Abstract
Macrorestriction analysis of SmaI-digested chromosomal DNA, using pulsed field gel electrophoresis (PFGE) was performed to type and estimate genetic relationships among 288 Staphylococcus aureus isolates recovered from 58 Eastern Canadian dairy herds. In addition, a subset of the collection was phage typed and evaluated for sensitivity to 10 antimicrobial compounds. Of 288 isolates recovered, 29 distinct PFGE types were identified. Based on estimates of genetic relationships, the PFGE types were assigned to six lineage groups, designated A through F. Of all of the isolates, ca. 93% were assigned to lineage groups A, D, or F. In 58.6% of herds, only a single PFGE type was recovered, while the remainder had two to four types. Of the 212 isolates evaluated for antimicrobial resistance, 24.5% were resistant to one or more antimicrobials. Resistance to penicillin (9.9%) was most common, followed by resistance to sulfadimethoxine (7.5%). Isolates resistant to multiple antibiotics were rare. A total of 63% of isolates responded to phages from groups 1 and 3, and 32.8% could not be typed with any of the phage strains used. The other 4.1% belonged to a variety of phage types. Most of the PFGE lineage group A and F isolates corresponded to phage groups 3 and 1, respectively, and most group D isolates were not typeable. PFGE typing had better discriminatory power than phage typing in defining the relatedness of the S. aureus isolates. Distribution of PFGE types and phage types was independent across regions and within herds.
Mastitis is an inflammation of the mammalian milk secretion gland caused by microbial infection. It is a major economic concern for the dairy industry worldwide. As many as 50% of all dairy cattle experience some form of mastitis at any given time (42). Although a large number of different bacteria, fungi, and mycoplasma can infect the bovine udder, Staphylococcus aureus has been implicated in intramammary infections (IMI) with a frequency ranging from 7 to 44% of clinical mastitis cases (32, 41). A survey of bulk tank milk from farms in Minnesota found the bacterium in 93 of 100 consecutive bulk tank cultures, indicating its almost ubiquitous presence in dairy herds (17). This high prevalence may be due to its ability to cause chronically recurring mastitis and its resistance to antibiotic treatment (43). Bacteriological cure rates for antibiotic treatment of S. aureus IMI are influenced by a variety of factors (33) and may range from 20 to 78% (12, 30, 36). S. aureus is also a significant pathogen involved in nosocomial and community-acquired infections in humans. Worldwide, the increasing prevalence of multidrug-resistant S. aureus is an additional problem (6), and resistance to antimicrobial compounds reduces their effectiveness and increases morbidity, mortality, and health care costs worldwide (10). In addition, S. aureus is an important food-borne pathogen (5, 9, 20, 29).
In the Canadian province of Ontario, clinical mastitis is a common disease in dairy cows, with approximately one in five cow lactations having at least one episode of clinical mastitis. In a study of Ontario dairy herds, S. aureus represented 6.7% of bacterial isolates from cows with clinical mastitis (32). In the development of an effective infection control strategy for mastitis, it is important to study the epidemiology of S. aureus in dairy herds and determine genetic types of isolates for monitoring the spread of the pathogen. Furthermore, knowledge of antimicrobial resistance properties of pathogens in dairy herds is necessary for the development of effective prevention and treatment strategies for the disease.
The objective of the present study was to determine the genetic structure of a population of S. aureus recovered from dairy cows experiencing clinical mastitis in three Eastern provinces within Canada and to survey their susceptibility to antimicrobial agents. A large proportion of Canadian dairy cattle are located in the provinces of Ontario, Quebec, and Prince Edward Island (11). To our knowledge, there are no previous data on the antimicrobial resistance profiles of bovine S. aureus isolates in this region, nor are there published reports regarding the distribution of bovine S. aureus strains or types in Canada as determined by modern molecular methods such as pulsed-field gel electrophoresis (PFGE). A variety of molecular methods have been used for typing and subtyping of S. aureus isolates with different degrees of discrimination, including ribotyping, multilocus enzyme electrophoresis (MLEE) and multilocus sequence typing (MLST) (18, 19, 37). Of these methods PFGE is, at present, the typing method of choice (7, 37). In the present study, the distribution of phage types of the isolates was also investigated.
MATERIALS AND METHODS
Bacterial strains.
A total of 288 S. aureus isolates from 179 cows with clinical mastitis before the scheduled cessation of lactation (“drying-off”) were collected between August 1999 and October 2000 and used for PFGE typing. Scheduled drying-off in the sample herds was typically ca. 60 days prior to expected calving. Isolates were collected from 58 commercial dairy herds in the Canadian provinces of Ontario (135 isolates from 87 cows in 30 herds), Quebec (136 isolates from 79 cows in 24 herds), and Prince Edward Island (PEI; 17 isolates from 13 cows in 4 herds). The number of cows sampled per herd ranged from 1 to 10, with an average of 3.1/herd. The number of isolates collected from a herd ranged from 1 to 24, with an average of 5.0/herd.
Of the 288 isolates, 252 were selected for evaluation of antimicrobial resistance. Bacterial isolates that originated from the same cow and exhibited identical PFGE and antimicrobial resistance profiles were considered to be duplicate representatives of the same strain, and only one such isolate was included in further analysis. After the removal of 40 duplicate isolates based on these criteria, 212 isolates from 58 farms in three provinces were included in the examination of resistance to antimicrobial agents. In the case of phage typing, multiple isolates collected from the same cow that also had the same PFGE type were considered duplicates, and only one was retained for analysis. Using these criteria, 195 isolates were selected for phage typing.
The dairy cows from which S. aureus isolates were collected were part of a dry-cow therapy trial (12), and the isolates were collected from animals before and/or after the antibiotic treatment had commenced. Antibiotics were administered by intramammary infusion after the cessation of milking, i.e., during the cow's “dry period” between lactations. Antibiotics used in treatment were either tilmicosin or benzathine cloxacillin. The cattle in this trial did not receive prophylactic antibiotics or antimicrobials for growth promotion. Cows also did not receive antibiotics in any form within the 28 days preceding trial enrollment. Milk samples were collected aseptically, and 10 μl was cultured on Columbia base agar containing 5% sheep blood for 24 to 48 h at 37°C. Putative S. aureus colonies were identified based on colony morphology and hemolysis; identity was confirmed with a tube coagulase test with rabbit plasma (Becton Dickinson, Franklin Lakes, N.J.). Bacterial isolates were stored in Luria-Bertani broth containing 15% glycerol and held at −86°C.
Preparation of genomic DNA.
Genomic DNA was prepared as described by Bannerman et al. (7), with modifications as suggested by the Centers for Disease Control pulse net laboratory protocol for molecular typing of S. aureus by PFGE (unpublished data). Briefly, whole cells from 200 μl of broth culture (optical density, 0.55 to 0.7 at 610 nm) were embedded in 600 μl of 0.9% agarose (SeaKem GTG; Cambrex Bioscience, Rockland, Maine) containing 4 μg of lysostaphin (Sigma-Aldrich, Oakville, Ontario, Canada). Cells were lysed in situ at 37°C for 4 h in EC lysis buffer (6 mM Tris-HCl [pH 8.0], 1 M NaCl, 100 mM EDTA, 0.5% Brij-58, 0.2% sodium deoxycholate, 0.5% sodium lauroylsarcosine). Subsequently, plugs were equilibrated against several changes of Tris-EDTA buffer and stored at 4°C. Slices taken from plugs were equilibrated in reaction buffer for 30 min at room temperature, followed by endonuclease digestion with SmaI (Promega, Madison, Wis.).
PFGE.
DNA fragments were separated by electrophoresis in 1% SeaKem GTG agarose gels (0.5× Tris-borate-EDTA running buffer) with a CHEF-DR3 apparatus (Bio-Rad Laboratories, Mississauga, Ontario, Canada). The parameters were as follows: switch times of 5 to 40 s at 6 V/cm for 21 h at 14°C. Concatameric bacteriophage lambda DNA molecules (New England Biolabs, Mississauga, Ontario, Canada) and SmaI fragments of the cellular DNA from S. aureus NCTC 8325 were used as standards. Macrorestriction fingerprint patterns were analyzed by using GelCompar II software (Version 2.4; Applied Maths, Kortrijk, Belgium), and dendrograms were created by using the Dice coefficient, the unweighted pair group method with arithmetic means, and a band position tolerance of 0.4%. Restriction patterns were assigned to types and lineage groups as previously described (38): isolates with identical restriction patterns were assigned to the same type, whereas types that differed from each other by three band differences or less were assigned to the same lineage group. Lineage groups were assigned alphabetically, and types within those groups were assigned numerically.
Phage typing.
Phage typing was conducted by using the method of Anderson and Williams (3) with some modifications with a set of 24 phages based on the recommended 21-phage typing set (8). The phages were divided into three main groups plus two miscellaneous phages. Group 1 included phages 29, 52, 52A, 79, and 80. Group 2 included phages 3A, 3C, 55, and 71. Group 3 included phages 6, 42E, 47, 53, 54, 75, 77, 83A, 84, and 85. The miscellaneous group phages were 81, 8290, 94932, 9588A, and 9683C. The phages were used at the routine test dilution and at 100× the routine test dilution (8). All S. aureus isolates were maintained at room temperature on Dorset Egg slants (QueLab, Inc., Montreal, Quebec, Canada). The isolates were plated on nutrient agar plates and incubated at 37°C for 18 h.
A single smooth colony was inoculated into 4.5 ml of nutrient broth (Oxoid, Nepean, Ontario, Canada) and incubated for 4 to 6 h in a shaking water bath at 37°C to attain a bacterial growth turbidity equivalent to 0.5 McFarland standard. Nutrient agar plates were flooded with 2 ml of culture, and excess culture was removed by using a Pasteur pipette. The seeded plates were allowed to dry for 15 min at room temperature, and ca. 20 μl of each of the typing phages was inoculated onto the bacterial lawn by a multiple-inoculation syringe method (16). The plates were incubated at 30°C overnight, and lytic patterns were observed (8, 31). To be regarded as distinct, two strains must have differed by at least two strong reactions (clear or turbid confluent plaques).
Antimicrobial susceptibility assay.
Susceptibility to a panel of 10 antibiotics was determined by using custom-ordered 96-well Sensititre plates (Trek Diagnostic Systems, Ltd., West Sussex, England). MICs for the following antibiotics were determined: pirlimycin, tetracycline, ceftiofur, tilmicosin, erythromycin, penicillin-novobiocin, cephalothin, oxacillin (with 2% NaCl), sulfadimethoxine, and penicillin. Wells in Sensititre plates were inoculated with Mueller-Hinton broth cultures of the test isolates and as recommended by the manufacturer in accordance with the National Committee for Clinical Laboratory Standards (NCCLS) (28, 44). After incubation, plates were read by eye with a Sensititre SensiTouch radiometer (Trek Diagnostic). Isolates were scored as antibiotic sensitive or resistant based on growth or no growth at the appropriate breakpoint MIC for a given antibiotic based on NCCLS guide M31-A2 (28); the breakpoints used are given in Table 4. Reference strain S. aureus ATCC 29213 served as the assay control.
TABLE 4.
Antimicrobials, breakpoint criteria, and frequency of resistance in 212 S. aureus isolates collected from Canadian dairy herds in three provinces
| Antimicrobial agent | Breakpoint (μg/ml) | No. of isolatesa in:
|
Total no.
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Ontario
|
PEI
|
Quebec
|
|||||||
| No. | % | No. | % | No. | % | No. | % | ||
| Tilmicosin | ≥32 | 2 | 2.1 | - | - | - | - | 2 | 0.9 |
| Penicillin | ≥0.25 | 12 | 12.5 | - | - | 9 | 8.6 | 21 | 9.9 |
| Erythromycin | ≥8 | 3 | 3.1 | - | - | - | - | 3 | 1.4 |
| Oxacillin plus 2% NaCl | ≥4 | 1 | 1.0 | 1 | 9.1 | - | - | 2 | 0.9 |
| Pirlimycin | ≥4 | 5 | 5.2 | - | - | - | - | 5 | 2.4 |
| Penicillin-novobiocin | ≥4/8 | - | - | - | - | - | - | - | - |
| Tetracycline | ≥16 | 1 | 1.0 | - | - | 2 | 1.9 | 3 | 1.4 |
| Cephalothin | ≥32 | - | - | - | - | - | - | - | - |
| Ceftiofur | ≥8 | - | - | - | - | - | - | - | - |
| Sulfadimethoxine | ≥512 | 5 | 5.2 | 1 | 9.1 | 10 | 9.5 | 16 | 7.5 |
| Resistant to none | 67 | 69.8 | 9 | 81.8 | 84 | 80.0 | 160 | 75.5 | |
| Total | 96 | 100.0 | 11 | 100.0 | 105 | 100.0 | 212 | 100.0 | |
A dash (-) indicates no isolates resistant to a given antimicrobial agent were isolated in this region.
Statistical analysis.
Data for the 195 isolates used for phage typing was analyzed with SAS version 8 (SAS Institute, Cary, N.C.) by using the Cochran-Mantel-Haenszel chi-square test (25). A type was comprised of three or more isolates, whereas any type of fewer than three isolates was pooled and represented as “others” in the analysis. Analysis was conducted to determine correlations between epidemiological markers (phage type and PFGE type) and geographical distribution.
RESULTS
PFGE.
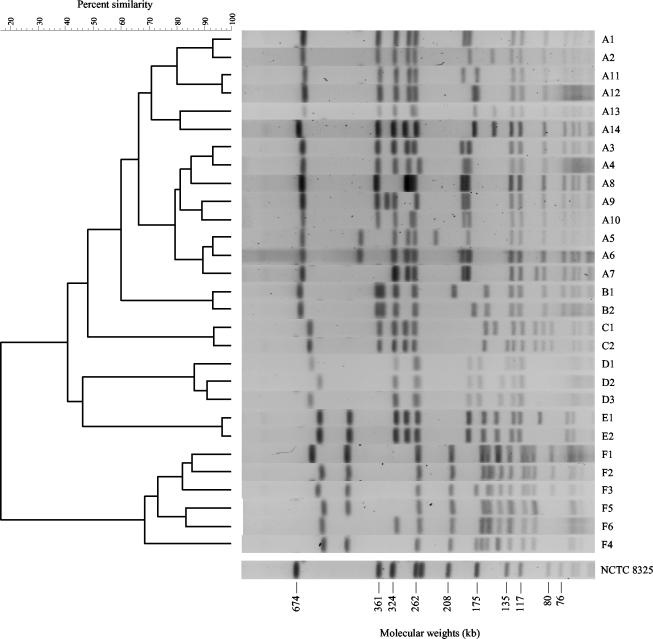
SmaI digestion of S. aureus chromosomal DNA produced 12 to 20 fragments ranging from 30 to 700 kb (Fig. 1). Of the 288 isolates collected from 58 herds, 29 distinct PFGE patterns were identified (Fig. 1). All of the isolates were typeable by the PFGE method. Based on estimates of genetic relationships, the 29 PFGE patterns were assigned to six major lineage groups, designated by the letters A through F, and individual patterns within these lineages were numbered. The dendrogram in Fig. 1 summarizes genetic relationships among 288 PFGE macrorestriction polymorphisms. Only the 12 types that had more than three isolates per herd were used for statistical analysis. The data are summarized in Table 1.
FIG. 1.
Pulsed-field gel electrophoresis of SmaI-digested total DNA from selected S. aureus isolates. The 29 PFGE patterns were assigned to six major lineage groups, designated by the letters A through F, and individual patterns within these lineages were numbered. The dendrogram shows estimates of genetic relatedness of the PFGE types. The PFGE pattern of standard S. aureus strain NCTC 8325 is shown unlinked in the dendrogram.
TABLE 1.
Distribution of molecular types of S. aureus isolates in three Canadian provinces
| PFGE type | Type distributiona in:
|
Total
|
||||||
|---|---|---|---|---|---|---|---|---|
| Ontario
|
Quebec
|
PEI
|
||||||
| No. | % | No. | % | No. | % | No. | % | |
| A1 | 25 | 18.5 | 71 | 52.2 | 3 | 17.6 | 99 | 34.4 |
| A2 | 1 | 0.7 | - | - | - | - | 1 | 0.3 |
| A3 | 1 | 0.7 | 5 | 3.7 | - | - | 6 | 2.1 |
| A4 | 1 | 0.7 | - | - | - | - | 1 | 0.3 |
| A5 | 4 | 3.0 | - | - | - | - | 4 | 1.4 |
| A6 | 9 | 6.7 | 13 | 9.6 | - | - | 22 | 7.6 |
| A7 | - | - | 2 | 1.5 | - | - | 2 | 0.7 |
| A8 | 1 | 0.7 | - | - | - | - | 1 | 0.3 |
| A9 | - | - | 2 | 1.5 | - | - | 2 | 0.7 |
| A10 | 6 | 4.4 | - | - | - | - | 6 | 2.1 |
| A11 | 29 | 21.5 | 1 | 0.7 | - | - | 30 | 10.4 |
| A12 | 1 | 0.7 | - | - | - | - | 1 | 0.3 |
| A13 | 1 | 0.7 | - | - | - | - | 1 | 0.3 |
| A14 | 2 | 1.5 | - | - | - | - | 2 | 0.7 |
| B1 | 2 | 1.5 | - | - | - | - | 2 | 0.7 |
| B2 | 1 | 0.7 | - | - | - | - | 1 | 0.3 |
| C1 | - | - | - | - | 1 | 5.9 | 1 | 0.3 |
| C2 | - | - | - | - | 1 | 5.9 | 1 | 0.3 |
| D1 | - | - | 4 | 2.9 | - | - | 4 | 1.4 |
| D2 | - | - | 5 | 3.7 | - | - | 5 | 1.7 |
| D3 | - | - | 17 | 12.5 | - | - | 17 | 5.9 |
| E1 | - | - | 1 | 0.7 | - | - | 1 | 0.3 |
| E2 | 2 | 1.5 | - | - | - | - | 2 | 0.7 |
| F1 | 15 | 11.1 | 3 | 2.2 | - | - | 18 | 6.3 |
| F2 | 25 | 18.5 | 6 | 4.4 | 12 | 70.6 | 43 | 14.9 |
| F3 | 1 | 0.7 | - | - | - | - | 1 | 0.3 |
| F4 | - | - | 2 | 1.5 | - | - | 2 | 0.7 |
| F5 | 6 | 4.4 | 4 | 2.9 | - | - | 10 | 3.5 |
| F6 | 2 | 1.5 | - | - | - | - | 2 | 0.7 |
| Total | 135 | 100.0 | 136 | 100.0 | 17 | 100.0 | 288 | 100.0 |
A dash (-) indicates there were no isolates of a given type isolated in this region.
Lineage group A was the largest group in the 288 isolates consisting of 14 different types, which together accounted for 61.8% of all of the isolates collected from the three provinces (Table 1). Lineage group F, which differed from group A, accounted for 26.4% of all of the isolates. Three A types (A1, A6, and A11) and three F types (F1, F2, and F5) together made up 77.1% of all isolates. These two lineage groups were also found in the three provinces surveyed. However, the remaining four groups, B, C, D, and E, comprising 11.8% of all isolates, were confined to one or two provinces. Of these lineage groups, 9% of isolates were D and the remaining 2.8% were associated with the minor lineage groups B, C, and E (Table 1).
Although certain PFGE types were predominant in the three provinces, statistical analysis of the 195 isolates selected for phage typing indicated that each herd and each province had a unique PFGE type distribution which was significant (P < 0.001). For example, whereas A1 and F2 types were detected in the three provinces with different frequencies, A3, A6, A11, F1, and F5 were present only in Ontario and Quebec. Types A5 and A10 were detected only in Ontario, and types D1, D2, and D3 were identified only in Quebec.
In the majority of all herds (58.6%), only a single PFGE type was recovered. However, the remainder (41.4%) contained two, three, and even four types (Table 2). There were six cases (three in Ontario and three in Quebec) in which different types were detected in different quarters of the same cow. Some of these belonged to the same lineage group (A5/A6, B1/B2, and F1/F2), whereas others were in different groups (A1/D1, A3/F2, and A3/F4).
TABLE 2.
Number of dairy herds containing S. aureus isolates with different PFGE types in three Canadian provinces
| No. of PFGE types/herd | No. of dairy herds in:
|
Total
|
||||||
|---|---|---|---|---|---|---|---|---|
| Ontario
|
Quebec
|
PEI
|
||||||
| No. | % | No. | % | No. | % | No. | % | |
| 1 | 16 | 53.3 | 15 | 62.5 | 3 | 75.0 | 34 | 58.6 |
| 2 | 9 | 30.0 | 5 | 20.8 | 0 | 0.0 | 14 | 24.1 |
| 3 | 2 | 6.7 | 3 | 12.5 | 1 | 25.0 | 6 | 10.3 |
| 4 | 3 | 10.0 | 1 | 4.2 | 0 | 0.0 | 4 | 6.9 |
| Total farms | 30 | 100.0 | 24 | 100.0 | 4 | 100.0 | 58 | 100.0 |
Phage types.
Of the 195 isolates tested, 63.1% were lysed by phage from groups 1 and 3, and 32.8% could not be typed with any of the phage strains used (NT). The remaining 4.1% belonged to a variety of phage types (“other”), with a frequency of <5 per region (Table 3). Only two isolates were found that reacted to any of the group 2 phages; these were placed in the phage type category “other.” Except for PFGE type A14, which responded to the miscellaneous phage set used, the other seven A types (A1, A3, A5, A6, A10, A11, and A12) corresponded to either phage group 3 (63.3%) or were NT (34%). Group F, except for type F3 (phage group “other”), corresponded to phage group 1 (98%). PFGE group D isolates, with the exception of a single D2 isolate (phage group 3), were NT. Other, less-common PFGE types (B1 and B2) were NT or responded to other phages not included in the standard typing set (C2) or to phage group 1 (E2). As with PFGE types, the phage type distribution and frequencies were significantly (P < 0.0001) associated with the dairy herd that the isolates were collected from.
TABLE 3.
Frequency and distribution of phage types in 195 S. aureus isolates collected in three Canadian provinces
| Phage type | No. of phage types in:
|
Total
|
||||||
|---|---|---|---|---|---|---|---|---|
| Ontario
|
Quebec
|
PEI
|
||||||
| No. | % | No. | % | No. | % | No. | % | |
| 1 | 34 | 38.6 | 10 | 10.3 | 8 | 80.0 | 52 | 26.7 |
| 3 | 18 | 20.5 | 52 | 53.6 | 1 | 10.0 | 71 | 36.4 |
| Nontypeable | 32 | 36.4 | 32 | 33.0 | 0 | 0.0 | 64 | 32.8 |
| Othera | 4 | 4.5 | 3 | 3.1 | 1 | 10.0 | 8 | 4.1 |
| Total | 88 | 100.0 | 97 | 100.0 | 10 | 100.0 | 195 | 100.0 |
Phage types with fewer than three isolates.
Susceptibility to antimicrobial agents.
Of the 288 S. aureus isolates typed by PFGE, 212 isolates from 58 farms in three provinces were examined for their resistance to antimicrobials (Table 4). A total of 24.5% of the isolates included in the analysis were resistant to at least one antimicrobial. Across all regions, resistance to penicillin was most common (9.9%), followed by resistance to sulfadimethoxine (7.5%). Four isolates (1.9%) were resistant to both penicillin and sulfadimethoxine, and two isolates (0.9%) were resistant to tilmicosin, erythromycin, and pirlimycin. All other resistant isolates were resistant to only one antimicrobial. Isolates collected in the province of Ontario exhibited the highest proportion of resistant isolates (30.2%), as well as resistance to the widest variety of antimicrobials (7 of the 10 antimicrobials tested). In Quebec, only resistance to penicillin, tetracycline, and sulfadimethoxine was found, although the frequency of sulfadimethoxine resistance exceeded that found in Ontario. Resistance to the penicillin-novobiocin combination, cephalothin, or ceftiofur was not found in any isolate. The levels of antimicrobial resistance in isolates collected from animals before and after antibiotic treatment were statistically similar (odds ratio = 0.93, P = 0.856).
DISCUSSION
A variety of subtyping techniques, such as phage typing, ribotyping, randomly amplified polymorphic DNA analysis, MLEE, and PFGE have been used to identify bacterial clones. PFGE has proven to have a satisfactory discriminatory ability and reproducibility for epidemiological survey of S. aureus variants (19, 37). MLST (19) has been used to detect variants of S. aureus (13). However, for cluster analysis of isolates, both PFGE and MLST methods produce similar results (13). For a fine-structure molecular analysis, a multienzyme typing method that includes MLEE has proven to be a more useful method (18).
Compared to phage typing, PFGE produced a much better discrimination between genetic types of S. aureus isolates. Approximately 20% of S. aureus isolates submitted to the Centers for Disease Control and Prevention for phage typing were nonreactive when the international phage typing set was used (37). Similar results were observed here, which shows that phage typing has a much lower discriminatory ability than PFGE typing (19).
Analysis of the restriction patterns revealed a reasonable amount of genetic heterogeneity among S. aureus isolates. There were herds (41.4%) with two, three, and even four types present (Table 2), as has been detected by others (18, 21, 26). However, in the majority (58.6%) of all herds, only a single PFGE type was recovered. Although prevalence of PFGE types was herd dependent, certain types were predominant in the three provinces (members of lineage groups A and F), whereas others, such as lineage group D types, were detected in a specific herd or provincial region. Although clones with different molecular types within the same herd have been reported, a common S. aureus clone associated with mastitis in dairy cows has been recovered from Ireland and the United States (4, 18). These observations support the previous findings in other regions that a limited number of bovine S. aureus clones that have broad geographic distribution are responsible for the majority of mastitis cases (18, 27, 34). A more detailed genetic analysis is needed to determine whether the S. aureus isolate types identified in the present study descended from a common ancestor or evolved independently.
Antimicrobial resistance was identified in 24.5% of the bacterial isolates analyzed. For some antibiotics, particularly penicillin and tetracycline, the frequency of resistance appears to be considerably lower in Canadian herds than is reported by workers in Europe and the United States. In almost all studies of this nature, penicillin is associated with the highest frequency of resistant isolates, as was found in the present study with 9.9% of isolates resistant. However, Erskine et al. reported that 46% of the S. aureus isolates were resistant to penicillin in a 1999 sample of Michigan dairy herds (15). Similarly, Makovec et al. reported 35.4% of isolates as resistant to penicillin in a survey of Wisconsin isolates (24). In studies concerning European herds penicillin resistance is more heterogeneous, ranging from only 2% (Norway) to more than 70% (Ireland), with an overall average of ca. 32% (39). In the case of tetracycline, 8.6% of S. aureus isolates are reported to be resistant in American herds versus 1.4% resistant isolates in the present study (24).
With regard to the other antibiotics tested, we found no resistance to the penicillin-novobiocin combination, cephalothin, and ceftiofur and little resistance (<2.5%) to tilmicosin, erythromycin, oxacillin, and pirlimycin. A relatively high level of sulfadimethoxine resistance (7.5%) was found in the Canadian dairy herds, making it the second most common type of antimicrobial resistance phenotype found. Unfortunately, sulfadimethoxine is not a commonly tested drug when the MICs of bovine S. aureus isolates are determined. The related sulfa compound sulfisoxazole has been associated with resistance in S. aureus at a frequency of 4.5% in America (24), and sulfamethoxazole-resistant isolates have a reported frequency of 0.8 to 13.5% in European herds (39).
The observed localized differences in the distribution of antibacterial resistance may reflect the use of specific antimicrobials that act as a selective agent for resistance. Although the rate of antimicrobial resistance for bacterial isolates varies in different geographic populations, it has been suggested that the main contributing factor is the frequent use of antimicrobials (35). Dairy cattle, however, are not routinely given antibiotics in the form of feed additives due to the detrimental effects of antibiotic residues in the milk. Dairy cattle are generally given antibiotics to treat specific ailments and for mastitis prophylaxis at drying-off. One recent survey suggests that penicillin, novobiocin, and cephalosporins (particularly ceftiofur) are the antibiotics of choice in Ontario dairy herds (23). This could partially explain the relatively high level of penicillin resistance found in this province (12.5%). However, a survey of producers participating in this trial indicated that use of penicillin-based dry-cow therapeutics is limited, with 2 of 77 respondents (2.5%) reporting their use. This may reflect differences in the sample groups, or it may indicate that penicillin-based products are used for purposes other than mastitis treatment. In terms of dry-cow mastitis therapy, benzathine cloxacillin, which is resistant to S. aureus penicillinase, is a commonly used product.
Analysis of antibiotic susceptibility patterns for various bacterial intramammary pathogens conducted over 15 years in New York state revealed a trend of decreasing susceptibility of Staphylococcus spp. to several antibiotics. Significant resistance of S. aureus strains to penicillin (64%) and ampicillin (46%) was reported in an Argentinian study (2). It is encouraging that no resistance was found to either novobiocin or ceftiofur in our survey. A recent review of the literature by Erskine et al. (14) suggests that there is no evidence of a widespread trend of increasing antimicrobial resistance in mastitis pathogens. There is an unfortunate lack in the literature regarding previous levels of antimicrobial resistance in Canadian dairy herds. Thus, we cannot determine whether the frequency of antimicrobial resistance reported here represents a decline or increase in resistance over previous levels.
Interestingly, antimicrobial resistance did not appear to be a marker that always spread clonally through a herd. In several instances, antimicrobial resistance was observed in only a small proportion of the S. aureus isolates taken from a given herd (data not shown). This finding underscores a potential pitfall in sampling herds, in that one or a few isolates taken from a given site will not necessarily represent the total antimicrobial resistance present at that site. Antibiotic resistance is usually linked to the acquisition of plasmids (40). A high percentage of plasmid-free S. aureus isolates from bovine mastitis have been reported (1, 22). An investigation into the type and distribution of plasmids within both the S. aureus and the general bacterial populations could determine the mode of inheritance of resistance and the relatedness of different PFGE types.
Acknowledgments
We thank X. Lu for help with statistical analysis of the data and Anna Bashiri for the initial collection of S. aureus isolates. We thank Marie Archambault and Grazyna Adamska-Jarecka for help with the antimicrobial resistance analysis. We also thank the producers who participated in this study.
J.J.G. received a graduate study grant from the Dairy Farmers of Ontario.
Footnotes
Agriculture and Agri-Food Canada Report S171.
REFERENCES
- 1.Aarestrup, F. M., H. C. Wegener, and V. T. Rosdahl. 1995. Evaluation of phenotypic and genotypic methods for epidemiological typing of Staphylococcus aureus isolates from bovine mastitis in Denmark. Vet. Microbiol. 45:139-150. [DOI] [PubMed] [Google Scholar]
- 2.Acuna, C. N., R. E. Chertcoff, G. Cisneros, E. Izak, and J. M. Nimo. 2000. Antimicrobial resistance of Staphylococcus aureus isolated from quarters with clinical mastitis in Argentina, p. 211-212. In National Mastitis Council meeting proceedings. National Mastitis Council, Inc., Madison, Wis.
- 3.Anderson, E. S., and R. E. O. Williams. 1956. Bacteriophage typing of enteric pathogens and staphylococci and its use in epidemiology. J. Clin. Pathol. 9:94-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annemuller, C., C. Lammler, and M. Zschock. 1999. Genotyping of Staphylococcus aureus isolated from bovine mastitis. Vet. Microbiol. 69:217-224. [DOI] [PubMed] [Google Scholar]
- 5.Asao, T., Y. Kumeda, T. Kawai, T. Shibata, H. Oda, K. Haruki, H. Nakazawa, and S. Kozaki. 2003. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol. Infect. 130:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayliffe, G. A. 1997. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 24:S74-S79. [DOI] [PubMed] [Google Scholar]
- 7.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blair, J. E., and R. E. O. Williams. 1961. Phage typing of staphylococci. Bull. W. H. O. 24:771-784. [PMC free article] [PubMed] [Google Scholar]
- 9.Blake, P. A. 1984. Staphylococcal food poisoning in the United States. JAMA 251:487-489. [PubMed] [Google Scholar]
- 10.Coast, J., and R. D. Smith. 2003. Solving the problem of antimicrobial resistance: is a global approach necessary? Drug Discov. Today 8:1-2. [DOI] [PubMed] [Google Scholar]
- 11.Dairy Farmers of Canada. 2000. Dairy facts and figures at a glance, 1999. Dairy Farmers of Canada, Ottawa, Ontario, Canada.
- 12.Dingwell, R. T., K. E. Leslie, T. F. Duffield, Y. H. Schukken, L. DesCosteaux, G. P. Keefe, D. F. Kelton, K. D. Lissemore, W. Shewfelt, P. Dick, and R. Bagg. 2003. Efficacy of intramammary tilmicosin and risk factors for cure of Staphylococcus aureus infection in the dry period. J. Dairy Sci. 86:159-168. [DOI] [PubMed] [Google Scholar]
- 13.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erskine, R., J. Cullor, M. Schaellibaum, B. Yancey, and A. Zecconi. 2004. Bovine mastitis pathogens and trends in resistance to antibacterial drugs, p. 400-414. In National Mastitis Council meeting proceedings. National Mastitis Council, Inc., Verona, Wis.
- 15.Erskine, R. J., R. D. Walker, C. A. Bolin, P. C. Bartlett, and D. G. White. 2002. Trends in antibacterial susceptibility of mastitis pathogens during a seven-year period. J. Dairy Sci. 85:1111-1118. [DOI] [PubMed] [Google Scholar]
- 16.Farmer, J. J. I., F. W. Hickman, and J. V. Sikes. 1975. Automation of Salmonella typhi phage typing. Lancet ii:787-790. [DOI] [PubMed] [Google Scholar]
- 17.Fetrow, J., S. Stewart, S. Eicker, R. Farnsworth, and R. Bey. 2000. Mastitis: an economic consideration, p. 3-47. In National Mastitis Council meeting proceedings. National Mastitis Council, Inc., Madison, Wis.
- 18.Fitzgerald, J. R., W. J. Meaney, P. J. Hartigan, C. J. Smyth, and V. Kapur. 1997. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol. Infect. 119:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grundmann, H., S. Hori, M. C. Enright, C. Webster, A. Tami, E. J. Feil, and T. Pitt. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 40:4544-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, T. F., M. E. Kellum, S. S. Porter, M. Bell, and W. Schaffner. 2002. An outbreak of community-acquired food-borne illness caused by methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 8:82-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapur, V., W. M. Sischo, R. S. Greer, T. S. Whittam, and J. M. Musser. 1995. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J. Clin. Microbiol. 33:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lange, C., M. Cardoso, D. Senczek, and S. Schwarz. 1999. Molecular subtyping of Staphylococcus aureus isolates from cases of bovine mastitis in Brazil. Vet. Microbiol. 67:127-141. [DOI] [PubMed] [Google Scholar]
- 23.Leger, D., D. Kelton, K. Lissemore, R. Reid-Smith, SW. Martin, and N. Anderson. 2003. Antimicrobial drug use by dairy veterinarians and free stall producers in Ontario, p. 318-319. In National Mastitis Council meeting proceedings. National Mastitis Council, Inc., Madison, Wis.
- 24.Makovec, J. A., and P. L. Ruegg. 2003. Antimicrobial resistance of bacteria isolated from dairy cow milk samples submitted for bacterial culture: 8,905 samples (1994-2001). J. Am. Vet. Med. Assoc. 222:1582-1589. [DOI] [PubMed] [Google Scholar]
- 25.Mantel, N. 1963. Chi-square tests with one degree of freedom: extensions of the Mantel-Haenszel procedure. J. Am. Stat. Assoc. 58:690-700. [Google Scholar]
- 26.Matthews, K. R., R. J. Harmon, and B. E. Langlois. 1992. Prevalence of Staphylococcus species during the periparturient period in primiparous and multiparous cows. J. Dairy Sci. 75:1835-1839. [DOI] [PubMed] [Google Scholar]
- 27.Musser, J. M., P. M. Schlievert, A. W. Chow, P. Ewan, B. N. Kreiswirth, V. T. Rosdahl, A. S. Naidu, W. Witte, and R. K. Selander. 1990. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc. Natl. Acad. Sci. USA 87:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.NCCLS. 2002. Performance standards for anti-microbial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 29.Olsen, S. J., L. C. MacKinon, J. S. Goulding, N. H. Bean, and K. Slutsker. 2000. Surveillance for food-borne disease outbreaks: United States, 1993-1997. Morb. Mortal. Wkly. Rep. CDC Surv. Summ. 49:1-51. [PubMed] [Google Scholar]
- 30.Osteras, O., V. L. Edge, and S. W. Martin. 1999. Determinants of success or failure in the elimination of major mastitis pathogens in selective dry cow therapy. J. Dairy Sci. 82:1221-1231. [DOI] [PubMed] [Google Scholar]
- 31.Parker, M. T. 1972. Phage-typing of Staphylococcus aureus. Methods Microbiol. 7B:1-28. [Google Scholar]
- 32.Sargeant, J. M., H. M. Scott, K. E. Leslie, M. J. Ireland, and A. Bashiri. 1998. Clinical mastitis in dairy cattle in Ontario: frequency of occurrence and bacteriological isolates. Can. Vet. J. 39:33-38. [PMC free article] [PubMed] [Google Scholar]
- 33.Schukken, Y. H. L., L. Tikovsky, D. Wilson, and O. Osteras. 2001. Factors affecting the success of antibiotic treatment at the dry-off, p. 80-87. In National Mastitis Council meeting proceedings. National Mastitis Council, Inc., Madison, Wis.
- 34.Smith, P., C. Spooner, R. Lyman, C. George, W. Kloos, and K. Anderson. 2002. Distribution of strains of Staphylococcus aureus isolated from milk of cows in North Carolina, p. 233-234. In National Mastitis Council meeting proceedings. National Mastitis Council, Inc., Madison, Wis.
- 35.Smith, R. D., and J. Coast. 2002. Antimicrobial resistance: a global response. Bull. W. H. O. 80:126-133. [PMC free article] [PubMed] [Google Scholar]
- 36.Sol, J., O. C. Sampimon, J. J. Snoep, and Y. H. Schukken. 1994. Factors associated with bacteriological cure after dry cow treatment of subclinical staphylococcus mastitis with antibiotics. J. Dairy Sci. 77:75-79. [DOI] [PubMed] [Google Scholar]
- 37.Tenover, F. C., R. Arbeit, G. Archer, J. Biddle, S. Byrne, R. Goering, G. Hancock, G. A. Hebert, B. Hill, R. Hollis, W. R. Jarvis, B. Kreiswirth, W. Eisner, J. Maslow, L. K. McDougal, J. M. Miller, M. Mulligan, and M. A. Pfaller. 1994. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vintov, J., F. M. Aarestrup, C. E. Zinn, and J. E. Olsen. 2003. Association between phage types and antimicrobial resistance among bovine Staphylococcus aureus from 10 countries. Vet. Microbiol. 95:133-147. [DOI] [PubMed] [Google Scholar]
- 40.Waage, S., J. Bjorland, D. A. Caugant, H. Oppegaard, T. Tollersrud, T. Mork, and F. M. Aarestrup. 2002. Spread of Staphylococcus aureus resistant to penicillin and tetracycline within and between dairy herds. Epidemiol. Infect. 129:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waage, S., T. Mork, A. Roros, D. Aasland, A. Hunshamar, and S. A. Odegaard. 1999. Bacteria associated with clinical mastitis in dairy heifers. J. Dairy Sci. 82:712-719. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, D. J., R. H. Gonzalez, and H. Das. 1997. Bovine mastitis pathogens in New York and Pennsylvania: prevalence and effects on somatic cell count and milk production. J. Dairy Sci. 80:2592-2598. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, D. J., R. N. Gonzalez, K. L. Case, L. L. Garrison, and Y. T. Grohn. 1999. Comparison of seven antibiotic treatments with no treatment for bacteriological efficacy against bovine mastitis pathogens. J. Dairy Sci. 82:1664-1670. [DOI] [PubMed] [Google Scholar]
- 44.Woods, G. L., and J. A. Washington. 1995. Antibacterial susceptibility tests: dilution and disk diffusion methods, p. 1327-1341. In P. Murray, E. Baron, M. Pfaller, F. Tenover, and R. Yolken (ed.), Manual of clinical microbiology. ASM, Washington, D.C.