Abstract
Cases of imported hepatitis E in industrialized countries infected with a genotype 1 hepatitis E virus (HEV) have been identified. We report a 56-year-old Japanese man who acquired infection with a genotype 4 HEV with 98.8% identity to a Vietnamese isolate after ingestion of uncooked shellfish while traveling in Vietnam.
CASE REPORT
A 56-year-old Japanese man visited an Internal Medicine Clinic in Tochigi, Japan, with complaints of dark urine and general malaise on 19 June 2003, 7 days after the onset of the illness. On that day, he was transferred to our hospital and was hospitalized with a clinical diagnosis of acute hepatitis. Physical examination on admission was essentially normal, except for jaundice. Laboratory data at admission showed an elevated total bilirubin level of 11.7 mg/dl, an aspartate aminotransferase level of 666 IU/liter, an alanine aminotransferase level of 972 IU/liter, an alkaline phosphatase level of 463 IU/liter, and a γ-glutamyl transpeptidase level of 258 IU/liter. The serum sample obtained at admission was negative for markers of hepatitis A, B, and C viruses. It was then tested for the immunoglobulin M (IgM) class of antibodies to hepatitis E virus (HEV) (anti-HEV IgM) by using an in-house enzyme immunoassay with purified recombinant open reading frame 2 (ORF2) protein that had been expressed in the pupae of silkworm (13) as the antigen probe, and it was also tested for HEV RNA by reverse transcription-PCR (RT-PCR) by a method described previously with primers targeting the ORF2 region (13). Based on positivity for anti-HEV IgM and HEV RNA, the patient was diagnosed as having sporadic acute hepatitis E. HEV RNA was detectable until the 12th hospital day. The maximum severity of the illness occurred at admission. After admission, he recovered rapidly and was discharged on the 13th hospital day.
The patient had no history of blood transfusion or liver disease and had no contact with pet animals or farm animals. Of note, he had traveled to Vietnam for sightseeing together with his wife and daughter from 21 April to 1 May 2003, and during their trip in Vietnam they consumed the same local Vietnamese cuisine or Western foods for every meal at restaurants. However, for lunch on 27 April 2003, the patient ingested only a raw shellfish (bivalve), i.e., a type of clam, which was served on the boat during a boat trip in Halong Bay, Vietnam. He developed hepatitis E 42 days after returning to Japan and 46 days after ingestion of the uncooked shellfish. His wife and daughter were negative for anti-HEV IgG and IgM and HEV RNA in the serum samples obtained on the 6th and 10th days of the patient's admission, respectively.
The presence of HEV RNA was confirmed by nested reverse transcription-PCR targeting a part of ORF1 (13). The amplified product of the ORF1 region and the amplified product of the ORF2 region from the serum sample that had been obtained at admission were sequenced directly on both strands. The HEV isolate (HE-JVN1) recovered from the infected patient was close to known human and swine genotype 4 HEV isolates, with 82.2 to 98.8% identity in a 326- or 412-nucleotide (nt) sequence of the ORF1 region (Table 1), and was most closely related to the V091 isolate (AB075967) of genotype 4 which had been isolated from a Vietnamese patient who contracted sporadic acute hepatitis E in Hanoi, Vietnam, in 2001, in a 326-nt sequence of the ORF1 region (7). In comparison with genotype 4 HEV strains reported from countries other than Vietnam, the HE-JVN1 isolate was only 87.4 to 89.8% similar to Chinese human isolates and 87.1 to 89.3% similar to Japanese human and swine isolates in the 412-nt ORF1 sequence. Upon comparison of a 241- to 412-nt sequence within ORF2, the HE-JVN1 isolate was closest to a Chinese swine HEV strain (SJ14 [AJ428856]), with 96.3% identity (20), and was only 83.4 to 94.0% similar to the remaining 78 human and swine isolates of Chinese, Indian, Indonesian, Japanese, or Taiwanese origin: no common ORF2 sequence of Vietnamese HEV isolates was available.
TABLE 1.
Comparison of the HEV isolate from the patient in the present study (HE-JVN1) with 104 human and swine HEV isolates of genotype 4 whose common 326- or 412-nt sequence in ORF1 or common 241- to 412-nt sequence in ORF2 is known
| Country | Host | ORF1
|
ORF2
|
||||
|---|---|---|---|---|---|---|---|
| No. of isolates compared | nt length compared | Identity (%) | No. of isolates compared | nt length compared | Identity (%) | ||
| China | Human | 2 | 412 | 87.4-89.8 | 22 | 301-412 | 83.4-94.0 |
| Swine | 0 | -a | - | 5 | 300 | 85.3-96.3 | |
| India | Swine | 0 | - | - | 12 | 241-263 | 85.4-88.7 |
| Indonesia | Swine | 0 | - | - | 1 | 412 | 90.8 |
| Japan | Human | 15 | 412 | 87.1-89.3 | 18 | 412 | 86.9-88.3 |
| Swine | 2 | 412 | 88.1 | 10 | 412 | 87.1-87.9 | |
| Taiwan | Human | 0 | - | - | 8 | 304-346 | 85.5-89.9 |
| Swine | 0 | - | - | 3 | 304-346 | 87.2-90.5 | |
| Vietnam | Human | 6 | 326 | 82.2-98.8 | 0 | - | - |
-, Not applicable.
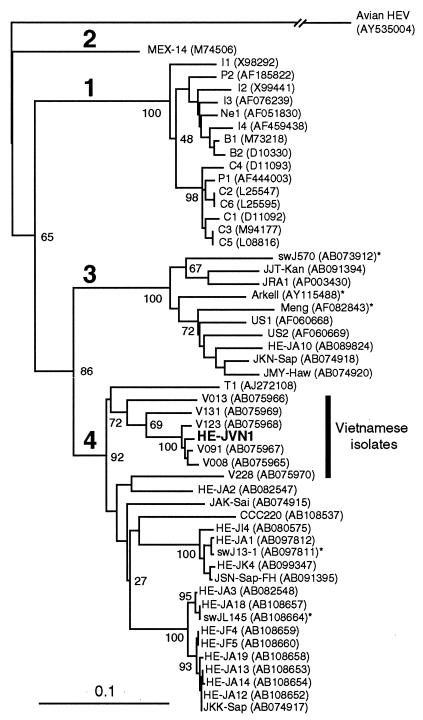
The phylogenetic tree constructed by the neighbor-joining method (15) based on the partial ORF1 sequence of 326 nt confirmed that the HE-JVN1 isolate belonged to genotype 4, and it segregated into a cluster consisting of six HEV strains, including V091, that had been isolated from six Vietnamese patients who had developed sporadic acute hepatitis E in Hanoi, Vietnam (Fig. 1).
FIG.1.
Phylogenetic tree constructed by the neighbor-joining method based on the partial nucleotide sequence (326 nt; nt 123 to 448 of the HE-JA10 genome, accession no. AB089824) of the ORF1 region of 52 human and swine HEV isolates using an avian HEV (AY535004) as an outgroup. In addition to 26 reported human and swine HEV isolates of genotypes 1 to 3 whose entire or nearly entire sequence is known, 26 reported isolates of genotype 4 whose common 326-nt sequence is available, as well as the HE-JVN1 isolate obtained in the present study, were included for comparison, with accession numbers in parentheses. The HE-JVN1 isolate is indicated by boldface type for visual clarity. The HEV strains of Vietnamese origin are shown by a vertical bar. The previously reported HEV sequences of genotype 1 are indicated with abbreviations in accordance with the recent review article by Schlauder and Mushahwar (16): B1 and B2 from Burma; C1, C2, C3, C4, C5, and C6 from China; I1, I2, I3, and I4 from India; Ne1 from Nepal; and P1 and P2 from Pakistan. Asterisks denote swine HEV strains. Bootstrap values are indicated for the major nodes as a percentage of the data obtained from 1,000 resamplings (4).
HEV infections are endemic and frequently epidemic in many developing countries in Asia, Africa, and Central America, where sanitation is suboptimal (14). Recent studies have documented that HEV-associated infection also occurs among individuals in industrialized countries with no history of travel to areas where HEV is endemic (6, 14, 16, 17). Although only one serotype has been recognized, extensive genomic diversity has been noted among HEV isolates, and HEV sequences have tentatively been classified into four genotypes (genotypes 1 to 4) (16). The majority of HEV infections are caused by genotype 1 in several developing countries in Asia and Africa, and one epidemic in Mexico caused by genotype 2 has been documented. Only isolated cases of infection with HEV of genotype 3 or 4 have been described in industrialized nations (16).
We encountered a patient with imported HEV who had traveled to Vietnam and ingested an uncooked shellfish and who was infected with an HEV strain of genotype 4 presumably indigenous to Vietnam, although many patients with imported HEV in industrialized countries are infected with a genotype 1 HEV which is prevalent in countries where the virus is hyperendemic (3, 8). In Japan, polyphyletic strains of HEV are circulating and genotype 4 HEV strains have been recovered from patients with sporadic acute or fulminant hepatitis E who have no history of traveling abroad (13) and from farm pigs (18). Of interest, pairwise comparison and phylogenetic analysis of HEV sequences indicated that the HE-JVN1 strain isolated from the patient in the present study was most closely related to the HEV isolates of Vietnamese origin and not to those of Japanese origin, suggesting that he was infected with HEV while traveling in Vietnam. As reviewed by Smith (17), it is likely that foods can act as vehicles for transmission of HEV. Increasing evidence has indicated that hepatitis E is a zoonosis (6, 11, 12, 17), and that the zoonotic food-borne mode of transmission of HEV to humans, through ingestion of uncooked or undercooked liver from pigs (21), meat from a wild deer (19), or liver from a wild boar (9), seems to play an important role. Furthermore, the occurrence of acute hepatitis E in individuals after consumption of raw or uncooked shellfish has been reported (1, 10). In this regard, HEV is similar to hepatitis A virus, which is an important pathogen responsible for many food-borne outbreaks: the foods implicated in outbreaks of hepatitis A include shellfish (2, 5). The patient's spouse and daughter, who had traveled with the patient to Vietnam, did not eat raw shellfish and did not contract hepatitis. Although we could not prove that the shellfish consumed by our patient was the source of HEV infection, it appears likely that this was the case.
Our study indicates that even an imported case of hepatitis E in Japan was infected with HEV of a genotype other than genotype 1, which is prevalent in areas where the virus is endemic; that consumption of uncooked shellfish contaminated with HEV may cause cryptic HEV infection; and that phylogenetic analysis of HEV strains may be useful for clinical surveys and for tracing infectious sources.
Nucleotide sequence accession numbers. Nucleotide sequence data have been deposited in the GenBank/DDBJ/EMBL databases under accession nos. AB168095 (ORF1) and AB168096 (ORF2).
REFERENCES
- 1.Cacopardo, B., R. Russo, W. Preiser, F. Benanti, G. Brancati, and A. Nunnari. 1997. Acute hepatitis E in Catania (eastern Sicily) 1980-1994: the role of hepatitis E virus. Infection 25:313-316. [DOI] [PubMed] [Google Scholar]
- 2.Desenclos, J. C. A., K. C. Klontz, M. H. Wilder, O. V. Nainan, H. S. Margolis, and R. A. Gunn. 1991. A multistate outbreak of hepatitis A caused by the consumption of raw oysters. Am. J. Public Health 81:1268-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donati, M. C., E. A. Fagan, and T. J. Harrison. 1997. Sequence analysis of full length HEV clones derived directly from human liver in fulminant hepatitis E, p. 313-316. In M. Rizzetto, R. H. Purcell, J. L. Gerin, and G. Verme (ed.), Viral hepatitis and liver disease. Edizioni Minerva Medica, Turin, Italy.
- 4.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 5.Halliday, M. L., L. Y. Kang, T. K. Zhou, M. D. Hu, Q. C. Pan, T. Y. Fu, Y. S. Huang, and S. L. Hu. 1991. An epidemic of hepatitis A attributable to the ingestion of raw clams in Shanghai, China. J. Infect. Dis. 164:852-859. [DOI] [PubMed] [Google Scholar]
- 6.Harrison, T. J. 1999. Hepatitis E virus—an update. Liver 19:171-176. [DOI] [PubMed] [Google Scholar]
- 7.Hijikata, M., S. Hayashi, N. T. Trinh, L. D. Ha, H. Ohara, Y. K. Shimizu, N. Keicho, and H. Yoshikura. 2002. Genotyping of hepatitis E virus from Vietnam. Intervirology 45:101-104. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa, K., K. Matsui, T. Madarame, S. Sato, K. Oikawa, and T. Uchida. 1995. Hepatitis E probably contracted via Chinese herbal medicine demonstrated by nucleotide sequencing. J. Gastroenterol. 30:534-538. [DOI] [PubMed] [Google Scholar]
- 9.Matsuda, H., K. Okada, K. Takahashi, and S. Mishiro. 2003. Severe hepatitis E virus infection after ingestion of uncooked liver from a wild boar. J. Infect. Dis. 188:944. [DOI] [PubMed] [Google Scholar]
- 10.Mechnik, L., N. Bergman, M. Attali, M. Beergabel, B. Mosenkis, N. Sokolowski, and S. Malnick. 2001. Acute hepatitis E virus infection presenting as a prolonged cholestatic jaundice. J. Clin. Gastroenterol. 33:421-422. [DOI] [PubMed] [Google Scholar]
- 11.Meng, X.-J., R. H. Purcell, P. G. Haubur, J. R. Lehman, D. M. Webb, T. S. Tsareva, J. S. Haynes, B. J. Thacker, and S. U. Emerson. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 94:9860-9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng, X.-J., P. G. Halbur, M. S. Shapiro, S. Govindarajan, J. D. Bruna, I. K. Mushahwar, R. H. Purcell, and S. U. Emerson. 1998. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 72:9714-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuo, H., K. Suzuki, Y. Takikawa, Y. Sugai, H. Tokita, Y. Akahane, K. Itoh, Y. Gotanda, M. Takahashi, T. Nishizawa, and H. Okamoto. 2002. Polyphyletic strains of hepatitis E virus are responsible for sporadic cases of acute hepatitis in Japan. J. Clin. Microbiol. 40:3209-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell, R. H., and S. U. Emerson. 2001. Hepatitis E virus, p. 3051-3061. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 15.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 16.Schlauder, G. G., and I. K. Mushahwar. 2001. Genetic heterogeneity of hepatitis E virus. J. Med. Virol. 65:282-292. [DOI] [PubMed] [Google Scholar]
- 17.Smith, J. L. 2001. A review of hepatitis E virus. J. Food Prot. 64:572-586. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi, M., T. Nishizawa, H. Miyajima, Y. Gotanda, T. Iita, F. Tsuda, and H. Okamoto. 2003. Swine hepatitis E virus strains in Japan form four phylogenetic clusters comparable with those of Japanese isolates of human hepatitis E virus. J. Gen. Virol. 84:851-862. [DOI] [PubMed] [Google Scholar]
- 19.Tei, S., N. Kitajima, K. Takahashi, and S. Mishiro. 2003. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362:371-373. [DOI] [PubMed] [Google Scholar]
- 20.Wang, Y., H. Zhang, N. Xia, G. Peng, H. Lan, H. Zhuang, Y. Zhu, S. Li, K. Tian, W. Gu, J. Lin, X. Wu, H. Li, and T. J. Harrison. 2002. Prevalence, isolation, and partial sequence analysis of hepatitis E virus from domestic animals in China. J. Med. Virol. 67:516-521. [DOI] [PubMed] [Google Scholar]
- 21.Yazaki, Y., H. Mizuo, M. Takahashi, T. Nishizawa, N. Sasaki, Y. Gotanda, and H. Okamoto. 2003. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J. Gen. Virol. 84:2351-2357. [DOI] [PubMed] [Google Scholar]