main categories: Names (e.g., patient and doctor names), Profession, Locations (e.g., street, city, zip code, organizations), Contacts (e.g., phone, fax, email), IDs (e.g., medical record, identification number), Age and Dates. The organizers provided a fully annotated mention-level training dataset, as well as a test dataset for the evaluation. This paper describes a hybrid method that integrates the results of knowledge-(dictionary- and rule-based components) and data-driven methods. We present the results and further discuss the challenges in the de-identification task.
2. Methodology
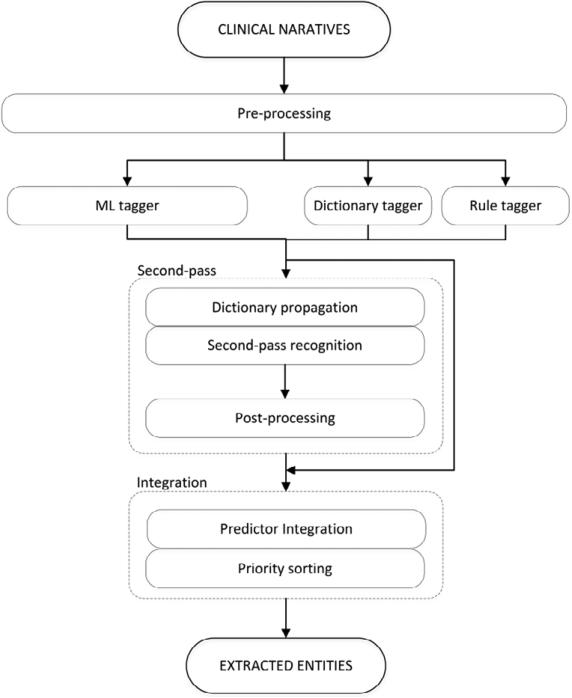
The training data (790 narratives, 460,164 tokens) was released in two batches by the organisers. We have used the first batch (521 narratives, 316,357 tokens) for the initial design of the methods, whereas the second batch (269 narratives, 143,807 tokens) was used as a development set for validation and tuning. The initial analysis of the training data confirmed that some of the entity types are more lexically closed (e.g. country and city names) or regularized (e.g. zip codes, phones, etc.) than the others (e.g. patient and doctor names). The methods developed have largely followed that observation, devising a hybrid approach aiming to combine different methods where appropriate. Figure 1 shows an overview of the system, and the steps are detailed below.
Figure 1.
System architecture.
(I) Pre-processing
The narratives were pre-processed with cTAKES18 and GATE19 to provide basic lexical and terminological features, including tokenization, sentence splitting, part-of-speech tagging and chunking.
(II) Dictionary- and rule-based taggers
The dictionary-based taggers were used for the Hospital, City, Country, State, Profession and Organization entity types. The dictionaries (see Supplementary material for the full list) were collected from open sources such as Wikipedia, GATE and deid2,20 We have merged the entity-specific term lists from these sources and then manually filtered the resulting dictionaries to exclude ambiguous terms.
The rule-based tagger included a set of rules that exploited several types of features including the output of the dictionary-based taggers to recognize entities. Five feature types were used in the rule engineering:
Orthographic features, which include word chracteristics such as allCapitals, upperInitial, mixedCapitals, or lowerCase; as well as token/word length.
Pattern features, which include common lexical patterns of specific entity types as derived from the training data set e.g., date (e.g., DD-MM-YYYY), zip (XXXX), telephone number (XXX-XXX-XXXX) and so forth.
Semantic/lexical cues or entity types. For example, Street names often include lexical cues such as ‘street’, ‘drive’, ‘lane’, State (e.g., “DC”, “CA” etc.), and so forth.
Contextual cues that indicate the presence of a particular entity type. They include specific lexical expressions (e.g., person and doctor titles, months, weekdays, seasons, holidays, common medical abbreviations, etc.), symbols (e.g., bracket and colon, e.g. used for Username and Medical record respectively), and other special characters such as white space and newline.
Negative contextual cues (e.g., lexical and orthographic) are used for disambiguation (e.g., for entity types that are similar e.g., phone and fax number, patient and doctor names).
Using the combination of these features enabled us to craft a relatively small rule set of 5 rules on average per entity type (the minimum of 1 for zip, fax and email, and the maximum of 11 for age). The rules were developed using Java Annotation Patterns Engine (JAPE)19 and Java regular expressions. An example rule is given in Table 1.
Table 1.
Example of a rule. Row 2 shows a rule for capturing Street mentions. The rule contains four types of components (pattern, orthographic indicators and semantic/lexical and contextual clues).
| Feature type | Pattern | Orthographic | Semantic/Lexical | Contextual |
| {RegEx} = [1-9][0-9]{0,3} | {ORTHO} = {upperInitial, allCapital, ...} | {STREET_CLUE} = {Street, St, Drive, Dr, ...} | {SYMBOL} = {Ø, ‘.’} | |
| A rule | {RegEx} {ORTHO} {STREET_CLUE}{SYMBOL} | |||
| In text | ... 62 Angora Dr . ... ... 1 Jefferson Road ... ... 55 Bury St ... |
|||
(III) ML-based tagger
As target entities comprise spans of text, we approached the task as a token tagging problem and trained separate Conditional Random Fields (CRF)21 models for each entity type. We used a token-level CRF with the Inside-Outside (I-O) schema22, for each of the entity types separately. In this schema, a token is labelled with I if it is inside the entity span and with O if it is outside of it. For example: in sentence “Saw Dr Oakley 4/5/67”, token “Oakley” will be tagged as I_Doctor (inside a doctor's name), whereas all other tokens will be annotated as O_Doctor (outside doctor's name). This schema provides more examples of “inside” tokens to learn from than the other schemas (e.g. the Beginning-Inside-Outside, B-I-O), and in our case, it also provided satisfactory results during trainig.
The feature vector consisted of 279 features for each token (see Supplementary material for the full list of features), representing the token's own properties (e.g. lexical, orthographic and semantic) and context features of the neighbouring tokens. Experiments on the development set with various context window sizes showed that two tokens on each side provide the best performance. The following features were engineered for each token:
Lexical features included the token itself, its lemma and POS tag, as well as lemmas and POS tags of the surrounding tokens. Each token was also assigned its location within the chunk (beginning or inside). All chunk types returned by cTAKES (see Supplementary material for the full list) were considered for this feature.
Orthographic features captured the orthographic patterns associated with gold-standard entity mentions. For example, a large percentage of hospital mentions are acronyms (e.g., DHN, EHMS), doctor and patient names are usually capitalized (e.g., Xavier Rush, Yosef Villegas), dates contain digits and special characters (e.g., “2069-04-07”, “04/07/69”) etc. We engineered two groups of orthographic features. The features in the first group captured standard orthographic characteristics (e.g., is the token capitalized, does it consist of only capital letters, does it contain digits, etc.). The second group aimed to further model the token's orthographic pattern using an abstract representation where each upper-case letter is replaced with “X“, lower-case letter with ”x”, a digit with “d” and any other character with “S“. Two features were created in this group: the first feature contained one mapping for each character in a token (e.g., BrightPoint was mapped to “XxxxxxXxxxx“); the second feature mapped a token to a four character string that contained (binary) indicators of a presence of a capital letter, a lower case letter, a digit or any other character (absence was mapped to a “_“), e.g., BrightPoint was mapped to “Xx_ _”.
Semantic features indicate if a given token represents an entity of a specific category. These features were extracted using dictionary matching for US states and cities, calendar months, professions and profession hints (e.g., “worked for”, “retired from” etc.). A feature that captures whether a token is likely to represent a US zip code was also extracted using a regular expression.
Positional features included the absolute position of the line containing the token (in order to utilize the semi-structured nature of clinical narratives) and a binary feature indicating the presence of a space character between the current and the next token (to capture the cases where a single annotation unit was tokenized in multiple tokens e.g., the following date ‘2069-04-07’ is tokenized in five tokens: ’2069’, ‘-’, ‘04’, ‘-’,’07’).
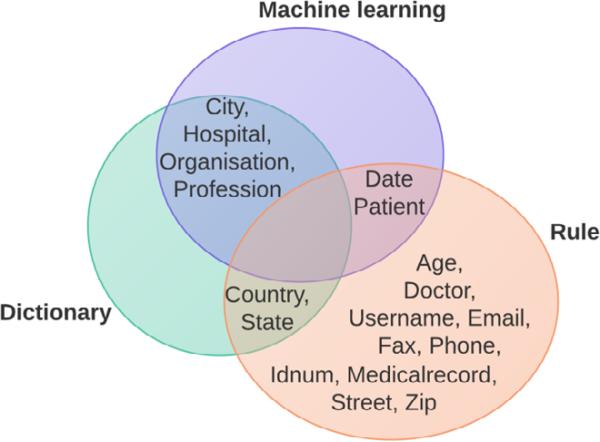
We initially constructed ML models for each entity category present in the training data. After the validation using the development dataset and comparison with the results of the rule-based component, we opted for the separate ML models for the following entity types: City, Date, Hospital, Organization, Profession and Patient. Each of the models was trained on a particular sub-set of features (determined by using a development set from the training data).
The output of the ML models was post-processed by a set of manually crafted rules with the goal of expanding the resulting tags (reducing false negatives) or removing them (reducing false positives). The rules were designed to capture the context (neighbouring tokens) of an ML tag. For example, if the token was tagged as a Hospital, its first letter was a capital letter and one of the nearby tokens is the word ‘Hospital’ then the whole window between that token and ‘Hospital’ was tagged as a Hospital (e.g., ‘Barney Convalescent Hospital’). Another type of rule is removing the Date tag of a token that has more than two ‘/’ characters in its neighbourhood e.g., ‘140/4.0/107/25.7/32/1’.
Along with our CRF model we have used another ML-based tagger i.e., the Stanford Named Entity Recognizer23 to obtain aditional annotations of Organizations; it was only applied on sentences that contained specific contextual indicators of the entity type (e.g., “works in”, “runs”, “church”, “lodge”, etc.). The output of the tagger was directly added to the final output of the system.
(IV) Second-pass recognition
In order to capture PHI mentions that lack local contextual cues implemented by steps II and III, we devised a ‘two-pass’ approach. Specifically, for each entity type, initial annotations were extracted at the patient-level (the dataset contained up-to five narratives per patient) using the methods described in steps II and III. These are then collected into a temporary, run-time patient-level dictionary, which is filtered to remove ambiguous terms and obvious false positives. This filtering was based on a set of terms obtained from the analysis performed on the development set. The patient-specific dictionary is then used for the ‘second-pass’ dictionary matching (using longest string matching) on the narratives belonging to that patient.
(V) Integration module
This component integrates the results from the previous steps into different submissions, merging the tags (at the mention-level) derived from different components (see below). The submission combinations were determined based on the performance achieved during development and specifically based on the ‘strict text matching’ results. Three different submissions were created.
In all of the submissions, we relied on rules only (i.e. no dictionaries or ML) for Age, Street, Zip, Email, Fax, Phone, Username, Identification number and Medical record. The dictionary and rules were combined for Country, and the dictionary and ML results were integrated for City, Hospital, Organization and Profession. The three submissions differed only in annotations used for Date, Doctor and Patient mentions (see Supplementary material for the complete submission schema; Fig. 2 illustrates Submission 3):
Submission 1 included only rule-based approaches for Date, Doctor and Patient; this submission aimed at optimizing precision.
Submission 2 targeted recall and integrated – on top of Submission 1 – all ML models for Date, Doctor and Patient.
Submission 3 aimed to optimize the F1-measure: it included Submission 1 and the ML model for Date and Patient.
Figure 2.
Proposed methods for NER of PHI
To deal with the integration of overlapping tags from multiple categories (i.e. conflicting annotations) we have developed a priority sorting approach. A frequent example were confusions between Doctor and Patient (given that both are personal names), and Age and Date (e.g., “80's”). Based on the results during development, we have defined specific priorities for each of the categories, for example, Doctor was “preferred” over Patient, while Age was “preferred” over Date etc. (see Supplementary material for the full list of priorities assigned to categories). During the integration, multiple categories assigned to a span were sorted by priority and the one with the highest priority was chosen as the final tag.
3. Results and discussion
The PHI entity recognition results on the test dataset (514 narratives, 297,459 tokens) are given in Table 2. There were three official evaluation measures based on different matching strategies: token level matching that requires at least one token of the gold standard span and the resulting span to match; text strict matching that requires an exact match of the gold standard span with the resulting span, and the HIPPA strict matching that considers only the categories that are in the strict interpretation of the HIPAA guidelines (see supplementary material for the full list). Our third run provided the best micro-average F1-measure (90.65%) along with the highest precision (93.06%) and was officially ranked second best in the challenge based on token level matching (both when considering all PHI and HIPAA only entity types) and third when considering all PHIs (text strict matching).
Table 2.
Micro-averaged results on the test data (514 narratives). (P = precision; R = recall; F = F1-measure)
| Token level matching | Text strict matching | HIPAA strict matching | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P% | R% | F% | P% | R% | F% | P% | R% | F% | |
| Submission 1 | 97.11 | 86.49 | 91.49 | 93.02 | 81.94 | 87.13 | 94.46 | 82.80 | 88.25 |
| Submission 2 | 96.55 | 93.16 | 94.82 | 91.73 | 88.50 | 90.09 | 93.11 | 92.00 | 92.55 |
| Submission 3 | 97.22 | 92.50 | 94.80 | 93.06 | 88.36 | 90.65 | 94.37 | 92.13 | 93.23 |
The results of our runs mostly concur with the aim of each of the submissions, with the exception of Submission 1 that had a slightly lower precision than Submission 3, which is likely due to the fact that the ML model for the Date category (Submission 3) provided slightly higher precision than the rule-based tagger included in Submission 1 (data not shown). The token-level matching scores were significantly higher (around 4% across all the measures). There are several reasons for this: there were fewer false negatives (1,334 for token-level vs. 1,728 for the strict matching), which indicate that both the ML and the rule-based approaches would benefit from a better method for boundary adjustment. Furthermore, the correctly recognized gold standard entities have 64% of terms of a length greater than one, which consequently resulted in the increase of true positives at the token-level evaluation.
We note that our results on the development set are fairly consistent (less than 1% F1-measure, see Table 3) compared to the test evaluation results shown in Table 2 (i.e., there was not much over-fitting present), indicating a generalizable methodology.
Table 3.
Micro-averaged results on the development set (269 narratives).
| Token level matching | Text strict matching | HIPAA strict matching | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P% | R% | F% | P% | R% | F% | P% | R% | F% | |
| Submission 1 | 97.56 | 88.71 | 92.92 | 94.26 | 84.03 | 88.85 | 96.63 | 84.06 | 89.91 |
| Submission 2 | 96.8 | 93.99 | 95.37 | 92.56 | 89.11 | 90.8 | 95.21 | 91.34 | 93.24 |
| Submission 3 | 97.63 | 93.11 | 95.31 | 93.73 | 88.68 | 91.13 | 95.65 | 90.94 | 93.23 |
The results for different PHI categories for Submission 3 (see Table 4) indicate that well-defined and structured categories such as Age, Date, Email, Idnum, Medical record, Phone, Street and Zip can be extracted with high F1-measure (over 94%). On the other hand, ambiguous (and potentially contextually dependent) categories such as City, Country, Doctor, Hospital, Patient and State were slightly more complex with F1-measure varying between 79% and 90%. Finally, the categories that are lexically variable and have low frequency (in both the training and test data) proved to be challenging, with the method achieving F-measures of 57% (Profession) and 27% (Organization). Organization, in particular, was a relatively infrequent (124 mentions in the 790 narratives in the training data) and broadly defined category (see below).
Table 4.
Per category performance on the test data, submission 3 (text strict matching).
| Category | Entity type | Frequency | Precision % | Recall % | F-measure % |
|---|---|---|---|---|---|
| AGE | Age | 764 | 97.49 | 91.62 | 94.47 |
| DATE | Date | 4980 | 95.52 | 95.58 | 95.55 |
| CONTACT | 1 | 100.00 | 100.00 | 100.00 | |
| Fax | 2 | 33.33 | 50.00 | 40.00 | |
| Phone | 215 | 96.57 | 91.63 | 94.03 | |
| LOCATION | City | 260 | 83.95 | 78.46 | 81.11 |
| Country | 117 | 83.65 | 74.36 | 78.73 | |
| Hospital | 875 | 81.88 | 76.46 | 79.08 | |
| Organization | 82 | 40.48 | 20.73 | 27.42 | |
| State | 190 | 92.00 | 84.74 | 88.22 | |
| Street | 136 | 96.92 | 92.65 | 94.74 | |
| Zip | 140 | 100.00 | 94.29 | 97.06 | |
| ID | Idnum | 195 | 90.53 | 78.46 | 84.07 |
| Medical record | 422 | 96.03 | 91.71 | 93.82 | |
| NAME | Doctor | 1912 | 96.56 | 83.79 | 89.80 |
| Patient | 879 | 88.14 | 84.53 | 86.30 | |
| Username | 92 | 100.00 | 95.65 | 97.78 | |
| PROFESSION | Profession | 179 | 59.17 | 55.87 | 57.47 |
Based on the experiments conducted during the development phase, the two-pass recognition method was found to be effective for the following entity types: in the rule-based components: Patient, Doctor, Zip, Medical record number, and Identification number; for the ML-based taggers: City, Hospital and Patient. We further evaluated the impact of the proposed two-pass recognition method on the test set for the relevant entity types (see Table 5). Five of the seven entity types on which we applied this method showed a gain of 2-7% F1-measure; four entity types showed a gain of 3-9% in recall and three entity types showed a gain of 5-7% precision. The notable gain in precision was unexpected. A closer examination showed that the dictionary filtering step was the main factor in this regard. These results are consistent with the training dataset (not shown), indicating that two-pass recognition can be a useful method for de-identification of longitudinal clinical notes.
Table 5.
Impact of the two-pass recognition method on the test set.
| Entity type | No two-pass | With two-pass | Δ | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P (%) | R (%) | F (%) | P (%) | R (%) | F (%) | P (%) | R (%) | F (%) | |
| Patient | 81.65 | 75.60 | 78.51 | 86.73 | 84.76 | 85.73 | +5.08 | +9.16 | +7.22 |
| City | 77.69 | 75.00 | 76.32 | 83.95 | 78.46 | 81.11 | +6.26 | +3.46 | +4.79 |
| Hospital | 81.29 | 70.51 | 75.52 | 80.96 | 76.80 | 78.83 | −0.33 | +6.29 | +3.31 |
| Doctor | 89.24 | 83.81 | 86.44 | 96.56 | 83.79 | 89.72 | +7.32 | −0.02 | +3.28 |
| Medicalrecord | 96.86 | 87.68 | 92.04 | 96.03 | 91.71 | 93.82 | −0.83 | +4.03 | +1.78 |
| Zip | 100.00 | 93.57 | 96.68 | 100.00 | 94.29 | 97.06 | 0.00 | +0.72 | +0.38 |
| Idnum | 89.47 | 78.46 | 83.61 | 90.00 | 78.46 | 83.84 | +0.53 | 0.00 | +0.23 |
Another characteristic of our method is the integration of knowledge- and data-driven methods. An analysis on the test dataset results showed notable gains for a number of entity types. We observed gains in F1-measure for Patient (+4.37%), City (+13%), Hospital (+1%), and Date (1.5%). Two special cases where our knowledge-driven component had the greatest impact were Organization (+25%) and Profession (+56%). Extremely poor performance of our ML models on these categories (~1% F-measure) is due to their low frequency in the training data and lexical broadness (see below). We note that despite expected low impact of the two ML models, we decided to include them in the final pipeline because our dictionary-based components for Profession and Organization also had low results during development (compared to other entity types).
We performed the error analysis on the whole test data set. Five major error categories have been identified. The first category comprises both FNs and FPs due to lack of representative features or training data. Typical examples are Organization and Profession as broadly defined, ambiguous, context dependent and infrequent (in terms of the gold standard mentions) entity types. Our features were not able to capture all possible variations of Organizations (’Vassar’, ‘army’, ‘catering business‘, ‘weight room' etc.) and Professions (‘Personnel Officer’, ‘mathematics’, ‘Ground Transit Operators Supervisor’, ‘model planes’, ’veteran’, ‘Craftperson’, ‘Justice of the peace’ etc.). FPs belonging to this category of errors were the consequence of context dependence, most evident with Profession type e.g., ‘with assistance from the plumber’, ‘use pill cutter’, ‘lab tech’.
Opting for the token level CRF contributed notably to drop in performance in terms of the strict measures. Large portion of FNs was due to the models correctly tagging only a subset of tokens of the gold standard annotations. This was the case for most of the entity types considered by ML e.g. (correctly tagged tokens are underlined): Doctor (‘Johnathan Kiefer’), Patient (‘Clarence H. HESS’), City (‘Cape Cod’), Profession (‘Ground Transit Operators Supervisor’) etc.
Specific feature groups generated a subset of FPs i.e., lexical features produced confusions between first names of doctors and patients, while orthographic features caused many medical abbreviations to be tagged as hospitals (‘PCP’, ‘LIMA’, ‘CHB’ etc.)
The forth error category comprised FNs and FPs that were the result of incorrect tokenization, which is mainly a direct consequence of data quality issues with the provided documents. For example, in a number of cases there was a missing space between two neighbouring tokens; examples include identification numbers and hospital abbreviations (‘45479406HBMC’); Hospitals (‘Roper Hospital NorthProblems’, ‘atNorth Mountain Hospital’); Patients (‘VivianLee Jorgenson’, ‘EarnestBranche’) etc.
As expected, some false positives and negatives were due to inconsistent gold-standard annotations. A prominent example includes mentions of the language spoken by a patient and the Country category. For example, 80% of the cases where a mention refers to patient speaking English were annotated as Country in the training data, while only 20% of such mentions were annotated in the test data.
4. Conclusion
Automated de-identification of clinical narrative data is a key for using EHR to facilitate large-scale evidence based research in medicine. In this paper we described and evaluated a hybrid approach for the identification of PHI from clinical narratives. Our approach is based on the combination of hand-crafted rules, focused dictionaries and various features used in the ML models. We have also proposed a novel two-pass recognition approach to address de-identification of longitudinal narratives by generating run-time and patient-specific PHI dictionaries that are used for identification of mentions that lacked specific clues considered by the initial entity extraction modules. A method integration approach proposed included a combination of initial taggers’ output (rule, dictionary, ML, and two-pass recognition) and a priority sorting approach used to select the categories in cases of overlapping text spans that are tagged as belonging to different PHI types.
The overall results showed good performance for frequent and well-scoped classes (e.g., Date, Email, Phone and Street); non-focused and context-dependent categories (e.g. City, Country, Doctor, Hospital and Patient) had reasonable performance, whereas infrequent and broadly scoped categories (Organization and Profession) proved to be challenging and will require further investigation for identifying additional local cues and/or modelling the contextual dependencies (e.g. taking into account inter-dependences between PHI mentions e.g., by applying data mining methods (association rule analysis, clustering etc.). We also plan to explore boundary adjustment techniques including alternative sequence label modelling to improve the identification of entity types.24
Supplementary Material
Highlights.
We present a method for automatic de-identification of clinical narratives
We propose and validate a two-pass tagging method to improve PHI entity recognition
We have shown that automated de-identification is comparable to human benchmark
Acknowledgement
This work has been partially supported Health e-Research Centre (HeRC), The Christie Hospital NHS Foundation Trust, KidsCan Charitable Trust, the Royal Manchester Children's Hospital and the Serbian Ministry of Education and Science (projects III44006; III47003).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Availability
The system is available at http://clinical-deid.sourceforge.net/
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Sweeney L. Replacing personally-identifying information in medical records, the Scrub system. AMIA Annu Fall Symp. 1996:333–337. [PMC free article] [PubMed] [Google Scholar]
- 2.Neamatullah I, Douglass MM, Lehman L-WH, et al. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak. 2008;8(32) doi: 10.1186/1472-6947-8-32. https://dx.doi.org/10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison FP, Lai AM, Hripcsak G. Repurposing the Clinical Record: Can an Existing Natural Language Processing System De-identify Clinical Notes? JAMIA. 2009;16(1):37–39. doi: 10.1197/jamia.M2862. https://dx.doi.org/10.1197/jamia.M2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandes AC, Cloete D, Broadbent MT, et al. Development and evaluation of a de-identification procedure for a case register sourced from mental health electronic records. BMC Med Inform Decis Mak. 2013;13(71) doi: 10.1186/1472-6947-13-71. https://dx.doi.org/10.1186/1472-6947-13-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guillen R. Automated De-Identification and Categorization of Medical Records. i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. 2006 [Google Scholar]
- 6.Aramaki E, Imai T, Miyo K, Ohe K. Automatic deidentification by using sentence features and label consistency. Paper presented at: i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. 2006 [Google Scholar]
- 7.Guo Y, Gaizauskas R, Roberts I, Demetriou G, Hepple M. Identifying personal health information using support vector machines. Paper presented at: i2b2 workshop on challenges in natural language processing for clinical data. 2006 [Google Scholar]
- 8.Szarvas G, Farkas R, Busa-Fekete R. State-of-the-art anonymization of medical records using an iterative machine learning framework. Journal of the American Medical Informatics Association. 2007;14(5):574. doi: 10.1197/j.jamia.M2441. https://dx.doi.org/10.1197/j.jamia.M2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner J, Xiong L. HIDE: An integrated system for health information DE-identification. Proceedings of the 21st IEEE International Symposium on Computer-Based Medical Systems. 2008:254–259. https://dx.doi.org/10.1109/CBMS.2008.129.
- 10.Aberdeen J, Bayer S, Yeniterzi R, et al. The MITRE Identification Scrubber Toolkit: Design, training, and assessment. JAMIA. 2010;79(12):849–859. doi: 10.1016/j.ijmedinf.2010.09.007. https://dx.doi.org/10.1016/j.ijmedinf.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Uzuner Ö , Sibanda TC, Luo Y, et al. A de-identifier for medical discharge summaries. Artif Intell Med. 2008;42(1):13–35. doi: 10.1016/j.artmed.2007.10.001. https://dx.doi.org/10.1016/j.artmed.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara K. Applying a SVM Based Chunker and a Text Classifier to the Deid Challenge. i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. 2006 [Google Scholar]
- 13.Wellner B, Huyck M, Mardis S, et al. Rapidly retargetable approaches to de-identification in medical records. JAMIA. 2007;14(5):564–573. doi: 10.1197/jamia.M2435. https://dx.doi.org/10.1197/jamia.M2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrández O, South BR, Shen S, et al. BoB, a best-of-breed automated text de-identification system for VHA clinical documents. JAMIA. 2013;20(1):77–83. doi: 10.1136/amiajnl-2012-001020. doi:10.1136/amiajnl-2012-001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uzuner Ö , Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. JAMIA. 2007;14(5):550–563. doi: 10.1197/jamia.M2444. https://dx.doi.org/10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stubbs A, Kotfila C, Xu H, Uzuner O. Practical applications for NLP in Clinical Research: the 2014 i2b2/UTHealth shared tasks. this issue. [Google Scholar]
- 17.Stubbs A, Ozlem Uzuner. De-identifying longitudinal medical records. this issue. [Google Scholar]
- 18.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. JAMIA. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham H, Maynard D, Bontcheva K, Tablan V. GATE: A Framework and Graphical Development Environment for Robust NLP Tools and Applications. ACL; 2002. pp. 507–513. [Google Scholar]
- 20.Goldberger AL, Amaral LAN, Glass L, et al. PhysioBank, PhysioToolkit, and Physionet: Components of a New Research Resource for Complex Physiologic Signals. Circulation. 2000;101(23):215–220. doi: 10.1161/01.cir.101.23.e215. https://dx.doi.org/10.1161/01.CIR.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 21.Lafferty JD, McCallum A, Pereira FCN. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. ICML; 2001. pp. 282–289. [Google Scholar]
- 22.Ramshaw LA, Marcus MP. Text chunking using transformation-based learning. 1995 arXiv preprint cmp-lg/9505040. [Google Scholar]
- 23.Finkel JR, Grenager T, Manning C. Incorporating Non-local Information into Information Extraction Systems by Gibbs Sampling. ACL; 2005. pp. 363–370. https://dx.doi.org/10.3115/1219840.1219885. [Google Scholar]
- 24.Dehghan A. Boundary identification of events in clinical named entity recognition. CoRR. 2013 arXiv:1308.1004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.