Abstract
Temporal genetic modification of mice using the ligand-inducible Cre/loxP system is an important technique that allows the bypass of embryonic lethal phenotypes and access to adult phenotypes. In this study, we generated a tamoxifen-inducible Cre-driver mouse strain for the purpose of widespread and temporal Cre recombination. The new line, named CM32, expresses the GFPneo-fusion gene in a wide variety of tissues before FLP recombination and tamoxifen-inducible Cre after FLP recombination. Using FLP-recombined CM32 mice (CM32Δ mice) and Cre reporter mouse lines, we evaluated the efficiency of Cre recombination with and without tamoxifen administration to adult mice, and found tamoxifen-dependent induction of Cre recombination in a variety of adult tissues. In addition, we demonstrated that conditional activation of an oncogene could be achieved in adults using CM32Δ mice. CM32Δ;T26 mice, which harbored a Cre recombination-driven, SV40 large T antigen-expressing transgene, were viable and fertile. No overt phenotype was found in the mice up to 3 months after birth. Although they displayed pineoblastomas (pinealoblastomas) and/or thymic enlargement due to background Cre recombination by 6 months after birth, they developed epidermal hyperplasia when administered tamoxifen. Collectively, our results suggest that the CM32Δ transgenic mouse line can be applied to the assessment of adult phenotypes in mice with loxP-flanked transgenes.
Keywords: FLP/FRT, mouse, SV40 large T antigen, tamoxifen-inducible Cre, tumorigenesis
Introduction
Temporally-controlled genetic modification in vivo is an indispensable technique in mouse reverse genetics. Embryonic lethal phenotypes caused by genetic modification in the germline or in embryonic cells provide information about the function of genes-of-interest that are essential in embryonic development; however, lethal phenotypes in embryos and pups prevent researchers from studying adult phenotypes, such as terminal differentiation of cells, regeneration, pathophysiology, and tumorigenesis. For instance, it is important for cancer biologists to be able to bypass embryonic lethal phenotypes caused by genetic modifications, because the expression of oncogenes or the inactivation of tumor suppressor genes often results in lethal phenotypes in embryos and young mice. One solution to circumvent this problem is to utilize ligand-induced site specific recombination (SSR). In this technique, a mutated ligand-binding domain (LBD) of a steroid receptor fused to a site-specific recombinase makes it possible to control recombinase activity in a tamoxifen-dependent manner [3]. Tamoxifen-inducible Cre recombinases consist of Cre fused to a mutated LBD of the estrogen receptor (ER). The mutated LBD of ER has a high affinity for the synthetic ER ligand tamoxifen and its active metabolite 4-hydroxytamoxifen, but is insensitive to endogenous estrogens. In the absence of tamoxifen, the Cre-ER fusion protein is retained in the cytoplasm. In the presence of tamoxifen, the protein bound with tamoxifen is translocated into the nucleus and recombines its loxP-flanked DNA substrate.
For high-throughput assessment of the function of a gene-of-interest in various cell types, it is preferable to use mice that constitutively express a tamoxifen-inducible Cre. Many tamoxifen-inducible Cre drivers under the control of constitutive promoters have been generated and are available from mouse resource banks and commercial suppliers. However, there is limited information about the Cre-drivers with respect to recombination efficiency, leakiness of tamoxifen-independent Cre activity, and the integration sites of randomly integrated transgenes. Without this information, it is difficult for researchers to select a driver line, design experiments, and evaluate Cre recombination-induced phenotypes of their flox mice.
In the present study, we generated, through embryonic stem cell (ESC) -mediated transgenesis, a transgenic mouse line that expresses tamoxifen-inducible Cre in various tissues in a FLP/FRT recombination-dependent manner. Using transgenic ESCs and mice, we demonstrated tamoxifen-dependent Cre recombination in vitro and in vivo, and evaluated recombination efficiency in adult tissues. Using the driver line, we found that the SV40 large T antigen (hereafter SV40 T-Ag) induces epidermal hyperplasia in adult mice.
Materials and Methods
Mice and Ethics Statement
We purchased C57BL/6J and MCH:ICR mice from CLEA Japan (Tokyo, Japan); ROSA26-loxP-stop-loxP-β-geo knock-in mice (Gt(ROSA)26Sortm1Sho) [13] from the Jackson Laboratory (Bar Harbor, ME, USA); and FLP66 transgenic (RBRC01252) [23] and CAG-FLPe36 transgenic mice (RBRC01834) [9] from RIKEN BRC (Tsukuba, Japan). CGE transgenic mice and T26 transgenic mice were described previously [7, 26]. All mice were housed under pathogen-free conditions. All mouse work in this study was approved by the Animal Care and Use Committee of the University of Tokyo and conducted in accordance with their guidelines (approval nos. 19-20, PA11-94, and PA11-95).
Generation of CAG-FRT-GEM-FRT-CreMer transgenic mice
The cDNA encoding a mutated version of GFPneo (GEM), followed by a BGH pA signal sequence, was constructed using the 5’ portion of GFPneo cDNA excised from pQBI PGK (Qbiogene, Carlsbad, CA, USA) and a 3’ portion of cDNA encoding a mutated neo with a BGH pA excised from pSA-βgeo (a gift from Dr. Hitoshi Niwa, RIKEN Center for Developmental Biology, Japan). The cassette was sandwiched with two synthetic FRT sequences cloned into pZErO2 (Invitrogen/Life Technologies, Carlsbad, CA, USA). The EcoRI-XhoI fragment harboring loxP-sandwiched βgeo with BGH pA in the pCGX vector [26] was replaced with the fragment harboring FRT-sandwiched GEM cDNA with BGH pA. The resulting vector was named pCFXFRT (F, fluorescent reporter). The cDNA encoding CreMer was PCR-amplified from pANMerCreMer [30] (a gift from Dr. Michael Reth, University of Freiberg, Germany), cloned into pZErO2 (Invitrogen), and sequence verified. The CreMer cDNA was subcloned into the SwaI site downstream of the second FRT site. A transgene cassette from SalI-digested pCFXFRT harboring the CreMer cDNA was gel-purified and dissolved in PBS. The transgene (10 μg) was introduced into E14.1 ESCs by electroporation. Transgene-expressing ESCs were selected by culturing in medium containing 400 μg/ml G418 (Invitrogen) for 7 days. G418-resistant colonies were picked and expanded for PCR genotyping and the formation of embryoid bodies (EBs). Of the 48 G418-resistant ESC clones isolated, several clones were analyzed for the presence of the CreMer cDNA and for widespread expression of GEM in EBs. One clone (no. 32) was then selected for further use.
For the production of transgenic mice, ESCs were injected into B6 blastocysts, which were transplanted into the uteri of pseudo-pregnant MCH:ICR female mice. Chimeric female mice were then crossed with C57BL/6J male mice. FRT-flanked GEM cassette-excised, CM32Δ mice were obtained using FLP66 mice (for Fig. 4) and CAG-FLPe36 mice (for Figs. 5–8). C57BL/6J-congenic CM32Δ mice were prepared and used for Figs. 5–8. For genotyping, we used the observation of the agouti coat color, GFP fluorescence in tail tips, and PCR genotyping for Cre.
Fig. 4.
Tamoxifen-inducible Cre recombination in CM32Δ;Rosa26-βgeo Cre indicator double-transgenic mice. A) X-gal-stained tissues of CM32Δ;Rosa26-βgeo Cre indicator double-transgenic mice administered with vehicle control (upper panels) or tamoxifen (TAM; lower panels). B) X-gal-stained skeletal muscle tissue cultures (left panels) and fibroblasts migrating from muscle tissues onto culture dishes (right panels) in the presence or absence of 4-hydroxytamoxifen (4OHT). Scale bars for right panels, 200 μm.
Fig. 5.
Cre-mediated EGFP expression in CGE;CM32Δ double-transgenic mice with or without tamoxifen administration. Tissues and immunostained sections of CGE transgenic mice are shown as negative controls. Scale bars, 1 mm and 100 μm for micrographs showing GFP fluorescence and GFP immunostaining, respectively.
Fig. 8.
Hyperplastic lesions in T26;CM32Δ double-transgenic mice on a congenic C57BL/6J background with or without tamoxifen administration. A) Kaplan-Meier survival curves of T26 mice and T26;CM32Δ mice. B) T26;CM32Δ mice develop pineoblastomas (arrow in top panel). Invasive pineoblastoma cells (right panels) express SV40 T-Ag and Crx, as shown by immunohistochemistry (red signal). Sections of the brain of a T26 mouse are also shown (left panels). HE, Haematoxylin and eosin stained section. Scale bars, 1 mm and 100 μm for whole-mount brains and sections, respectively. C) T26;CM32Δ mice exhibit enlarged thymuses (left panel). Enlarged thymuses contained SV40 T-Ag-expressing cells, as shown by immunohistochemistry (red signal in right panel). Scale bars, 1 mm and 100 μm for whole-mount thymuses and a section, respectively. D) T26;CM32Δ mice at 3 months after tamoxifen administration develop epidermal hyperplasia (lower panels). Sections of the ear are shown. Immunostaining shows that SV40 T-Ag is detected in epidermal cells of the double-transgenic mice after tamoxifen administration (lower-right panel). A small number of SV40 T-Ag-expressing cells are present in the dermis of CM32Δ mice in the absence of tamoxifen (upper-right panel). Red signals of cartilage are background signals (right panels). Scale bar, 100 μm.
Identification of the CM32 transgene integration site
To isolate the insertion site of the CM32 transgene, splinkerette-PCR and sequencing were performed, according to a previously described procedure [24] with a minor modification. Briefly, tail DNA from the transgenic mouse was digested with PstI and used for adaptor ligation. AmpliTaq360 master mix (Life Technologies) was used for PCR. The oligonucleotides for a splinkerette adaptor are 5’-CGAAGAGTAACCGTTGCTAGGAGAGACCGTGGCTGAATGAGACTGGTGTCGACACTAGTGGtgca-3’ and 5’-CCACTAGTGTCGACACCAGTCTCTAATTTTTTTTTTCAAAAAAA-3’. Primers used for splinkerette-PCR are as follows: 5’-CGAAGAGTAACCGTTGCTAGGAGAGACC-3’ (Splink1) and 5’-CATAATGCCAGGCGGGCCATTTACC-3’ (CMVIE-4R) for primary PCR amplification; and 5’-GTGGCTGAATGAGACTGGTGTCGAC-3’ (Splink2) and 5’-GGGCGTACTTGGCATATGATACACTTGATG-3’ (CMVIE-5R) for secondary PCR amplification.
The 3’ portion of the integrated transgene was PCR-amplified using CM32 Tg genomic DNA and the primers 5’-TGCTCCTGGAGATGTTGGATG-3’ and 5’-TCAGTGAAGCCACAGTCCTC-3’. The agarose gel-purified PCR amplicon was directly sequenced using both primers.
Assessment of tamoxifen-induced Cre activity
For Fig. 1, recombinant adenovirus (rAd) infection of mouse ESCs was performed as described previously [10]. CAG-GEM-CreMer ESCs were infected with a FLP-expressing rAd, AxCAFLP [15], and the FLP-recombined ESCs, verified by PCR genotyping and the absence of GFP signal, were termed CAG-CreMer ESCs and used for further study. For the assessment of tamoxifen-induced Cre recombination in vitro, the CAG-CreMer ESCs were infected with a Cre-target rAd, AxCALNLG [15], which conditionally expresses EGFP after Cre-mediated removal of a floxed neo cassette, at an MOI of 0 or 10. Half of the rAd-infected ESCs were then cultured in medium containing 800 nM 4-hydroxytamoxifen (4OHT; H7904; Sigma-Aldrich, St Louis, MO, USA), while the rest were cultured in 4OHT-free medium. EGFP expression was determined at 24 hours after infection. For Fig. 4B, small pieces of the skeletal muscle were removed from the hind legs of adult mice aseptically and then cultured in high glucose (4.5 g/l) Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Several days later, tissues and fibroblasts were cultured in medium containing 800 nM 4OHT. Tissues and cells were X-gal-stained 72 h after 4OHT administration.
Fig. 1.

Tamoxifen-inducible Cre recombination in CM32Δ ESCs. Cre-target recombinant adenovirus conditionally expressing EGFP (AxCALNLG) was infected in CM32Δ ESCs at MOI 10 prior to 4-hydroxytamoxifen (4OHT) treatment. Scale bar, 200 μm.
For inducing Cre recombination in vivo, tamoxifen administration was performed according to procedures described previously [1, 5], with slight modifications. Briefly, for Fig. 4, tamoxifen (T5648; Sigma-Aldrich) dissolved in corn oil (Sigma-Aldrich) was administered by a single intraperitoneal injection of 9 mg/40 g (225 mg/kg). Two weeks after injection, tissues were fixed and subjected to X-gal staining. Tamoxifen dissolved in peanut oil (Sigma-Aldrich) containing 10% ethanol was administered to 6-week-old mice once weekly for 3 weeks for Figs. 5 and 7, or once for Fig. 8, by oral gavage of 200 mg/kg. Mice were sacrificed for Figs. 5 and 7 more than 1 week after the last administration of tamoxifen. For Fig. 8, mice were sacrificed 5 months after birth.
Fig. 7.
Ligand-dependent and -independent, CreMer-mediated recombination rates on the Cre reporter CGE allele in adult mice. Recombination rates in tissues were quantitated by real-time PCR using the relative standard curve method. Relative quantities of recombined DNAs were normalized to that of Gapdh. Error bars, SD; n=4.
For Fig. 7, embryonic fibroblasts were obtained from CGE;CM32Δ double-transgenic embryos at E13.5 and cultured in DMEM supplemented with 10% FBS. Cells were cultured in medium containing 50 nM or 250 nM 4OHT and used for assays 24, 48, and 72 h after 4OHT administration.
Observation of βgeo, GFPneo, and EGFP expression in cells and whole-mount mouse tissues
X-gal staining was performed overnight at room temperature, according to a previously described procedure [18]. Tissues were immersion fixed and subjected to X-gal staining. GFPneo and EGFP signals were observed in cultured cells and freshly isolated tissues. Bright-field and fluorescence micrographs were acquired using Olympus microscopes equipped with digital cameras (IX70/DP70 and SZX12/DP72; Olympus, Tokyo, Japan). Micrographs shown in the figures are representative of three independent specimens. Micrographs were acquired using the same equipment settings as the micrographs of controls were. Adjustment of the brightness and contrast levels using Adobe Photoshop was applied equally across entire images and was applied equally to controls.
Immunohistochemistry and immunocytochemistry
GFP immunostaining of sections was performed as previously described [7]. Rat anti-GFP (Nacalai Tesque, Kyoto, Japan) was used as a primary antibody. For immunostaining for SV40 T-Ag and Crx, paraffin sections (3–5 μm) were prepared and antigen-retrieved in ImmunoSaver solution (Wako Pure Chemical Industries, Osaka, Japan) at 95°C for 45 min, or in PBS containing 0.25% trypsin and 0.5 mM EDTA at room temperature for 1 h. All sections were incubated with 3% H2O2 in PBS or methanol prior to immunostaining. Rabbit anti-SV40T-Ag and anti-Crx (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used. For sections, the Histofine reagent (Nichirei Biosciences, Tokyo, Japan), and the TSA/TSA PLUS HRP Detection System (NEN/PerkinElmer, Waltham, MA, USA) were used for detection. For embryonic fibroblasts, rat anti-GFP and anti-rat secondary antibody labeled with Alexa Fluor 546 (Molecular Probes/Life Technologies) were used. 4,6-diamidino-2-phenylindole (DAPI; Molecular Probes) was used for nuclear staining. Fluorescence micrographs were acquired with a BioRevo BZ-9000 microscope (Keyence, Osaka, Japan). Micrographs in figures are representative of two independently-stained specimens from two or more mice. We verified that the background staining in sections of wild-type mouse tissues by anti-GFP, or false-positive staining by an isotype control antibody, did not affect the staining results. Micrographs and control micrographs were acquired using the same equipment settings. Adjustment of the brightness and contrast levels using Adobe Photoshop was applied equally across entire images and was applied equally to controls.
Quantitative real-time PCR
Quantitative real-time PCR was performed using the StepOne Real-time PCR system with PowerSYBR Master Mix (Life Technologies). Genomic DNAs were isolated and purified from cells and tissues using a ZR-96 Quick-gDNA kit (Zymo Research, Irvine, CA, USA).
Transgene copy number was estimated by the relative standard curve method. The relative quantity of the neo gene was normalized to that of Gapdh. As a PCR template for relative standard curves, tail genomic DNA from a mouse homozygous for the Rosa26 locus harboring the neo gene was used. Ct values were determined using 80 ng of tail genomic DNA from an Rprd1b+/CM32mouse and a Cd6CGE/CGE mouse (that is homozygous for the neo gene) and plotted onto relative standard curves. Data represent technical triplicates of genomic DNA from each mouse. Primer sets used for this assay were 5’-TCCTGCCGAGAAAGTATCCA-3’ and 5’-TGATGCTCTTCGTCCAGATC-3’ for neo, and 5’-TACTCGCGGCTTTACGGGTG-3’ and 5’-TGGAACAGGGAGGAGCAGAGAGCAC-3’ for Gapdh.
Cre-mediated recombination rates were determined by the relative standard curve method. For in vitro studies, genomic DNAs were obtained from 4OHT-treated and non-treated primary embryonic fibroblasts. For in vivo studies, genomic DNAs were obtained from tissues of tamoxifen-treated and non-treated mice (one female and one male per group). Two specimens were excised from each mouse. For the testis, four specimens were excised from one male mouse. Primer sets used in this study were as follows: 5’-ACGTGCTGGTTATTGTGCTG-3’ and 5’-TCTCTCGATCAGGGTGCTCTC-3’ for the CGEΔ allele; 5’-TACTCGCGGCTTTACGGGTG-3’ and 5’-TGGAACAGGGAGGAGCAGAGAGCAC-3’ for Gapdh. Relative quantitation of recombined genomic DNAs was performed using the relative standard curve method. As PCR templates for relative standard curves, tail genomic DNA from Cd6+/CGEΔ mice was used. Relative quantities of recombined alleles were normalized to that of Gapdh. PCR amplification (amplicon-specific peaks in melt curves) was not detected, and no Ct value was determined, when genomic DNA from non-recombined Cd6+/CGE mice was used.
Results
Generation of a FLP recombination-activated, tamoxifen-inducible Cre-driver mouse ESC line and its derivative mouse strain
We sought to generate a transgenic mouse line that expresses tamoxifen-inducible Cre in a variety of tissues under the control of FLP/FRT recombination. We designed a CAG promoter [17]-driven, FLP/FRT-based binary transgene that expresses a GFPneo-fusion protein and a CreMer protein [30] before and after FLP/FRT recombination, respectively. We exploited ESC-mediated transgenesis technology [11] because it is an effective method for obtaining transgenic lines in which single or low-copy transgenes are integrated into loci permissive for transgene expression [11, 21]. When constructing the transgene, we introduced a hypomorphic mutant of the neo-resistance gene [28] into the GFPneo gene, which leads to G418-resistant ESC clones strongly expressing the transgene, as described in previous studies [4, 22]. The mutated GFPneo is referred to as GEM (the GFPneo gene encoding a mutated neomycin phosphotransferase). Among G418-resistant clones, we selected one clone (clone 32, hereafter CreMer (CM) 32) that had GFP expression in both undifferentiated and differentiated states and carried a CreMer cDNA, followed by a polyadenylation core signal sequence, as determined by PCR (data not shown). To determine whether tamoxifen-driven CreMer-mediated Cre/loxP recombination would work properly in ESCs, we obtained ESCs lacking the FRT-flanked GEM cassette (hereafter CM32Δ) by using a FLP-expressing adenovirus vector (AxCAFLP [15]) before monitoring CreMer activity using a Cre reporter adenovirus vector (AxCALNLG [15]) in the presence or absence of hydroxytamoxifen, according to a previously described procedure [10]. Ligand-induced CreMer activity was verified by hydroxytamoxifen-induced EGFP expression (Fig. 1), and we then generated a mouse line using that ESC clone. The CM32 transgenic mouse line was obtained through germline transmission from a chimeric female mouse, but not from chimeric male mice, indicating that the CM32 ESCs lack the Y chromosome. The hemizygotes grew normally with no obvious defect and were fertile. For instance, there was no significant difference in mean body weight (mean ± S.D.) between C57BL/6J congenic hemizygous females (22.5 ± 0.7 g, n=6) and wild-type females (22.2 ± 1.1 g, n=6) at 8–12 weeks of age (P>0.5).
Identification of the transgene integration site as the Rprd1b gene on chromosome 2 of CM32 transgenic mice
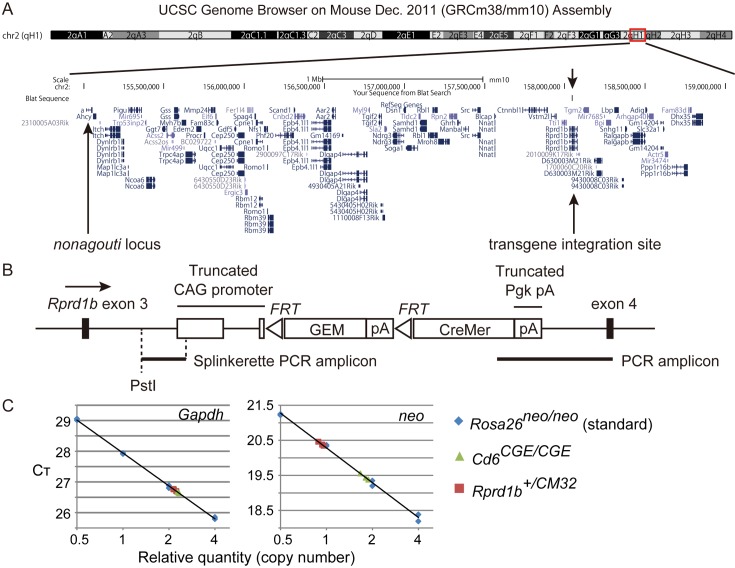
We found that almost all the hemizygous transgenic mice kept the agouti coat color despite serial backcrossing to C57BL/6J, suggesting that the CM32 transgene inserted close to the wild-type Agouti gene on chromosome 2 of 129P2/OlaHsd, which is the parental mouse strain of E14.1 ESCs. Indeed, we found that the transgene is located within an intron between exon 3 and exon 4 of the Rprd1b gene, 2–3 Mb away from the wild-type Agouti gene (Fig. 2A), as determined by Splinkerette-PCR, conventional PCR, and DNA sequencing (Fig. 2B). Moreover, the 5’ and 3’ portions (CMV enhancer region and Pgk pA region downstream of the AATAAA sequence) of the introduced transgene were deleted (Fig. 2B). The intronic repeat sequence was split, but apparently not deleted, by the integration event. Based on PCR-based relative quantitation, we estimated that a single transgene integrated into the Rprd1b locus (Fig. 3C). RPRD1B (regulation of nuclear pre-mRNA domain-containing protein 1B), also known as CREPT (cell cycle-related and expression elevated protein in tumor), is a binding partner of RNA polymerase II [16] and is a regulator of the transcription of cell cycle-related genes [12]. According to the Guidelines for Nomenclature of Mouse and Rat Strains (http://www.informatics.jax.org/mgihome/nomen/strains.shtml), the CM32 transgenic allele was formally named Rprd1bTg(CAG-GFPneo*, -Cre/Esr1*)32Ichi. In the text below, we described this allele as Rprd1bCM32. Transgenic mice homozygous for the Rprd1bCM32 allele have not been found in the offspring from mating pairs of Rprd1b+/CM32mice at weaning (data not shown). The insertion of the transgene may disrupt Rprd1b and lead to a recessive lethal phenotype, probably due to aberrant RPRD1B-mediated transcriptional control. By contrast, the hemizygotes grow normally and are fertile, as described above. In addition, the International Mouse Phenotyping Consortium (IMPC) has generated Rprd1b heterozygous mutant mice and performed systematic phenotyping of the heterozygotes. Judging from the data presented by the IMPC at the present time, the heterozygotes are normal in terms of morphology and physiology, even though mean corpuscular hemoglobin has been reported to be higher in heterozygous mutant females than in wild-type females (for details, visit the IMPC website at http://www.mousephenotype.org/). A potential artifact of Rprd1b haploinsufficiency on CM32Δ-harboring mice can be eliminated by using floxed mice that harbor one GFPneo-expressing CM32 allele, or CreMer-expressing CM32Δ hemizygous mice without floxed alleles, as negative controls.
Fig. 2.
Location of the transgene in the CM32 transgenic mouse line. A) Graphical representation of a BLAT sequence alignment between the splinkerette-PCR amplicon sequence and the B6 genome sequence on the UCSC genome browser. The transgene is mapped distal to the nonagouti (a) locus on chromosome 2 of C57BL/6J. A counterpart for the a locus in 129P2/OlaHsd-derived E14.1 ESCs is the wild-type agouti (Aw) locus. B) The transgene is integrated into the intron between exon 3 and exon 4 of the Rprd1b gene. The splinkerette-PCR amplicon that contains a 5’ portion of the transgene and the PCR amplicon that contains the boundary between the 3’ portion of the transgene and the endogenous gene is indicated. GEM, a mutated version of GFPneo. C) Estimation of transgene copy number within the CM32 allele by real-time PCR. Semi-log graphs represent relative standard curves for the Gapdh and neor genes. Rosa26neo/neo and Cd6CGE/CGE mice have two copies of the neor gene.
Fig. 3.
GFP expression in CM32 transgenic mice. A) GFP expression in a 6-week-old CM32 transgenic male mouse (right panels). Auto-fluorescent background signals in a wild-type mouse are also shown (left panels). Scale bar, 1 mm. B) GFP immunostaining of tissue sections of a 2-day-old CM32 transgenic mouse. Scale bar, 100 μm.
Expression pattern of the GFPneo-fusion gene in CM32 mice
To determine the expression pattern of the transgene, we examined the GFP signal in postnatal and young mice using fluorescence microscopy and immunohistochemistry. GFP expression was observed in the skin, heart, skeletal muscle, pancreas and testis of 6-week-old CM32 male mice under a fluorescence microscope (Fig. 3A). Weak GFP signals were also found in the brain, thymus, lung, intestine, liver, and kidney (data not shown). Immunostaining of tissue sections using anti-GFP demonstrated that the transgene is expressed in a wide variety of tissues, including the brain, thymus, heart, lung, intestine, pancreas, spleen, liver, kidney, and skin (Fig. 3B) in 2-day-old mice. These results suggest that the CM32 transgene may express CreMer in various tissues after FLP-mediated excision of the FRT-flanked GEM cassette. Strong CreMer expression is particularly likely in the skin, heart, skeletal muscle, pancreas, and testis.
Cre recombinase activity in FLP-recombined CM32Δ mice
To examine CreMer activity across the whole body, we next generated CM32Δ mice that lack the FRT-flanked GEM cassette by using germline FLP-deleter mice (Suppl. Fig. 1) crossed to Cre reporter strains.
Initially, we used the Rosa26-βgeo Cre reporter line, Gt(ROSA)26Sortm1Sho [13], which expresses βgeo under the control of the Rosa26 gene after Cre recombination (Suppl. Fig. 1). The double-transgenic mice that had been injected intraperitoneally with tamoxifen, but not those injected with vehicle, showed βgeo expression in tissues such as the liver, skin, and skeletal muscle (Fig. 4A). CM32;Gt(ROSA)26Sortm1Sho double-heterozygous mice that were administered tamoxifen were negative for X-gal staining, indicating that CreMer expression is blocked by the floxed GEM cassette (data not shown). Tamoxifen-induced recombination was also seen in skeletal muscle explant culture and fibroblasts migrating from muscle tissues onto culture dishes in the presence of hydroxytamoxifen (Fig. 4B).
To examine Cre recombination pattern and efficiency in more detail, we used CGE transgenic mice as a Cre reporter. CGE transgenic mice express EGFP after Cre-mediated excision of floxed cassettes under the control of the CAG promoter in the Cd6 locus [7] (Suppl. Fig. 1). Tamoxifen-induced recombination was achieved in CGE;CM32Δ double-transgenic mice, although background Cre recombination was found in pancreatic acini in the absence of tamoxifen (Fig. 5).
To determine background and ligand-induced recombination rates in CGE;CM32Δ mice, we measured Cre recombination rates in embryonic fibroblasts and adult tissues by quantitative PCR using a relative standard curve method. In vitro assays revealed that tamoxifen-induced recombination was induced in approximately 70% of CGE;CM32Δ double-transgenic embryonic fibroblasts during 3 day culture in the presence of 50 nM hydroxytamoxifen, whereas background recombination occurred in approximately 1% of cells prior to hydroxytamoxifen treatment (Fig. 6). PCR-based quantitation of Cre recombination in adult tissues revealed that tamoxifen administration induces recombination in many tissues, although we also found that the liver, pancreas, heart, and skeletal muscle were susceptible to background Cre activity in the absence of tamoxifen (Fig. 7). These results suggest that, using CM32Δ mice, we can obtain mosaic mice that consist of both non-Cre-recombined and Cre-recombined cells in adult tissues, and can increase the ratio of the recombined cells to non-recombined cells via tamoxifen administration.
Fig. 6.

Cre-mediated recombination in CGE;CM32Δ double-transgenic embryonic fibroblasts with or without 4-hydroxytamoxifen administration. A) CGE;CM32Δ double-transgenic embryonic fibroblasts were cultured in the presence of the indicated concentration of 4OHT for the indicated hours. Error bars, SD; n=3. Recombination rates were determined by quantitative real-time PCR using the relative standard curve method. The relative quantity of recombined DNAs was normalized to that of Gapdh. B) Micrographs show GFP immunostaining of the cells. Scale bar, 100 μm.
Conditional activation of the SV40 large T antigen using the CM32Δ allele induces hyperplastic lesions in adult mice
SV40 T-Ag transforms cells by functionally inactivating the p53 protein and retinoblastoma family proteins. SV40 tsA58 is a SV40 mutant that encodes a thermolabile, weakly-acting large T antigen. While the tsA58 T-Ag has been utilized for establishing immortalized cell lines that retain the cellular characteristics of terminally differentiated cells [8, 27], tsA58 T-Ag can also act as an oncogenic protein in particular settings. Previous studies report that tsA58 T-Ag can lead to hyperplastic phenotypes in vivo when expressed in particular types of cells [8, 14, 27]. Previously, we generated the transgenic mouse line T26, which conditionally expresses tsA58 T-Ag in a Cre-recombination-dependent manner [26] (Suppl. Fig. 1). We established immortalized endothelial cells using T26;Tie2-Cre double-transgenic mice without observing hyperplastic changes in the tsA58 T-Ag-expressing endothelial cells in vivo. By contrast, we could not obtain T26 mice in which Cre had recombined in the germline. These observations suggested that expression of tsA58 T-Ag might cause hyperplastic changes in cells other than endothelial cells and lead to a lethal phenotype during development.
To circumvent the lethal phenotype of tsA58 T-Ag-expressing mice and examine the implications of using tsA58 T-Ag-expressing adult mice as experimental tumor models, we used CM32Δ mice as a partner for T26 mice. T26 homozygous mice were viable and fertile. The mating between CM32Δ hemizygous mice and T26 homozygous mice yielded CM32Δ;T26 double hemizygous transgenic mice at the expected ratio. The double transgenic mice were viable, fertile, and exhibited no gross phenotypes until approximately 3 months of age (Fig. 8A). However, they developed aggressive and invasive brain tumors with high penetrance (8 out of 10 mice were moribund or dead at 19–27 weeks after birth; Fig. 8B). Immunostaining against a pineal cell marker Crx showed that the tumors were pineoblastomas (Fig. 8B). Some of those mice also showed respiratory distress caused by enlarged thymuses occupying the thoracic cavity (7 out of 10 mice; Fig. 8C). The pineoblastomas and enlarged thymuses were SV40 T-Ag-positive (Figs. 8B and 8C), indicating that the lesions were the result of SV40 T-Ag expression caused by background Cre recombination. On the other hand, tamoxifen administration was effective in inducing the hyperplastic phenotype in those mice. Tamoxifen administration activated the expression of SV40 T-Ag in the epidermis and the double transgenic mice developed epidermal hyperplasia after tamoxifen treatment (in 4 out of 4 mice, 5 months after birth and approximately 3 months after tamoxifen administration; Fig. 8D), in addition to pineoblastomas and thymic hyperplasia.
Collectively, our results indicate that the CM32Δ transgenic mouse line can be applied to the temporal genetic modification of adult tissues, and demonstrate that CM32Δ mice in combination with T26 mice can be used to study SV40 T-Ag-driven hyperplastic and tumorigenic phenotypes in adulthood.
Discussion
The use of FLP-activated CM32Δ mice depends on either ligand-inducible Cre activity or ligand-independent, background Cre activity, as demonstrated by hyperplastic phenotypes of T26;CM32Δ mice (Fig. 8). This transgenic mouse line would be effective for temporal genetic modification in tissues with low background Cre recombination. For example, FLP-activated CM32Δ mice would be useful for temporal activation and inactivation of floxed genes in adult male germ cells and epidermal cells, because recombination of floxed alleles in these cells is highly ligand-dependent, as described above (Figs 5–8). On the contrary, in tissues exhibiting high background Cre recombination such as the skeletal muscle, it is difficult to distinguish between cells that Cre-recombined due to background Cre activity and those that did so in response to tamoxifen-activated Cre. However, cells that Cre-recombined due to background Cre activity appeared to be distributed in a scattered pattern in adult tissues, as observed in the pancreas (Fig. 5). Notably, background Cre activity was low enough to prevent early lethality of Cre-recombined T26 mice, even though T26;CM32Δ mice developed tumors 3–5 months after birth (Fig. 8). Therefore, we postulate that the background Cre activity of CM32Δ mice does not prevent the bypass of embryonic lethal phenotypes in floxed mice.
SV40 T-Ag-induced lesions of the pineal gland and thymus have been reported previously. The interstitial retinol binding protein promoter-driven SV40 T-Ag transgenic mice developed pineal tumors by as early as 2 weeks of age [6]. H-2Kb-tsA58 transgenic mice, which express the tsA58 T-Ag under the control of the interferon-inducible promoter, exhibit thymic enlargement [8]. On the other hand, inactivation of Trp53 and Rb1 genes also causes similar phenotypes in these organs. p53-deficient mice are known to develop enlarged thymuses [2]. Williams et al. showed that 40% of Trp53-/-;Rb1+/- mice developed pineoblastomas, in which the remaining wild-type Rb allele was inactivated by spontaneous mutations [25]. Moreover, K14-Cre-driven, epidermis-specific inactivation of Rb1 causes epidermal hyperplasia [19]. Therefore, it is likely that pineoblastoma, thymic enlargement, and epidermal hyperplasia in T26;CM32Δ mice were the result of functional inactivation of p53 and/or Rb proteins. Our results suggest that T26;CM32Δ mice may be used to study the hyperproliferation and tumorigenesis of cell types that are susceptible to inactivation of p53 and Rb family proteins.
While the background recombination rate was low in the brain and thymus (below 0.1%; Fig. 7), T26;CM32Δ mice exhibited pineal gland tumors and thymic enlargement (Fig. 8). It is likely that sporadic cancer development is triggered by a single cell that harbors an oncogenic mutation. In the case of T26;CM32Δ mice, cells that are highly sensitive to oncogenic functions of SV40 T-Ag may acquire an oncogenic property and growth advantage once the T26 allele is excised by Cre. Background recombination in such cells may result in formation of tumorigenic/hyperplastic lesions, regardless of background recombination rates in the cell population. This explanation may be supported by the fact that the pineal gland and thymus are prone to tumorigenic/hyperplastic disorders caused by inactivating mutations of p53 and Rb proteins, as described above. In future studies using CM32Δ mice, there would be also a possibility that defects might be caused by background recombination in cells that are few in number in organs but play critical roles in organ development, function, and homeostasis. It would be necessary to pay attention to background recombination-induced effects of a Cre-target gene-of-interest.
We developed the CM32 mouse line for use in the dual SSR system. In this system, spatial regulation of CreMer depends on tissue-specific FLP expression, which activates CreMer expression by removing a FRT-flanked GFPneo cassette. Although a limited number of tissue-specific FLP-driver strains are currently available, it is likely that an increasing number of tissue-specific FLP and inducible FLP-driver strains, as well as FRT-flanked mutant mice, will be generated in the near future. In tissues with low background Cre recombination in CM32Δ mice such as testis and skin, dual recombinase-based studies of male germ cells and epidermal cells would be practical. By contrast, sequential induced mutation is difficult in the liver and pancreas of FLP-driver- and CM32 double-transgenic mice because leaky Cre activity in these tissues could result in recombination of the target alleles shortly after FLP recombination. However, the mosaic pattern of Cre recombination would facilitate study of the function of Cre-target genes in FLP-recombined cells. For instance, gain- or loss-of-function mutations could be induced by leaky Cre activity in subsets of FLP-induced oncogene-expressing cells in these tissues. Recently, two dual SSR system-compatible mouse lines were reported. Both lines have CAG promoter-driven, FLP/FRT-based binary transgenes that expresses a CreERT2 protein after FLP/FRT recombination. Schönhuber et al. and Zhang et al. introduced transgenes into the Rosa26 locus on chromosome 6 and the Col1a1 locus on chromosome 11, respectively [20, 29]. CM32 mice, whose transgene is located on chromosome 2 (Fig. 2), may be more effective than the reported lines in modifying homozygous floxed loci on chromosome 6 and 11. When combined with a tissue-specific FLP-expressing mouse line, our transgenic mouse line as well as the reported lines will be valuable tools for the establishment of dual recombination system-based experiments.
Supplementary Material
Acknowledgments
We thank Reiko Sakamoto and Mio Kikuchi for technical assistance; Drs. Hitoshi Niwa and Michael Reth for providing pSA-βgeo and pANMerCreMer, respectively; and RIKEN BRC for providing FLP-deleter mice (RBRC01252 and RBRC01834). This work was supported by grants from the Japan Society for the Promotion of Science (to H.I., T.I. and N.Y.; 15500297 and 22500384) and the Ministry of Education, Culture, Sports, Science and Technology, Japan (to H.I., T.I. and N.Y.; 18013020).
References
- 1.Anastassiadis K., Glaser S., Kranz A., Berhardt K., Stewart A.F.2010. A practical summary of site-specific recombination, conditional mutagenesis, and tamoxifen induction of CreERT2. Methods Enzymol. 477: 109–123. doi: 10.1016/S0076-6879(10)77007-5 [DOI] [PubMed] [Google Scholar]
- 2.Donehower L.A., Harvey M., Slagle B.L., McArthur M.J., Montgomery C.A., Jr, Butel J.S., Bradley A.1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215–221. doi: 10.1038/356215a0 [DOI] [PubMed] [Google Scholar]
- 3.Feil R., Brocard J., Mascrez B., LeMeur M., Metzger D., Chambon P.1996. Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. USA 93: 10887–10890. doi: 10.1073/pnas.93.20.10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedrich G., Soriano P.1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5: 1513–1523. doi: 10.1101/gad.5.9.1513 [DOI] [PubMed] [Google Scholar]
- 5.Hayashi S., McMahon A.P.2002. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244: 305–318. doi: 10.1006/dbio.2002.0597 [DOI] [PubMed] [Google Scholar]
- 6.Howes K.A., Lasudry J.G., Albert D.M., Windle J.J.1994. Photoreceptor cell tumors in transgenic mice. Invest. Ophthalmol. Vis. Sci. 35: 342–351. [PubMed] [Google Scholar]
- 7.Ichise H., Ichise T., Sasanuma H., Yoshida N.2014. The Cd6 gene as a permissive locus for targeted transgenesis in the mouse. Genesis 52: 440–450. doi: 10.1002/dvg.22779 [DOI] [PubMed] [Google Scholar]
- 8.Jat P.S., Noble M.D., Ataliotis P., Tanaka Y., Yannoutsos N., Larsen L., Kioussis D.1991. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proc. Natl. Acad. Sci. USA 88: 5096–5100. doi: 10.1073/pnas.88.12.5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanki H., Suzuki H., Itohara S.2006. High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp. Anim. 55: 137–141. doi: 10.1538/expanim.55.137 [DOI] [PubMed] [Google Scholar]
- 10.Kondo S., Takahashi Y., Shiozawa S., Ichise H., Yoshida N., Kanegae Y., Saito I.2006. Efficient sequential gene regulation via FLP-and Cre-recombinase using adenovirus vector in mammalian cells including mouse ES cells. Microbiol. Immunol. 50: 831–843. doi: 10.1111/j.1348-0421.2006.tb03850.x [DOI] [PubMed] [Google Scholar]
- 11.Lobe C.G., Koop K.E., Kreppner W., Lomeli H., Gertsenstein M., Nagy A.1999. Z/AP, a double reporter for cre-mediated recombination. Dev. Biol. 208: 281–292. doi: 10.1006/dbio.1999.9209 [DOI] [PubMed] [Google Scholar]
- 12.Lu D., Wu Y., Wang Y., Ren F., Wang D., Su F., Zhang Y., Yang X., Jin G., Hao X., He D., Zhai Y., Irwin D.M., Hu J., Sung J.J., Yu J., Jia B., Chang Z.2012. CREPT accelerates tumorigenesis by regulating the transcription of cell-cycle-related genes. Cancer Cell 21: 92–104. doi: 10.1016/j.ccr.2011.12.016 [DOI] [PubMed] [Google Scholar]
- 13.Mao X., Fujiwara Y., Orkin S.H.1999. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc. Natl. Acad. Sci. USA 96: 5037–5042. doi: 10.1073/pnas.96.9.5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.March K.L., Sandusky G., Fan L.1999. Hyperplasia in multiple smooth muscle tissues in transgenic mice expressing a temperature-sensitive SV40 T-antigen under the control of smooth muscle alpha-actin regulatory sequences. Oncogene 18: 3773–3782. doi: 10.1038/sj.onc.1202994 [DOI] [PubMed] [Google Scholar]
- 15.Nakano M., Odaka K., Ishimura M., Kondo S., Tachikawa N., Chiba J., Kanegae Y., Saito I.2001. Efficient gene activation in cultured mammalian cells mediated by FLP recombinase-expressing recombinant adenovirus. Nucleic Acids Res. 29: E40. doi: 10.1093/nar/29.7.e40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni Z., Olsen J.B., Guo X., Zhong G., Ruan E.D., Marcon E., Young P., Guo H., Li J., Moffat J., Emili A., Greenblatt J.F.2011. Control of the RNA polymerase II phosphorylation state in promoter regions by CTD interaction domain-containing proteins RPRD1A and RPRD1B. Transcription 2: 237–242. doi: 10.4161/trns.2.5.17803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niwa H., Yamamura K., Miyazaki J.1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108: 193–199. doi: 10.1016/0378-1119(91)90434-D [DOI] [PubMed] [Google Scholar]
- 18.Nonchev S.G., Maconochie M.K.2000, Spatial analysis of gene expression., In Mouse genetics and transgenics: A practical approach., Jackson I.J. and Abbott C.M. eds., Oxford University Press. [Google Scholar]
- 19.Ruiz S., Santos M., Segrelles C., Leis H., Jorcano J.L., Berns A., Paramio J.M., Vooijs M.2004. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development 131: 2737–2748. doi: 10.1242/dev.01148 [DOI] [PubMed] [Google Scholar]
- 20.Schönhuber N., Seidler B., Schuck K., Veltkamp C., Schachtler C., Zukowska M., Eser S., Feyerabend T.B., Paul M.C., Eser P., Klein S., Lowy A.M., Banerjee R., Yang F., Lee C.L., Moding E.J., Kirsch D.G., Scheideler A., Alessi D.R., Varela I., Bradley A., Kind A., Schnieke A.E., Rodewald H.R., Rad R., Schmid R.M., Schneider G., Saur D.2014. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat. Med. 20: 1340–1347. doi: 10.1038/nm.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada H., Kaname T., Suzuki M., Hitoshi Y., Araki K., Imaizumi T., Yamamura K.1999. Comparison of ES cell fate in sandwiched aggregates and co-cultured aggregates during blastocyst formation by monitored GFP expression. Mol. Reprod. Dev. 52: 376–382. [DOI] [PubMed] [Google Scholar]
- 22.Skarnes W.C., Moss J.E., Hurtley S.M., Beddington R.S.1995. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc. Natl. Acad. Sci. USA 92: 6592–6596. doi: 10.1073/pnas.92.14.6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi T., Nomura T., Tsujita M., Suzuki M., Fuse T., Mori H., Mishina M.2002. Flp recombinase transgenic mice of C57BL/6 strain for conditional gene targeting. Biochem. Biophys. Res. Commun. 293: 953–957. doi: 10.1016/S0006-291X(02)00321-2 [DOI] [PubMed] [Google Scholar]
- 24.Uren A.G., Mikkers H., Kool J., van der Weyden L., Lund A.H., Wilson C.H., Rance R., Jonkers J., van Lohuizen M., Berns A., Adams D.J.2009. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat. Protoc. 4: 789–798. doi: 10.1038/nprot.2009.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams B.O., Remington L., Albert D.M., Mukai S., Bronson R.T., Jacks T.1994. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat. Genet. 7: 480–484. doi: 10.1038/ng0894-480 [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi T., Ichise T., Iwata O., Hori A., Adachi T., Nakamura M., Yoshida N., Ichise H.2008. Development of a new method for isolation and long-term culture of organ-specific blood vascular and lymphatic endothelial cells of the mouse. FEBS J. 275: 1988–1998. doi: 10.1111/j.1742-4658.2008.06353.x [DOI] [PubMed] [Google Scholar]
- 27.Yanai N., Suzuki M., Obinata M.1991. Hepatocyte cell lines established from transgenic mice harboring temperature-sensitive simian virus 40 large T-antigen gene. Exp. Cell Res. 197: 50–56. doi: 10.1016/0014-4827(91)90478-D [DOI] [PubMed] [Google Scholar]
- 28.Yenofsky R.L., Fine M., Pellow J.W.1990. A mutant neomycin phosphotransferase II gene reduces the resistance of transformants to antibiotic selection pressure. Proc. Natl. Acad. Sci. USA 87: 3435–3439. doi: 10.1073/pnas.87.9.3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M., Kirsch D.G.2015. The generation and characterization of novel Col1a1FRT-Cre-ER-T2-FRT and Col1a1FRT-STOP-FRT-Cre-ER-T2 mice for sequential mutagenesis. Dis. Model. Mech. 8: 1155–1166. doi: 10.1242/dmm.021204 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Zhang Y., Riesterer C., Ayrall A.M., Sablitzky F., Littlewood T.D., Reth M.1996. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res. 24: 543–548. doi: 10.1093/nar/24.4.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.