Abstract
Protocols were designed for quantification and detection of hepatitis C virus (HCV) RNA by the use of an analyte-specific reagent (ASR) (Roche COBAS TaqMan48 [CTM48] HCV) after manual and automated RNA extraction. The purposes were to determine (i) assay performance characteristics using manual and automated RNA extraction methods, (ii) whether measurable range and limit of detection (LOD) of the ASR assay were influenced by genotype, and (iii) correlation of quantification by CTM48 HCV ASR and COBAS Monitor HCV v. 2.0. For HCV genotype 1 (Gt1), the lower limits of quantification after manual extraction were slightly lower than those for automated extraction (1.0 versus 1.5 log10 IU/ml). Results were linear up to the highest concentration tested after extraction by both methods (manual, 6.1 log10; automated, 6.4 log10). Similar results were obtained for Gt2 (1.8 to 6.8 log10 IU/ml) and Gt3 (1.6 to 6.8 log10 IU/ml) after automated extraction. The LOD of Gt1 virus was 10 IU/ml after manual extraction and between 25 and 37.5 IU/ml after automated extraction. Results with Gt2 and Gt3 viruses were similar after automated extraction (Gt2, between 25 and 50 IU/ml; Gt3, 25 IU/ml). Variability (intrarun and interrun, at concentrations throughout the range of quantification) was ≤13% for both extraction methods. Clinical specimens tested by Monitor were quantified using the CTM48 HCV ASR assay. Characteristics of the regression line included a slope of 0.98 and y intercept of −0.23. Quantification by the two methods was correlated (r = 0.97). CTM48 HCV ASR assay values were on average twofold lower than those for Monitor HCV v. 2.0. These data suggest that our assay combines the characteristics of qualitative and quantitative PCR platforms into a single test.
Hepatitis C Virus (HCV) was identified by molecular methods in 1989, and culture assays for detection are still not available. Diagnosis of chronic HCV infection and monitoring for efficacy of therapy are performed with molecular assays for detection and quantification of HCV RNA. Qualitative and quantitative assays have been developed, and each test format has a different utility. The low limit of detection (LOD) of qualitative assays makes these tests suitable for diagnosis of chronic or acute infection and assessment of virus clearance after treatment. Viral load determination with quantitative assays is used prior to treatment to establish a baseline level of viremia and can be used (in combination with genotype) to predict likelihood of response (2, 3, 5). During treatment, viral load testing is useful in predicting the outcome of continued therapy (2).
Most laboratorians have become familiar with qualitative and quantitative molecular assays for HCV. However, health care practitioners often request these tests inappropriately and fail to interpret results correctly, in part due to an incomplete understanding of the performance characteristics and the utility of different assay formats. The potential for mismanagement of patients exists in this setting.
In response, many laboratories have implemented diagnostic and therapeutic algorithms. In a typical diagnostic algorithm, a qualitative assay is performed to ascertain a diagnosis of chronic HCV infection. If the result is positive, the specimen is then tested by a quantitative assay to determine baseline viral load. In therapeutic algorithms, a quantitative test is performed to assess viral load during treatment. If no HCV RNA is detected, a qualitative assay is performed to assess whether viral clearance has occurred.
Algorithms have been effective for clinicians, but they are cumbersome for laboratories that must also offer individual tests. Laboratories that offer algorithms must implement a system for storage and tracking of specimens and results to ensure that the correct tests are performed in a timely manner. However, the potential for errors exists, even with an optimally designed system.
The development of a single test platform that combines the performance characteristics of qualitative and quantitative assays would greatly simplify HCV diagnostics for health care practitioners and clinical laboratories. The advent of quantitative PCRs in which amplification and detection occurs simultaneously (real-time PCR) offers a potential solution. The broad measurable range and increased sensitivity of real-time PCR make it an ideal platform for the consolidation of HCV molecular assay platforms.
We have investigated the performance of the Roche COBAS TaqMan48 (CTM48) HCV analyte-specific reagent (ASR) for the detection and quantification of HCV RNA in plasma. These reagents utilize TaqMan chemistry to amplify and detect sequences in the 5′ untranslated region (UTR) of HCV. The aim of this study was (i) to determine the measurable range, LOD, and precision of quantification of HCV Gt1 after extraction by manual column and automated instrumentation, (ii) to determine whether genotype bias occurred by comparing the LOD and measurable range of quantification of Gt1, -2, and -3 viruses after RNA extraction by automated instrumentation, and (iii) to determine the correlation of quantification by Monitor HCV v. 2.0 and our CTM48 HCV ASR assay using patient specimens.
MATERIALS AND METHODS
Viruses and clinical specimens.
Viruses for instrument calibration and analytical performance studies (measurable range, LOD, and precision) were obtained from AcroMetrix (Benicia, Calif.) and Teragenix (Ft. Lauderdale, Fla.), respectively. Six virus preparations of a Gt1 virus were used to calibrate the assay (6.7, 5.7, 4.7, 3.7, 2.7, and 1.7 log10 IU/ml). The calibration panel members were manufactured by serially diluting (10-fold) the panel member with the highest concentration.
Accuracy of HCV quantification can vary widely due to a number of reasons, including variability in testing proficiency by individual laboratories and the lack of a “gold standard” method for quantification. Therefore, viruses were requantified using the method employed by the manufacturer to verify the stated concentrations (Monitor HCV v. 2.0 for quantifiable calibrators and Teragenix Gt3 virus; COBAS Taqman HCV ASR for Teragenix Gt1 and Gt2 viruses). New values were assigned if the concentration observed after requantification differed from the expected value by a factor of 1.5-fold or greater. Viruses were diluted in defibrinated human plasma (Boston Biomedica, Inc., West Bridgewater, Mass.) for measurable range and LOD experiments. For LOD determinations, viruses were diluted to 100 IU/ml (in 10-fold increments) and then further diluted to 50, 37.5 (for Gt1 automated extraction only), 25, 10, and 5 IU/ml. A sufficient volume of each dilution was prepared to allow extraction of 20 replicates per extraction method.
Plasma samples that had been previously tested by Monitor HCV v. 2.0 were extracted and tested with the CTM48 HCV ASR assay. Specimens with concentrations greater than Monitor's upper limit of quantification (5.7 log10 IU/ml) were diluted 100-fold and retested by Monitor HCV v. 2.0. Samples were not tested simultaneously on both platforms but were frozen at −70°C until CTM48 HCV ASR testing. These studies were performed following guidelines for human subject experimentation. Approval was received from the Johns Hopkins Medicine Institutional Review Board to conduct these experiments.
RNA extraction methods.
HCV RNA was extracted by an automated method (MagAttract viral RNA M48 reagents on a BioRobot M48 instrument; QIAGEN, Valencia, Calif.) and a manual method (QIAamp MinElute virus vacuum kit; QIAGEN). Plasma input and elution volumes for the automated method were 300 and 65 μl, respectively, as per the manufacturer's recommendations.
For manual extraction, the manufacturer's protocol was followed with two exceptions: the protease was rehydrated in distilled water instead of the supplied protease resuspension buffer, and carrier RNA was reconstituted in AVE buffer to which quantitative standard reagent (QS) was added. Plasma input and elution volumes were 500 and 60 μl, respectively. For each extraction method, QS was added to each sample such that the final concentration after elution was approximately 1,100 copies in 50 μl of eluate.
QS contains a synthetic virus (“armored RNA”) comprised of MS2 bacteriophage bearing RNA sequences complementary to the primers used to amplify HCV and an intervening unique sequence that is used to detect QS during the real-time PCR. QS is used to correct for the presence of reaction inhibitors and other variables that can affect the efficiency of real-time reverse transcription-PCRs. QS was added to the lysis buffer to prevent degradation of the armored RNA.
HCV RNA amplification and detection.
The Monitor HCV v. 2.0 test was performed according to the manufacturer's instructions. For the CTM48 HCV assay, working Master Mix was prepared by combining manganese (170 μl) and ASR Master Mix reagents (1.4 ml). Master Mix contains deoxynucleoside triphosphates, oligonucleotide primers (upstream and downstream) containing sequences from the HCV 5′ UTR, fluorescent-labeled probes (complementary to 5′ UTR and QS-specific sequences), Z05 polymerase (thermostable polymerase with reverse transcriptase activity), and uracil-N-glycosylase (to prevent amplification of contaminants from previous reactions). The working Master Mix (50 μl) was added to each K-tube in the K-Carrier, followed by 50 μl of test sample. The K-Carrier was placed in the CTM48 instrument for reverse transcription, amplification, and detection. The HCV PCR profile used for amplification is shown in Table 1. Results from the six virus preparations used for calibration (1.7 to 6.7 log10 IU/ml in 10-fold increments, four to six replicates per concentration) were imported into ASR external calibration software v. 2.1 to derive the three coefficients that defined the calibration curve for each ASR lot. Data from a minimum of three replicates at each concentration were used to derive the three calibration coefficients. Calibrator data that caused the standard deviation of the QS crossing threshold to exceed 0.5 were excluded from the coefficient calculation. Quantification was performed in subsequent runs by loading the calibration coefficients into the CTM48 instrument software (AmpliLink v. 3.0.1). The calibration coefficients are specific to each ASR lot, therefore the calibration protocol was repeated and new coefficients were calculated for each new lot of ASR reagents.
TABLE 1.
Instrument settings for CTM48 ASR assaya
| Step | Ramp slope | Temperature (°C) | Cycle time (s) | Cycle delay (s) |
|---|---|---|---|---|
| Cover heating | 3 | 100.0 | — | — |
| Precycle | 12 | 50.0 | 300 | 0 |
| Precycle | 12 | 59.0 | 1,800 | 0 |
| Sequence start | — | — | — | — |
| Denaturation | 12 | 95.0 | 15 | 0 |
| Annealing | 12 | 58.0 | 25 | 25 |
| Sequence end | 2 | — | — | — |
| Sequence start | — | — | — | — |
| Denaturation | 12 | 91.0 | 15 | 0 |
| Annealing | 12 | 58.0 | 25 | 25 |
| Sequence end | 60 | — | — | — |
| Postcycle | 12 | 40.0 | 120 | 0 |
—, program step with no required parameter.
Design of analytical performance and correlation experiments. (i) Measurable range.
The measurable range of quantification using the CTM48 ASR reagents was determined by testing undiluted viruses (representing maximum concentrations) and serial dilutions of virus stocks. In experiments using automated RNA extraction, the starting concentrations of viruses were 6.7 (Gt1), 6.8 (Gt2), and 6.2 (Gt3) log10 IU/ml. The number of replicates tested at each concentration was 10 for Gt1 and 5 for Gt2 and -3. In manual extraction experiments, the starting concentration of virus (Gt1) was 6.1 log10 IU/ml, and 10 replicates were tested at each dilution. The criteria of linearity and acceptable variability (standard deviation, ≤0.4) were used to define the upper and lower limits of quantification.
(ii) LOD.
To determine the LOD, viruses were serially diluted to concentrations ranging from 50 to 5 IU/ml. Twenty replicates were tested at each concentration, except for Gt1 at 5 IU/ml extracted by the automated method (n = 10) and Gt3 at 50 IU/ml (n = 5). The LOD was defined as the concentration at which 95% of replicates were detected (7).
(iii) Precision.
Precision of the CTM48 ASR assay after automated and manual extraction was assessed using a Gt1 virus at 2.0, 4.0, and 6.0 log10 IU/ml. For intra-assay precision, 20 replicates were tested at each concentration. To calculate interassay precision, 10 additional replicates were tested on two additional runs performed on two separate days, and the data were combined with the initial run of 20 replicates.
(iv) Correlation between quantification using COBAS Monitor v. 2.0 and CTM48 HCV ASR.
To assess whether quantification by the real-time PCR ASR and quantitative conventional PCR methods correlated, 106 specimens that had been quantified by COBAS Monitor HCV v. 2.0 were quantified using the CTM48 HCV ASR assay. Specimens throughout the range of quantification were chosen for analysis. Data were logarithmically transformed prior to analysis. The distribution of genotypes was as follows: Gt1, n = 76; Gt2, n = 8; Gt3, n = 6; Gt4, n = 1. Genotypes were determined by direct sequencing of core-E1 sequences (1) or Versant HCV genotyping assay (LiPA; Bayer, Tarrytown, N.Y.). Genotype could not be determined for 15 specimens.
Statistical Methods.
Regression lines and their characteristics were determined using Microsoft Excel (Microsoft Office 2001; Microsoft Corp., Redmond, Wash.).
RESULTS
Measurable range using CTM48 HCV ASR.
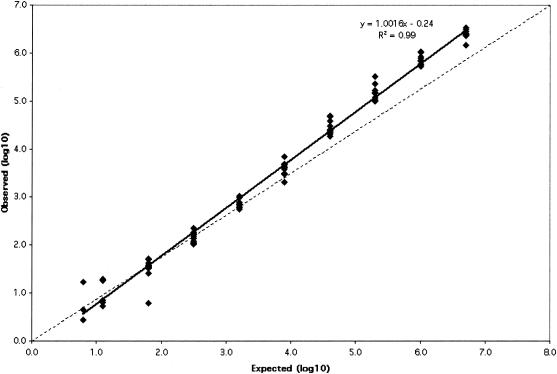
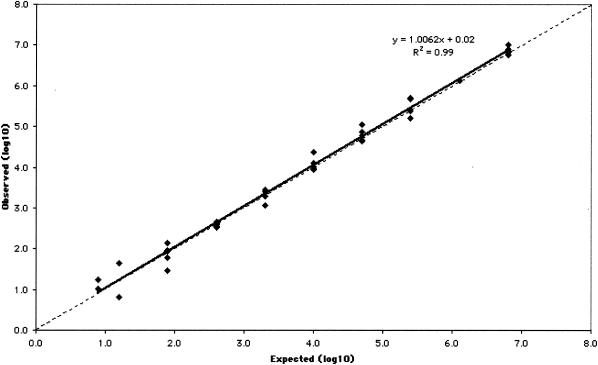
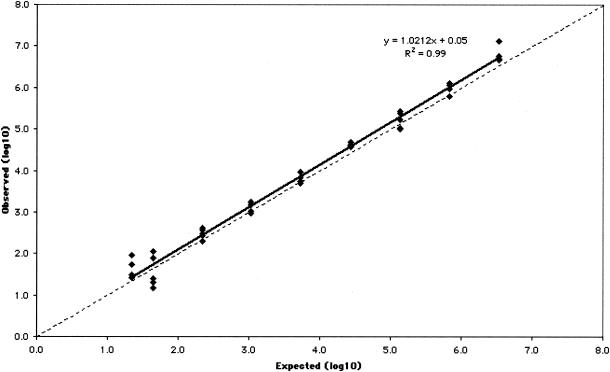
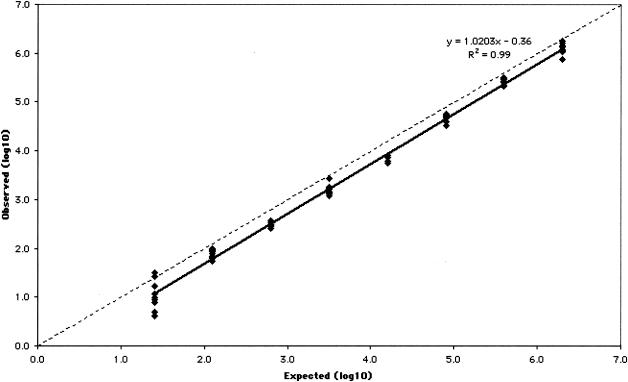
The measurable range of the CTM48 HCV ASR assay after automated extraction was determined for the three HCV genotypes (Gt1, -2, and -3) that are most prevalent in our patient population. Measurable ranges, as log10 (IU/ml), were 1.5 to ≥6.4 for Gt1 (Fig. 1), 1.8 to ≥6.8 for Gt2 (Fig. 2), and 1.6 to ≥6.8 for Gt3 (Fig. 3). Some replicates were not detected at the lowest concentrations of Gt1 (0.8 and 1.1 log10 IU/ml), Gt2 (0.9 and 1.2 log10 IU/ml), and Gt3 (1.3 log10 IU/ml). As a result, these concentrations failed to meet the criterion of acceptable variability used to define the limits of quantification. The lower limit of quantification after manual extraction of Gt1 (1.0 log10 IU/ml) was less than that observed after automated extraction (1.5 log10 IU/ml), and quantification was linear up to the highest concentration of virus tested (6.1 log10 IU/ml; Fig. 4).
FIG. 1.
Measurable range (IU/ml) of Gt1 quantification after automated RNA extraction. The dashed line indicates theoretical trend line of complete agreement between expected and observed measurements.
FIG. 2.
Measurable range (IU/ml) of Gt2 quantification after automated RNA extraction. The dashed line indicates theoretical trend line of complete agreement between expected and observed measurements.
FIG. 3.
Measurable range (IU/ml) of Gt3 quantification after automated RNA extraction. The dashed line indicates theoretical trend line of complete agreement between expected and observed measurements.
FIG. 4.
Measurable range (IU/ml) of Gt1 quantification after manual extraction. The dashed line indicates theoretical trend line of complete agreement between expected and observed measurements.
LODs for HCV Gt1, -2, and -3 using CTM48 ASR.
For all three genotypes, 100% of replicates were detected at 50 IU/ml (Table 2). For Gt1, after automated extraction all replicates were detected at 37.5 IU/ml and 90% of replicates were detected at 25 IU/ml, suggesting that the LOD was between 25 and 37.5 IU/ml. For Gt2, 80% of replicates were detected at 25 IU/ml and 100% of replicates were detected at 50 IU/ml, suggesting that the LOD was between 25 and 50 IU/ml (Table 2). For Gt3, 95% of replicates were detected at 25 IU/ml. After manual extraction, the LOD of Gt1 was 10 IU/ml, compared to 25 to 37.5 IU/ml for the automated platform (Table 2).
TABLE 2.
LODs of CTM48 HCV ASR
| Concentration (IU/ml) | % Replicates detected (no. positive/total no. of replicates) with:
|
|||
|---|---|---|---|---|
| Gt1
|
Gt2b | Gt3b | ||
| Automated extraction | Manual extraction | |||
| 50 | 100 (20/20) | 100 (20/20) | 100 (20/20) | 100 (5/5) |
| 37.5 | 100 (20/20) | ND | ND | ND |
| 25 | 90 (18/20) | 100 (20/20) | 80 (16/20) | 95 (19/20) |
| 12 | NDa | ND | ND | 65 (13/20) |
| 10 | 60 (12/20) | 95 (19/20) | ND | ND |
| 5 | 40 (4/10) | 80 (16/20) | ND | ND |
ND, not done.
Only automated extraction values given for Gt2 and Gt3.
Precision of the CTM48 HCV ASR assay.
Precision with the automated and manual extraction methods was determined (Table 3). The data demonstrate that interassay and intra-assay precision after extraction by both methods were similar, although at 2.0 log10 IU/ml, interrun variability was greater than intrarun variability after extraction by automated instrument (7 versus 13%; Table 3). Variability was greatest at 2.0 log10 IU/ml after extraction by both methods.
TABLE 3.
Precision of CTM48 HCV ASR
| Variability (log10) | Mean (log10 IU/ml)a | SDa | CV (%)ab |
|---|---|---|---|
| Intrarun | |||
| 2.0 | 1.8/1.8 | 0.1/0.1 | 7.0/8.1 |
| 4.0 | 3.7/3.8 | 0.1/0.1 | 3.0/2.2 |
| 6.0 | 5.8/6.0 | 0.1/0.1 | 2.0/1.5 |
| Interrun | |||
| 2.0 | 1.8/1.8 | 0.2/0.1 | 13.0/8.1 |
| 4.0 | 3.7/3.9 | 0.2/0.1 | 4.0/2.1 |
| 6.0 | 5.8/6.0 | 0.1/0.1 | 1.0/1.5 |
Values given for automated extraction/manual extraction.
CV, coefficient of variation.
Correlation between quantification using COBAS Monitor v. 2.0 and CTM48 HCV ASR.
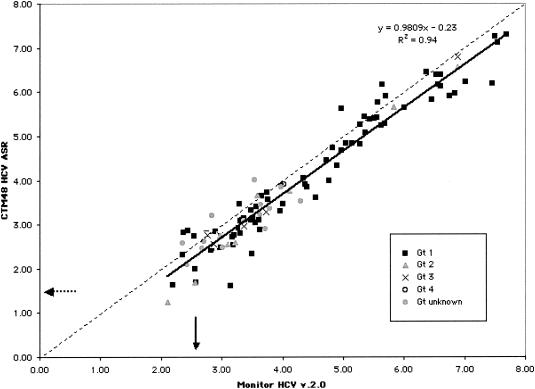
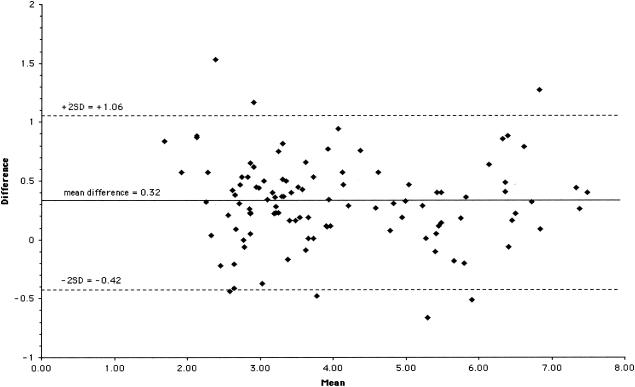
The data obtained by the two assays were linearly related (slope = 0.98) and correlated (r = 0.97) but not identical (Fig. 5). Values obtained with the CTM48 HCV ASR assay were usually lower than those for Monitor HCV v. 2.0 (slope approaching 1.0, y intercept = −0.23). Most specimens contained Gt1 HCV. Twelve of the 15 specimens containing viruses other than Gt1 had viral loads <5.0 log10 IU/ml by Monitor HCV v. 2.0. An analysis of the difference between Monitor HCV v. 2.0 and CTM48 HCV ASR assay results (Monitor HCV v. 2.0 log10 IU/ml minus CTM49 HCV ASR log10 IU/ml) demonstrated that almost all (100 of 106) results fell within two standard deviations from the mean difference (Fig. 6). No bias in relation to concentration was observed.
FIG. 5.
Correlation between quantification with CTM48 HCV ASR assay and Monitor HCV v. 2.0. Solid arrow, lower limit of quantification for Monitor HCV v. 2.0; dashed arrow, lower limit of quantification for CTM48 HCV ASR. The dashed line indicates the theoretical trend line of complete agreement in quantification between the two assays.
FIG. 6.
Difference in quantification between CTM48 HCV ASR and Monitor HCV v. 2.0 (CTM48 HCV ASR log10 IU/ml minus Monitor HCV v. 2.0 log10 IU/ml) throughout the range of quantification. Mean, average of CTM48 HCV ASR and Monitor HCV v. 2.0 log10 values; SD, standard deviation (0.37).
DISCUSSION
One of the major differences between Monitor HCV v. 2.0 and the CTM48 HCV ASR assay is that the latter employs external calibrators rather than an internal quantitative reference standard to measure viral loads. Calibration is performed once, when a new reagent lot is first used. Calibration reagents are therefore critical to the performance of the CTM48 HCV ASR assay. We chose to use calibrators obtained from a commercial source because they were well characterized and large supplies were available. Despite their advantages, care should be taken to ascertain that stated viral loads of commercially obtained calibrators are correct.
User-defined calibrators are an alternative to commercial reagents. These are typically residual plasma (typically 2 to 3 ml) or a large-volume specimen from a patient with a high viral load. Each of these two approaches has disadvantages. Namely, new calibrators must be extensively recharacterized when residual plasma volumes have been depleted and consent from an institutional human subject review board may be required in order to perform large-volume phlebotomy.
The use of external calibrators raises additional issues. One question is whether it can be assumed that assays performed after calibration meet the same performance standard achieved during calibration. This may not be true, and laboratories should consider quality assurance measures such as tracking crossing thresholds (or elbow values, as designated for the CTM48 HCV ASR assay) of controls and analyzing individual controls within a run.
A second question is whether quantification is accurate at concentrations greater and less than the assay calibrators. At the low concentrations, our data suggest that quantification is accurate at concentrations two- to fivefold less than the lowest calibrator (50 IU/ml). We could not assess whether the upper limit of quantification exceeded the concentration of the highest calibrator because the concentrations of virus reagents were equivalent to calibrators. We observed high viral loads in patient specimens (see Fig. 5, with the caveat that these may not be accurate measurements), suggesting the need for accurate measurement in this range.
Our data demonstrate that our CTM48 ASR assay has a measurable range of at least 5 orders of magnitude, at least 10-fold greater than that of Monitor HCV v. 2.0 (4). An accurate determination of the upper limit of quantification will require the use of calibrators with viral loads greater than 7.0 log10 IU/ml. Users will need to define such reagents since they are not currently available commercially.
The potential for genotype-dependent bias in quantification exists, as demonstrated by the decreased efficiency of quantification of non-1 genotypes observed with the first version of HCV Monitor (6). This bias was corrected in Monitor v. 2.0 (4), and we did not observe any evidence of its occurrence in our assay with the CTM48 ASR reagents. The measurable range also appeared to be independent of extraction method, since linearity observed with manual and automated RNA preparation was largely similar.
QS is provided with the CTM48 ASR. In this assay, the QS is used to assess amplification inhibition in individual specimens. We found that addition of QS to the lysis buffer during extraction was critical for assay performance. Although the QS is an armored RNA product, it may be subject to nuclease attack in native specimens.
The LODs of our CTM48 HCV ASR assay were lower than those for Monitor HCV v. 2.0 (600 IU/ml) and comparable to those of the conventional qualitative PCR platform COBAS Amplicor HCV v. 2.0 (4). No significant genotype bias was observed after automated HCV RNA extraction. The LOD after manual extraction with the MinElute vacuum manifold protocol was lower than after automated extraction with the BioRobot M48. One explanation for this might be the greater effective concentration of plasma after manual versus automated extraction (a 1.8-fold increase in concentration after manual compared to automated extraction). It is also possible that the inclusion of protease K in the lysis buffer of the MinElute reagents (not included in MagAttract viral RNA reagent) improves the efficiency of HCV RNA extraction.
Precision throughout the range of quantification was within an acceptable range for a real-time PCR method. Variability in quantification was greatest at low concentrations as expected for a PCR-based amplification method. We expected that precision using an automated extraction instrument would be better than a manual method, but our data demonstrate that reproducibility was comparable between the two methods.
The major benefit of manual extraction using the MinElute reagents and vacuum manifold setup was an apparent increase in sensitivity and a decrease in the lower limit of quantification of our CTM48 HCV ASR assay. Automated extraction could be implemented for routine testing, given the comparable performance characteristics and obvious labor savings. However, extraction with MinElute could be incorporated selectively, to provide an ultrasensitive assay for HCV RNA detection.
We chose to determine the correlation between Monitor HCV v. 2.0 and CTM48 HCV ASR using the automated RNA extraction method because we planned to use the latter assay to replace Monitor in routine diagnostic testing. We characterized the correlation throughout the range of quantification because many clinicians are now requesting viral load measurement (rather than testing with a qualitative HCV RNA detection assay) for patients receiving therapy. Consequently, viral loads in the range of 2.0 to 4.0 log10 IU/ml are now commonly observed. Our data demonstrated excellent correlation throughout the range of concentrations tested.
Interestingly, CTM48 HCV ASR yielded values (∼0.2 log10 IU/ml) for Gt1 lower than those with Monitor HCV v. 2.0, as demonstrated by regression analysis results of Gt1 measurable range and quantification correlation of clinical specimens (primarily Gt1). Similar bias for Gt2 and -3 was not observed in measurable range experiments. The existence of bias in quantification of Gt2 and -3 in clinical samples could not be determined because of the small number of non-1 genotype specimens.
The potential sources of variation that could account for quantification differences between the two assays include extraction method and assay format (real-time PCR with external calibrator versus conventional PCR with internal QS). It was recently noted that HCV Monitor v. 2.0 overestimated an international (Gt1) standard concentration by approximately 0.5 log10 IU/ml (8). The authors hypothesized that the discrepancy was due to a potential flaw in QS conversion from copies to international units per milliliter, lending support to the possibility that assay format might explain quantification differences between the two tests. If Monitor v. 2.0 truly overestimates HCV Gt1 viral load, our data suggest that quantification by CTM48 HCV ASR may be more accurate.
The advantages of the CTM48 HCV ASR assay include increased throughput (48 positions per CTM versus 24 positions per COBAS instrument) and decreased turnaround time (∼3 versus 6 h) compared to COBAS Monitor HCV v. 2.0. In addition, the incorporation of automated RNA extraction should result in substantial labor savings due to decreased hands-on time.
The disadvantages of our CTM48 HCV ASR assay include calibrators, the need to provide our own assay controls (manufacturers cannot provide positive and negative controls for ASRs, in accordance with United States Food and Drug Administration regulation 21 CFR 809.10[e][1][x]), and the need to perform extensive in-house verification. Another potential disadvantage is the need to perform additional quality assurance steps (on each run, as described above). One comparatively smaller concern is that the volume of extracted material is sufficient for only a single round of testing.
Our data suggest that our CTM48 HCV ASR assay has the combined performance characteristics of qualitative and quantitative platforms. Laboratories that wish to consolidate testing into a single platform must weigh the benefits of streamlining with the added burden of providing quality control and rigorously performing quality assurance.
Acknowledgments
We give many thanks to our colleagues in the Division of Microbiology, especially John Ticehurst and Bill Merz, for their suggestions and comments on this work.
ASR materials were provided by Roche Diagnostics Corporation.
REFERENCES
- 1.Corbet, S., J. Bukh, A. Heinsen, and A. Fomsgaard. 2003. Hepatitis C virus subtyping by a core-envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J. Clin. Microbiol. 41:1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 3.Hadziyannis, S. J., H. Sette, Jr., T. R. Morgan, V. Balan, M. Diago, P. Marcellin, G. Ramadori, H. Bodenheimer, Jr., D. Bernstein, M. Rizzetto, S. Zeuzem, P. J. Pockros, A. Lin, and A. M. Ackrill. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346-355. [DOI] [PubMed] [Google Scholar]
- 4.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Gutekunst, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 6.Mellor, J., A. Hawkins, and P. Simmonds. 1999. Genotype dependence of hepatitis C virus load measurement in commercially available quantitative assays. J. Clin. Microbiol. 37:2525-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NCCLS. 2003. Quantitative molecular methods for infectious diseases. Approved guideline. NCCLS document MM6-A. NCCLS, Wayne, Pa.
- 8.Pisani, G., K. Cristiano, M. Wirz, G. M. Bisso, and G. Gentili. 2002. Overestimation of the hepatitis C virus RNA content of reference preparations by the AMPLICOR HCV monitor test, version 2.0. J. Clin. Microbiol. 40:4765-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]