Abstract
Screening for the Escherichia coli O serotype is the traditional test for identification of E. coli clones. The O-antigen gene cluster of the E. coli O114 type strain was sequenced, and 12 open reading frames were assigned functions on the basis of homology. By screening against all 186 E. coli and Shigella O serotypes, five genes specific to E. coli O114 were identified. A PCR assay based on the O-antigen-specific genes was developed and tested on 41 clinical isolates of E. coli O114. The PCR assay was shown to be highly specific and sensitive. When tested with pork and water samples, as few as 0.12 CFU of E. coli O114 g−1 were detected. Thus, the PCR assays established in this study can be used to reliably identify E. coli O114 strains and may also be used to detect E. coli O114 strains in food, water, and other environmental samples.
Escherichia coli strains causing diarrhea in humans express different virulence factors and are accordingly divided into five major pathotypes: enteropathogenic, enterotoxigenic, enteroinvasive, enteroaggregative, and enterohemorrhagic (14, 15). These pathotypes consist of genetic clones that often correspond to distinct O:H serotypes (17, 31).
Serogroup O114 belongs to the traditional set of enteropathogenic E. coli-associated O groups and strains isolated from infants with diarrhea and from septicemic calves (9, 18). Serogroup O114 was later found to include enteropathogenic, enterotoxigenic, and enterohemorrhagic E. coli strains as well as uropathogenic and apathogenic groups (8, 25). Serotype O114:H2 strains were classified as typical (E. coli adhesin factor positive) and atypical (E. coli adhesin factor negative) enteropathogenic E. coli (29), E. coli O114:H21 and O114:H49 strains were identified as enterotoxigenic E. coli producing heat-labile or heat-stable enterotoxins (8, 20, 25, 34), some E. coli O114:H4 strains were identified as enterohemorrhagic E. coli producing Shiga toxin 1 (8, 33), and a clone of O114:H9 strains showed properties of uropathogenic E. coli and expressed P-fimbriae, aerobactin, and alpha-hemolysin (8).
Conventional O-serotyping of E. coli strains in clinical specimens, food, and environmental samples is laborious and time-consuming and not practical for analysis of large numbers of specimens. Moreover, the serological assay cannot be performed on strains with a rough O antigen, which are frequently isolated from clinical and environmental samples. Here, we were interested in developing a PCR-based method for sensitive and reliable detection of genes coding for the O114 serogroup, which would be useful for rapid screening of clinical and environmental samples, such as E. coli-contaminated food and water.
The O antigen (O-specific polysaccharide), which consists of many repeats of an oligosaccharide unit (O unit), is the outer component of lipopolysaccharide on the surface of gram-negative bacteria (22). There are 186 O-antigen forms recognized in E. coli (including Shigella).
Genes for O-antigen synthesis are normally located in a gene cluster which maps between galF and gnd on the E. coli chromosome. The O-antigen genes generally fall into three main classes: (i) genes for synthesis of nucleotide sugar precursors, (ii) genes for transfer of sugars to build the O unit, and (iii) genes carrying out specific assembly or processing steps in conversion of the O unit to the O antigen as part of the complete lipopolysaccharide. Genes belonging to the second two groups are specific to different O antigens. In the present study, the O-antigen gene cluster of the E. coli O114 type strain was sequenced, and genes in the cluster were identified. By screening with all 186 E. coli (including Shigella) type strains and 41 clinical isolates of E. coli O114, five genes specific for E. coli O114 were identified. A PCR assay based on the specific genes was developed for the rapid detection and identification of E. coli O114 from various sources. The sensitivity of the specific PCR assay was also tested.
Nucleotide sequence analysis of E. coli O114 O-antigen gene cluster.
E. coli O114 type strain G1088 (O114:H32) (12) was obtained from the Institute of Medical and Veterinary Science, Adelaide, Australia, and grown under aeration for 12 h in Luria-Bertani broth at 37°C. Chromosomal DNA was prepared as described previously (4). Long-range PCR was performed with the Expand Long Template PCR system (Roche Applied Science) with primers 1523 (5′-ATT GTG GCT GCA GGG ATC AAA GAA AT-3′) and 1524 (5′-TAG TCG CGT GNG CCT GGA TTA AGT TCG C-3′), which were designed based on galF and gnd, respectively (30). PCR was performed as follows: denaturation at 94°C for 10 s, annealing at 60°C for 30 s, and extension at 68°C for 15 min for 30 cycles. The PCR products were digested with DNase I, and the resulting DNA fragments were cloned into pGEM-T Easy (Promega) to produce a shotgun bank as described previously (30). To minimize nucleotide sequence errors from the PCR assay, five individual PCR products were combined before construction of the bank. Nucleotide sequencing was carried out with an ABI 3773 automated DNA sequencer. Sequence data were assembled with the Staden package (26).
The program Artemis (23) was used for gene annotation. BLAST and PSI-BLAST (3) were used for searching databases, including GenBank, COG, and Pfam (5, 27). The program BlockMaker (13) was used for searching conserved motifs. The algorithm described by Eisenberg (11) was used to identify potential transmembrane segments. Sequence alignment was performed with the program ClustalW (28).
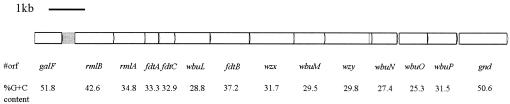
A sequence of 13,272 bases from galF (positions 1 to 765) to gnd (positions 11935 to 13272) was obtained, which contained 12 open reading frames (ORFs) with the same transcriptional direction from galF to gnd. In E. coli O114, ORFs had a low G+C content of 25.3 to 42.6% (Fig. 1), significantly lower than that of the E. coli genome (50%) in all reported E. coli O-antigen gene clusters. All of the putative genes were assigned functions based on their similarity to genes in the databases (Table 1).
FIG. 1.
O-antigen gene cluster of E. coli O114. All genes are transcribed in the direction from galF to gnd.
TABLE 1.
Putative genes in E. coli O114 O-antigen gene cluster
| Gene | Location in sequence | % G + C | Conserved domain(s) | Similar proteins (accession no. of amino acids) | % Identical/% similar | Putative function |
|---|---|---|---|---|---|---|
| rmlB | 1138-2214 | 42.6 | NAD dependent epimerase/dehydratase family PF01370, E 2.3×e−211 | dDTDP-glucose 4,6-dehydratase E. coli O91 (AAK60448/358) | 96/98 | dTDP-glucose 4,6-dehydratase |
| rmlA | 2211-3089 | 34.8 | Nucleotidyl transferase PF00483, E 1.2×e−109 | d-Glucose-1-phosphate thymidylyltransferase E. coli O91 (AAK60449/287) | 80/89 | d-Glucose-1-phosphate thymidylyltransferase |
| fdtA | 3094-3486 | 33.3 | WxcM-like, C-terminal PF05523, E 1×e−67 | dTDP-6-deoxy-3,4-keto-hexulose isomerase A. thermoaerophilus strain L420-91T (AAO06351/139) | 47/64 | Isomerase |
| fdtC | 3479-3925 | 32.9 | Acetyltransferase (GNAT) family PF00583, E 8.3×e−14 | dTDP-d-Fucp3N acetylase A. thermoaerophilus strain L420-91T (AAO06352/193) | 43/57 | Acetyltransferase |
| wbuL | 3925-4923 | 28.8 | MurN Streptococcus mutans UA159 (AAN58445/410) | 23/47 | Glycosyltransferase | |
| fdtB | 4925-6031 | 37.2 | DegT/DnrJ/EryC1/StrS aminotransferase PF01041, E 7.8×e−136 | dTDP-6-deoxy-d-xylo-hex-3-ulose aminase A. thermoaerophilus strain L420-91T (AAO06353/363) | 55/70 | Aminotransferase |
| wzx | 6028-7281 | 31.7 | Polysaccharide biosynthesis protein PF01943, E 1.5×e−4 | Wzx, E. coli O91 (AAK60454/421) | 34/56 | O-antigen flippase |
| wbuM | 7274-8143 | 29.5 | Glycosyl transferase PF00535, E 1.4×e−26 | Putative glycosyltransferase WbgO E. coli O55:H7 (AAL67559/265) | 33/54 | Glycosyltransferase |
| wzy | 8137-9468 | 29.8 | Antigen polymerase E. coli O6 (CAD19991/447) | 21/43 | O-antigen polymerase | |
| wbuN | 9486-10172 | 27.3 | Phosphoserine phosphatase Clostridium tetani E88(AAO36271/220) | 28/48 | Phosphoserine phosphatase | |
| wbuO | 10251-11030 | 25.3 | CDP-diacylglycerol-serine O-phosphatidyltransferase Methanosarcina acetivorans strain C2A (AAM03570/247) | 12/29 | Serine transferase | |
| wbuP | 11039-11851 | 31.5 | Glycosyl transferase PF00535, E 2.4×e−18 | Putative glycosyltransferase Bacteroides fragilis strain NCTC 934 (AAK68920/268) | 38/57 | Glycosyltransferase |
Genes for dTDP-d-Qui3NAc.
The E. coli O114 O antigen consists of repeating units of a pentasaccharide, which has the structure (→3)-α-d-GlcNAc-(1→4)-β-d-Qui3NAcyl-(1→3)-β-d-ribofuranose-(1→4)-β-d-Gal-(1→) (where acyl = N-acetyl-l-seryl) (10). GlcNAc, ribofuranose, and Gal are common sugars, and their synthesis is independent of the O-antigen gene cluster. The genes for the synthesis of the rare sugar Qui3NAc (3-acetamido-3,6-dideoxy-d-glucose) were expected in the O-antigen gene cluster. ORF1 and ORF2 showed 96 and 80% identity to RmlB and RmlA, respectively, of the E. coli K-12 (O16) O-antigen gene cluster. ORFs 3, 4, and 6 showed 47, 43, and 55% identity, respectively, to FdtA (dTDP-6-deoxy-3,4-keto-hexulose isomerase), FdtC (dTDP-d-Fuc3N acetylase), and FdtB (dTDP-6-deoxy-d-xylo-hex-3-ulose aminotransferase), respectively, of Aneurinibacillus thermoaerophilus strain L420-91T. In A. thermoaerophilus, RmlA, RmlB, FdtA, FdtB, and FdtC are involved in the biosynthesis of dTDP-d-Fuc3NAc (dTDP-3-acetamido-3,6-dideoxy-alpha-d-galactose), which is an epimer of dTDP-d-Qui3NAc (dTDP-3-acetamido-3,6-dideoxy-α-d-glucose) (21). Thus, orf1, orf2, orf3, orf4, and orf6 were identified as genes responsible for the biosynthesis of UDP-Qui3NAc in E. coli O114 and named rmlB, rmlA, fdtA, fdtC, and fdtB, respectively.
Genes for O-unit processing.
Wzx and Wzy are typical inner membrane proteins with more than nine transmembrane segments. Wzy also typically contains a large periplasmic loop of more than 30 amino acids. ORF7 had 12 predicted transmembrane segments, which is the typical number for Wzx proteins. It also showed 34% identity and 56% similarity to the putative Wzx of E. coli O91 (Table 1) and showed 52% to 54% similarity to putative Wzx proteins of other E. coli and Salmonella strains (data not shown). Therefore, orf7 was identified as wzx, encoding the O-unit flippase, and was named accordingly. ORF9 had 11 predicted transmembrane segments with a large periplasmic loop of 49 amino acid residues, the typical topological characteristics of Wzy proteins. It also showed 43% similarity to the Wzy protein of E. coli O6, which has been well characterized. Therefore, orf9 was identified as wzy, encoding an O-antigen polymerase, and named accordingly.
Genes encoding sugar transferases.
In E. coli O114, GlcNAc is present as the first sugar and as such is generally transferred by wecA, which is located outside the O-antigen gene cluster (1). Genes encoding transferases for the other three sugars were expected in the O-antigen gene cluster of E. coli O114. ORFs 5, 8, and 12 showed different levels of similarity to putative glycosyltransferases (Table 1). ORFs 8 and 12 were also related to glycosyltransferase family 2 described by Wiggins (32). Therefore, orf5, orf8, and orf12 were proposed to encode glycosyltransferases and named wbuL, wbuM, and wbuP, respectively.
Genes for synthesis and transfer of the seryl group.
ORF10 showed 48% similarity to phosphoserine phosphatase of Clostridium tetani E88, which converts the phosphoserine precursor to serine. ORF11 showed 29% similarity to CDP-diacylglycerol-serine O-phosphatidyltransferase of Methanosarcina acetivorans strain C2A. It also showed similarity to the same enzyme of many other bacterial strains (data not shown). These serine transferases and ORF11 all had six predicted transmembrane segments and shared homology to permeases. Modification of the O-antigen backbone commonly takes place in the periplasm, and some of the membrane proteins involved in the modification process share activity of permeases, such as those involved in the modification process of the acetyl group and glucose residue (2). Therefore, we propose that orf10 and orf11 are involved in the synthesis and transfer of the seryl group to the O114 antigen, respectively, and named them wbuN and wbuO, respectively.
Screening for E. coli O114 serogroup-specific genes.
Primers were designed based on the proposed O-unit processing genes wzx and wzy, two putative transferase genes wbuM and wbuP, and the serine group transferase gene wbuO (Table 2). Two pairs of primers for each gene were used to screen DNA pools comprising all 186 E. coli (including Shigella) type strains, in which type strains for O serogoups 5, 11, 23, 48, 70, 71, and 82 were reported to show serological cross-reactivity with the O114 antigen (12, 16).
TABLE 2.
PCR specificity test with E. coli O114 genes
| Gene | Base positions | Forward primers (base positions), oligonucleotide sequence | Reverse primers (base positions), oligonucleotide sequence | Length of PCR fragment (bp) | No. of pools giving correct band | Annealing temp (°C) of PCR |
|---|---|---|---|---|---|---|
| wzx | 6028-7281 | wl-750(6278-6295), 5′-CAGGTTTAAGTTGGGTAT-3′ | wl-751(6863-6880), 5′-AAGAAGAAAGTCTGGGTA-3′ | 603 | 0a | 50 |
| wl-752(6289-6308), 5′-TGGGTATGTATAATATCAGC-3′ | wl-753(7198-7216), 5′-AATATGCGTAAGTAACTCC-3′ | 928 | 0 | 56 | ||
| wbuM | 7274-8143 | wl-459(7568-7585), 5′-GATAGATTAACCACGCAG-3′ | wl-460(7792-7808), 5′-CCTCCTTATACCCTCCT-3′ | 241 | 0 | 56 |
| wl-461(7641-7658), 5′-AGGTGGATGATTCTAATG-3′ | wl-462(8081-8098), 5′-ACTATCGAGCCTATGTAA-3′ | 458 | 0 | 56 | ||
| wzy | 8137-9468 | wl-754(8367-8384), 5′-TTTTGGCGGTTCGTTGAT-3′ | wl-755(9190-9207), 5′-TGCCCATGCTTCTGAAAT-3′ | 841 | 0b | 56 |
| wl-756(8567-8584), 5′-CTTTCCCAAGCCCATTAT-3′ | wl-757(9160-9177), 5′-AACATTCCATCCACCTAA-3′ | 611 | 0 | 60 | ||
| wbuO | 1025-11030 | wl-463(10612-10629), 5′-TGTGGGCTATCTGGTTTA-3′ | wl-464(10794-10811), 5′-TTTCTCCGACATCCTTTC-3′ | 200 | 0 | 56 |
| wl-465(10451-10468), 5′-AAATGATTGCGAGACGAT-3′ | wl-466(10736-10753), 5′-GTGACCTGATAATTCCCT-3′ | 303 | 0 | 56 | ||
| wbuP | 11039-11851 | wl-467(11426-11443), 5′-AGTGATTGTTCGCTACCT-3′ | wl-468(11615-11632), 5′-CCCAAACTTCAGCCCTAA-3′ | 207 | 0 | 56 |
| wl-469(11343-11359), 5′-TAGACCGGCTGGAACGA-3′ | wl-470(11797-11814), 5′-AGCGGCATAAAGTGGGAT-3′ | 472 | 0 | 56 |
Three pools gave a band of the wrong size.
All pools gave a band of the wrong size.
DNA from each type strain was prepared as described previously (30). A total of 13 DNA pools were prepared, each containing DNA from 12 to 19 strains (Table 3). The PCR cycles used were as follows: denaturation at 95°C for 15 s, annealing for 30 s, and extension at 72°C for 1 min for 30 cycles. The annealing temperatures of the primer pairs are listed in Table 2. With either pair of primers used, no expected PCR products were observed for any of the DNA pools except the DNA pool containing E. coli O114, which gave PCR products of the correct sizes. Therefore, wzx, wzy, wbuM, wbuO, and wbuP are all specific to E. coli O114 strains. Two primer pairs (wl-750/751, targeting wzx, and wl-754/755, targeting wzy) produced bands of the incorrect size in some or all of the sample pools. This may be due to chance priming elsewhere on the chromosome. This problem may be avoided by redesigning primers for those genes. A combination of the specific genes or primers binding to adjacent specific genes may also achieve improved specificity.
TABLE 3.
E. coli and Shigella type strains and PCR pools used for testing of E. coli O114-specific primers
| Pool no. | Chromosomal DNAs included in the pool | Sourcea |
|---|---|---|
| 1 | E. coli type strains for O serotypes 1, 2, 5, 7, 12, 13, 14, 15, 16, 17, 19ab, 20, 21, 22, 23, 24, 59, 3, 11 | IMVS |
| 2 | E. coli type strains for O serotypes 25, 26, 27, 28, 29, 30, 32, 31, 33, 35, 36, 37, 38, 40, 41, 42, 43, 39, 59 | IMVS |
| 3 | E. coli type strains for O serotypes 44, 45, 46, 48, 49, 50, 51, 52, 54, 55, 56, 57, 58, 60, 61, 62, 64, 73 | IMVS |
| 4 | E. coli type strains for O serotypes 63, 65, 66, 69, 70, 71, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 96, 95 | IMVS |
| 5 | E. coli type strains for O serotypes 84, 85, 86, 87, 88, 89, 91, 92, 98, 99, 101, 102, 103, 104, 105, 106, 100, 151 | IMVS |
| 6 | E. coli type strains for O serotypes 107, 108, 109, 110, 111, 112ab, 112ac, 113, 115, 116, 118, 120, 123, 125, 126, 128 | IMVS |
| 7 | E. coli type strains for O serotypes 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145 | IMVS |
| 8 | E. coli type strains for O serotypes 146, 147, 148, 150, 152, 154, 156, 157, 158, 159, 160, 161, 163, 164, 165, 166 | IMVSb |
| 9 | E. coli type strains for O serotypes 168, 169, 170, 171, 172, 173, 155, 124 and S. dysenteriae type strains for O serotypes D1, D2, D3, D4, D5, D6, D7, D8, D9, D10, D11, D12 | IMVSc |
| IEM | ||
| 10 | S. boydii type strains for O serotypes B1, B2, B3, B4, B6, B7, B8, B9, B10, B11, B12, B13, B14, B15, B16, B17, B18 | IEM |
| 11 | S. flexneri type strains for O serotypes F1a, F1b, F2a, F2b, F3, F4b, F5(v:4), F5(v:7), F6, FX variationsFY variation and S. sonnei type strains for O serotypes DS, DR, | IEM |
| 12 | E. coli type strains for O serotypes 3, 11, 39, 59, 64, 73, 96, 95, 100, 114, 151, 167, 162, 121, 127, 149, 119 | IMVSd |
| 13 | Same as pool 12 but lacks E. coli O114, used as a control | IMVS |
IMVS, Institute of Medical and Veterinary Science, Adelaide, Australia; IEM, Institute of Epidemiology and Microbiology, Chinese Academy of Preventive Medicine, Beijing, People's Republic of China.
O165 and O166 from Statens Serum Institute, Copenhagen, Denmark, the rest from IMVS.
O155 and O124 from IMVS, the rest from Statens Serum Institute, Copenhagen, Denmark.
O167 from Statens Serum Institute, Copenhagen, Denmark, the rest from IMVS.
We further tested 57 E. coli clinical isolates as well as seven E. coli type strains and a Citrobacter freundii strain from Germany, which are reported to show serological cross-reactivity with E. coli O114. An O-rough laboratory K-12 strain was also tested. In the 57 E. coli clinical isolates, 41 were identified as O114 strains by O-serotyping at the Robert Koch Institute in Berlin. A total of 66 strains described above were tested by PCR with all of 10 primer pairs specific to the E. coli O114 type strain (Table 4). A double-blind test was performed with the following conditions. Each strain was cultured in Luria broth at 200 rpm at 37°C for 12 h, and 3 ml of culture was centrifuged at 5,000 × g for 5 min. The pellet containing E. coli O114 was mixed with 100 μl of Milli-Q water, boiled at 100°C for 15 min, and centrifuged at 12,000 × g for 8 min. The supernatant was used as the template in the PCR, which was performed as follows: denaturation at 95°C for 30 s, annealing for 45 s, and extension at 72°C for 1 min for 30 cycles. PCRs were carried out in a total volume of 25 μl, including 1 μl of template DNA.
TABLE 4.
E. coli isolates used for evaluation of E. coli O114-specific PCR
| Culture no. | Strain | O group | H type | Origin and yr of isolationa | Sourceb | Virulence markersc |
|---|---|---|---|---|---|---|
| G1331 | C 319-58 | O114 | H10 | UK, 1957 | HF | None |
| G1358 | C 556-59 | O114 | H10 | Sweden, 1959 | Calf septicemia | None |
| G1376 | C 288-63 | O114 | H2 | Germany, 1959 | HF, D | LA, eaf, bfp, eae |
| G1378 | C 276-63 | O114 | H2 | Germany, 1960 | HF, D | LA, eaf, bfp, eae |
| G1366 | C 280-63 | O114 | H2 | Germany, 1961 | HF, D | LA, eaf, bfp, eae |
| G1367 | C 281-63 | O114 | H2 | Germany, 1961 | HF, D | LA, eaf, bfp, eae |
| G1365 | C 290-63 | O114 | H2 | Germany, 1959 | HF, D | LA, eaf, bfp, eae |
| G1339 | C 315-60 | O114 | H2 | UK, 1960 | HF, D | LA, eaf, bfp, eae |
| G1377 | C 289-63 | O114 | H2 | Germany, 1958 | HF, D | LA, eaf, bfp, eae |
| G1379 | C 275-53 | O114 | H21 | Egypt, 1953 | HF | ST |
| G1361 | C 339-54 | O114 | H21 | Israel, 1954 | HF | LT |
| G1336 | C 1003-63 | O114 | H21 | Iran, 1963 | HF | ST |
| G1359 | 1098/63 | O114 | H32 | Germany, 1963 | Pig feces | None |
| G1360 | C 240-58 | O114 | H32 | Denmark, 1958 | Pig feces | None |
| G1356 | 26 W | O114 | H32 | Sweden, 1944 | calf septicemia | None |
| G1351 | CB 9421 | O114 | H34 | Brazil, 2002 | HF, D | eae |
| G1373 | C 2870/67 | O114 | H4 | Germany, 1967 | Monkey | None |
| G1374 | C 3142/71 | O114 | H4 | Germany, 1971 | HF | None |
| G1375 | C 3143/71 | O114 | H4 | Germany, 1971 | HF | None |
| G1341 | 3075/69 | O114 | H4 | Germany, 1969 | HF, D | Stx1 |
| G1349 | C 637-62 | O114 | H4 | Italy, 1962 | Calf septicemia | None |
| G1340 | 707/59 | O114 | H4 | UK, 1959 | Pig feces | None |
| G1343 | CB 177 | O114 | H4 | New Zealand, 1986 | HF | None |
| G1313 | C 2924-68 | O114 | H4 | Germany, 1968 | HF | None |
| G1362 | C 340-53 | O114 | H49 | Israel, 1953 | HF | LT |
| G1325 | IP 831 | O114 | H49 | Tunesia, 1984 | HF | LT |
| G1371 | C 3813 | O114 | H9 | Germany, 1978 | HF | F12-fim, alpha-hly |
| G1372 | C 4011 | O114 | H9 | Germany, 1983 | HF | F12-fim |
| G1350 | C 4155 | O114 | H9 | Germany, 1984 | HF | F12-fim, alpha-hly |
| G1353 | C 311-58 | O114 | H9 | Sweden, 1958 | HF | F12-fim |
| G1330 | 539/83 | O114 | H9 | Germany, 1980 | HF, D | F12-fim, alpha-hly |
| G1337 | CB 176 | O114 | H9 | New Zealand, 1986 | HF | F12-fim |
| G1305 | C 2837-67 | O114 | H9 | Germany, 1967 | HF | F12-fim |
| G1342 | C 4462 | O114 | ND | Germany, 1989 | HF | None |
| G1338 | C 4519 | O114 | ND | Germany, 1990 | HF | eae |
| G1355 | C 4557 | O114 | ND | Germany, 1990 | HF | None |
| G1333 | 3140 | O114 | NM | Germany, 1971 | HF | None |
| G1364 | 4154 | O114 | NM | Germany, 1984 | HF, D | F12-fim, alpha-hly |
| G1347 | 3008/69 | O114 | NM | Germany, 1969 | HF, D | Alpha-hly |
| G1321 | CB 9605 | O114 | NM | Germany, 2003 | HF, D | eae |
| G1308 | C 1962 | O114 | NM | Germany, 1962 | HF | None |
| G1345 | CB 9723 | O145 | H28 | Germany, 2003 | HF, D | EHEC-hly, eae |
| G1328 | CB 9805 | O145 | NM | Germany, 2003 | HF, D | eae |
| G1334 | CB 9759 | O157 | H7 | Germany, 2003 | HF, D | EHEC-hly, Stx1+2, eae |
| G1329 | CB 9777 | O157 | H7 | Germany, 2003 | HF, D | EHEC-hly, Stx1+2, eae |
| G1363 | CB 9468 | O157 | NM | Germany, 2003 | HF, D | EHEC-hly, Stx1+2, eae |
| G1324 | CB 4364 | O172 | ND | Germany, 1995 | HF | ND |
| G1323 | CB 7132 | O172 | ND | Switzerland, 1997 | Unknown | ND |
| G1322 | CB 7133 | O172 | ND | Switzerland, 1998 | Unknown | ND |
| G1309 | 2533-54 | O175 | ND | ND | ND | ND |
| G1326 | CB 9776 | O180 | H2 | Germany, 2003 | HF, D | eae |
| G1311 | E39a | O23 | H15 | — | Reference strain | ND |
| G1344 | CB 9720 | O26 | H11 | Germany, 2003 | HF, D | EHEC-hly, eae |
| G1354 | CB 9748 | O26 | H11 | Germany, 2003 | HF, D | EHEC-hly, eae |
| G1352 | CB 9752 | O26 | H11 | Germany, 2003 | HF, D | EHEC-hly, eae |
| G1332 | CB 9772 | O28 | NM | Germany, 2003 | HF, D | eae |
| G1303 | U8-41 | O48d | NM | — | Reference strain | ND |
| G1302 | U1-41 | O5d | H4 | — | Reference strain | ND |
| G1315 | P9c | O70d | H42 | — | Reference strain | ND |
| G1319 | P10a | O71d | H12 | — | Reference strain | ND |
| G1304 | H14 | O82d | NM | — | Reference strain | ND |
| G1314 | C 600 | O-rough | H48 | E. coli K-12 | Laboratory strain | None |
| G1335 | CB 9767 | O100 | H25 | Germany, 2003 | HF, D | eae |
| G1327 | CB 9801 | O103 | H2 | Germany, 2003 | HF, D | eae |
| G1306 | Bi 623-42 | O11 | H10 | — | Reference strain | ND |
| G1318 | CB 6911 | C. freundiid | ND | Germany, 1997 | HF, D | None |
—, E. coli serotype reference strain (19).
HF, human feces; D, diarrhea; ND, no data.
The virulence markers LT (heat-labile enterotoxin), ST (heat-stable enterotoxin), presence of the EAF plasmid, localized adherence to HEp-2 cells (LA), alpha-hemolysin, and presence of P (F12-related) fimbriae of E. coli O114 strains were tested by DNA hybridization (LT, ST, and EAF), colony immunoblot (alpha-hemolysin), and latex agglutination tests (P-fimbriae) as described previously (7, 8). The presence of the Shiga toxins, enterohemorrhagic E. coli (EHEC) hemolysin, the intimin gene (eae), and the bundle-forming pili (bfp) gene was investigated by PCR as described previously (6, 24). None, indicates that none of the virulence factors investigated in this study were detected.
All 41 E. coli O114 strains were specifically detected, while none of the non-O114 strains produced bands of the expected size (data not shown). The primer pairs wl-461-wl-462, wl-465-wl-466, and wl-752-wl-753 gave the expected bands for all O114 strains, while they gave no nonspecific bands on the gel for any of the pools. They may be preferable for for the identification and detection of E. coli O114 strains.
Detection of E. coli O114 strains in pork and water samples by PCR.
A 10-fold serial dilution of E. coli O114 type strain G1088 DNA ranging from 1 ng to 0.1 pg was amplified with primer pairs wl-461-wl-462, wl-465-wl-466, and wl-752-wl-753. The PCR method used was as described above. A sensitivity of 1 pg μl−1 was obtained for each of the three replicates with all the primers.
Primer pairs wl-461/wl-462, wl-465-wl-466, and wl-752-wl-753 were used to screen E. coli O114 in pork and water samples. Raw pork was purchased from three local butchers, weighed into 20-g portions, and stored at −40°C before use. Serial 10-fold dilutions (10−3 to 10−9) from the full-grown culture of E. coli O114 strain G1313 were added to each portion of pork or 20 ml of Milli-Q water. The concentration of the O114 cells in pork or water samples was determined by checking the CFU on agar plates. Samples spiked with different concentrations of E. coli O114 cells were homogenized in 200 ml of Luria broth culture. The homogenized samples were cultured at 200 rpm at 37°C for 12 h and chilled at 4°C. The culture was passed through a six-chamber filter bag, and 3 ml of filtrate was collected for use as the template DNA in the PCR as described above. With each primer pair used, as few as 0.12 CFU g−1 in pork or water could be detected (data not shown). This result indicates that the O-antigen-specific PCR assay developed in this study is useful for fast and sensitive detection of E. coli O114 in food and environmental samples such as pork and water.
Conclusion.
The O-antigen gene cluster of E. coli O114 contains genes for the synthesis of dTDP-d-Qui3NAc, genes encoding glycosyltransferases, genes responsible for the synthesis and transfer of the seryl group, and O-unit-processing genes wzx and wzy. By PCR screening all 186 E. coli and Shigella O serogroups, genes wzx, wzy, wbuM, wbuO, and wbuP were shown to be highly specific to E. coli O114. PCR assays based on the O-antigen-specific genes were developed to detect and identify E. coli O114 strains. In addition to the E. coli O114 type strain, 41 E. coli O114 strains belonging to different pathotypes from humans and animals were also identified by the PCR method. We also tested pork and water samples contaminated with E. coli O114, and as few as 0.12 CFU g−1 of O114 cells can be picked up by the PCR method. The cell lysate is used directly as the PCR template, saving the time spent on DNA purification. Although detection of O114 serogroup strains in a sample does not necessarily reflect the presence of pathogenic E. coli O114, the PCR assays established in this study can be used as a fast and sensitive alternative to O-serotyping for the detection and identification of E. coli O114 strains.
Nucleotide sequence accession number.
The DNA sequence of the E. coli O114 O-antigen gene cluster has been deposited in GenBank under accession number AY573377.
Acknowledgments
This work was supported by the Chinese National Science Fund for Distinguished Young Scholars (30125001), NSFC General Program (30270029), the 863 Program (2002AA2Z2051), and funding from the Science and Technology Committee of Tianjin City (013181711) to L.W. Gladys Krause (Berlin, Germany) was supported by funds from the European Commission project “Attaching and Effacing Escherichia coli Infections” (QLK2-CT-2000-00600).
REFERENCES
- 1.Alexander, D. C., and M. A. Valvano. 1994. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J. Bacteriol. 176:7079-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, G. E., and N. K. Verma. 2000. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 8:17-23. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3398-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastin, D. A., and P. R. Reeves. 1995. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene 164:17-23. [DOI] [PubMed] [Google Scholar]
- 5.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin, L., O. Marches, K. A. Bettelheim, K. Gleier, S. Zimmermann, H. Schmidt, and E. Oswald. 2003. HEp-2 cell adherence, actin aggregation, and intimin types of attaching and effacing Escherichia coli strains isolated from healthy infants in Germany and Australia. Infect. Immun. 71:3995-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutin, L., M. A. Montenegro, I. Orskov, F. Orskov, J. Prada, S. Zimmermann, and R. Stephan. 1989. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 27:2559-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutin, L., I. Orskov, F. Orskov, S. Zimmerman, J. Prada, H. Gelderblom, R. Stephan, and T. S. Whittam. 1990. Clonal diversity and virulence factors in strains of Escherichia coli of the classic enteropathogenic serogroup O114. J. Infect. Dis. 162:1329-1334. [DOI] [PubMed] [Google Scholar]
- 9.Charter, R. E. 1956. Escherichia coli type O114 isolated from infantile diarrhoea and calf scours. J. Pathol. Bacteriol. 72:33-38. [DOI] [PubMed] [Google Scholar]
- 10.Dmitriev, B. A., V. Lvov, N. V. Tochtamysheva, A. S. Shashkov, N. K. Kochetkov, B. Jann, and K. Jann. 1983. Cell-wall lipopolysaccharide of Escherichia coli O114:H2. Structure of the polysaccharide chain. Eur. J. Biochem. 134:517-521. [DOI] [PubMed] [Google Scholar]
- 11.Eisenberg, D., E. Schwarz, M. Komaromy, and R. Wall. 1984. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179:125-142. [DOI] [PubMed] [Google Scholar]
- 12.Ewing, W. H. 1986. Edwards and Ewing's identification of the Enterobacteriaceae, 4th ed. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 13.Henikoff, S., J. G. Henikoff, W. J. Alford, and S. Pietrokovski. 1995. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene 163:GC17-GC26. [DOI] [PubMed] [Google Scholar]
- 14.Levine, M. M. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic and enteroadherent. J. Infect. Dis. 155:377-389. [DOI] [PubMed] [Google Scholar]
- 15.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orskov, F., and I. Orskov. 1984. Serotyping of Escherichia coli. Academic Press, London, England.
- 17.Orskov, F., T. S. Whittam, A. Cravioto, and I. Orskov. 1990. Clonal relationships among classic enteropathogenic Escherichia coli (EPEC) belong to different O groups. J. Infect. Dis. 162:76-81. [DOI] [PubMed] [Google Scholar]
- 18.Orskov, I., and F. Orskov. 1966. Episome-carried surface antigen K88 of Escherichia coli. I. Transmission of the determinant of the K88 antigen and influence on the transfer of chromosomal markers. J. Bacteriol. 91:69-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orskov, I. F., F. B. Orskov, B. Jann, and K. Jann. 1977. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol. Rev. 41:667-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peruski, L. F. J., B. A. Kay, R. A. El-Yazeed, S. H. El-Etr, A. Cravioto, T. F. Wierzba, M. Rao, N. El-Ghorab, H. Shaheen, S. B. Khalil, K. Kamal, M. O. Wasfy, A. M. Svennerholm, J. D. Clemens, and S. J. Savarino. 1999. Phenotypic diversity of enterotoxigenic Escherichia coli strains from a community-based study of pediatric diarrhea in periurban Egypt. J. Clin. Microbiol. 37:2974-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfoestl, A., A. Hofinger, P. Kosma, and P. Messner. 2003. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-d-galactose in Aneurinibacillus thermoaerophilus L420-91T. J. Biol. Chem. 278:26410-26417. [DOI] [PubMed] [Google Scholar]
- 22.Reeves, P. R., and L. Wang. 2002. Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 264:109-135. [PubMed] [Google Scholar]
- 23.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualisation and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt, H., L. Beutin, and H. Karch. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scotland, S. M., N. P. Day, A. Cravioto, L. V. Thomas, and B. Rowe. 1981. Production of heat-labile or heat-stable enterotoxins by strains of Escherichia coli belonging to serogroups O44, O114, and O128. Infect. Immun. 31:500-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 27.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W—impoving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trabulsi, L. R., R. Keller, and T. A. Tardelli Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O-antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiggins, C. A., and S. Munro. 1998. Activity of the yeast MNN1 alpha-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc. Natl. Acad. Sci. USA 95:7945-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willshaw, G. A., S. M. Scotland, H. R. Smith, and B. Rowe. 1992. Properties of Verocytotoxin-producing Escherichia coli of human origin of O groups other than O157. J. Infect. Dis. 166:797-802. [DOI] [PubMed] [Google Scholar]
- 34.Wolf, M. K. 1997. Occurrence, distribution, and association of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin. Microbiol. Rev. 10:569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]