Abstract
The bacterial microfloras of 8 healing and 10 nonhealing chronic venous leg ulcers were compared by using a combination of cultural analysis and denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA gene products. Cultural analysis of the microflora revealed that the majority of both wound types carried the aerobes Staphylococcus and Pseudomonas spp. (89 and 80%, respectively). Sequencing of 16S ribosomal DNAs selected on the basis of DGGE profiling allowed the identification of strains not detected by cultural means. Of considerable interest was the finding that more than 40% of the sequences represented organisms not cultured from the wound from which they were amplified. DGGE profiles also revealed that all of the wounds possessed one apparently common band, identified by sequencing as Pseudomonas sp. The intensity of this PCR signal suggested that the bacterial load of nonhealing wounds was much higher for pseudomonads compared to healing wounds and that it may have been significantly underestimated by cultural analysis. Hence, the present study shows that DGGE could give valuable additional information about chronic wound microflora that is not apparent from cultural analysis alone.
Chronic wounds, such as those evident in significant numbers of patients with venous leg ulceration, are a repository of complex polymicrobial populations, including both aerobic and anaerobic species (6). There is evidence that the microfloras of these wounds play a role in the healing process, although there is still considerable debate as to the importance of individual species or microbial density in relation to healing and subsequently to chronic wound management (4, 19, 27, 42). The predominant bacteria isolated from these wounds are the well-characterized staphylococci and pseudomonads, although many other bacterial groups have been documented (5, 6, 19, 39). Direct comparison between the numerous cultural investigations undertaken to evaluate the role of bacteria in chronic wounds is difficult since the studies have been based on different patient populations, used a diversity of sampling and culturing methodologies, and often only selected for specified bacterial groups. In addition, all previous studies have relied on a traditional cultural analysis, and there are likely to be biases in such an approach.
There have been significant advances in recent years in the characterization and identification of bacteria by molecular techniques. This has facilitated a shift from conventional phenotypic methods for the detection of pathogens in clinical samples to the increased use of molecular approaches for both taxonomic and phylogenetic analysis (13). Conventional analysis of chronic wound communities has relied on traditional bacteriological culture methods (5, 6, 11, 12, 16, 18), thereby completely missing the unculturable population that may be present (53) and thus strengthening the need for the application of such molecular techniques to wound ecology studies.
We applied molecular methods to the analysis of the microflora of a single chronic venous leg ulcer wound (23). Direct sequencing of PCR-amplified 16S rRNA genes demonstrated significantly greater bacterial diversity than that revealed by culture alone. Furthermore, sequences which may possibly represent novel species of bacteria, were retrieved. Hence, the study revealed that a far more comprehensive analysis of the wound microflora was possible by using molecular techniques. However, due to the labor-intensive nature of direct cloning and sequencing to identify the unculturable portion of the microflora, the study was limited to the analysis of a single clinical sample.
In environmental microbiology, denaturing gradient gel electrophoresis (DGGE) has been used as a tool for profiling complex microbial populations without the biases of cultural analysis for many years (30, 32, 51, 52), and the technique has now been applied to the study of a limited number of human microbial populations (17, 21, 28, 45, 55). The advantage of this approach is that it creates a genetic fingerprint or profile of total community diversity by separating mixed 16S rRNA PCR amplification products on the basis of their sequence melting behavior. Subtractive analysis can then be used to identify specific bands of interest such as those sequences that are present only by molecular means (the so-called uncultured fraction) by screening sequences amplified from clinical wound samples alongside those derived from bacteria cultured from these same wounds. These bands can then be cloned and sequenced, without the need to sequence amplification products from cultured organisms as well. DGGE also has other advantages in that direct comparisons can be made between wound samples run on the same gels.
In the present study, we describe the use of DGGE to determine the extent of bacterial diversity within 18 chronic venous leg ulcers, with particular emphasis on the unculturable microflora. Comparison of healing and nonhealing wounds was undertaken to determine whether specific bacterial species were important in the nonhealing phenotype of this painful and often debilitating condition.
MATERIALS AND METHODS
Patients.
With local ethical research committee approval and after we obtained patient informed written consent, patients with newly diagnosed venous ulceration attending the outpatient clinic in the Wound Healing Research Unit at University Hospital of Wales, Cardiff, United Kingdom, and the Royal Gwent Hospital, Newport, United Kingdom, were recruited (mean age, 68.4 years; range, 36 to 89). Venous ulceration was diagnosed according to previously defined criteria (43). Diagnosis of venous incompetence was confirmed by duplex ultrasonography, and arterial disease was excluded by measurement of the ankle brachial pressure index. All patients included in the study had an ankle brachial pressure index of ≥0.8. Patients were also screened to exclude the presence of systemic disease (including diabetes), and patients who had received systemic or topical antimicrobial therapy in the previous month were also excluded. All patients in the study were treated with a standard regimen of compression therapy.
Patients were followed up for more than 6 months postbiopsy to determine healing rates over this time. Ulcers with 100% closure of the wound were considered healed, but only wounds which did not reoccur within 1 month of wound closure were included in the present study. The study group selected for molecular analysis included 10 healers and 10 nonhealers, although two patients in the healing group were later excluded because the wounds recurred within 1 month of complete healing.
Swab and tissue sample collection.
The wound surface was irrigated with 5 ml of sterile saline (0.9% [wt/vol] NaCl), and the surface microflora was sampled by swabbing a 1-cm2 area of the wound by using a cotton tipped swab. The swab was immediately inoculated into 2 ml of transport medium (TM) (3). This medium had been prereduced by incubation in an anaerobic environment (10% CO2, 10% H2, and 80% N2) at 37°C for 24 h. After the swabbing step, local anesthetic, 5 ml of Lidocaine Hydrochloride BP (Phoenix Pharma, Ltd., Gloucester, United Kingdom) was injected subcutaneously. After 5 to 12 min, a specimen from the central region of the ulcer bed (where no healing was observed) was obtained by using a 6-mm disposable sterile punch biopsy (Stiefel Laboratories, Ltd., Sligo, Ireland) and placed immediately into a second vial containing 2 ml of TM. Both samples were transported to the laboratory at room temperature within 2 h of collection for processing. The tissue sample was weighed and bisected with one portion used for traditional cultural analysis and the second was stored at −20°C for subsequent molecular analysis.
Cultural analysis of the wound microflora.
The swab sample was vortex mixed in TM for 5 min. The tissue for cultural analysis was cut up finely with a sterile scalpel and also vortex mixed in TM for 5 min. Serial dilutions in 0.85% saline were performed to obtain quantitative culture counts by plating onto the following media (LabM, Bury, United Kingdom): blood agar and Fastidious Anaerobe Agar (FAA), both supplemented with 5% (vol/vol) horse blood, MacConkey no. 3, and Sabouraud medium. All plates except FAA were incubated aerobically at 37°C. FAA plates were incubated under anaerobic conditions at 37°C. Prolonged incubation of the macerated tissue was undertaken in Fastidious Anaerobe Broth for 7 days prior to plating on FAA to allow recovery of fastidious and slow-growing anaerobic species. Primary isolation plates were initially examined after 48 h and then incubated for at least 10 days. Identification of bacteria followed standard microbiological schemes by examination of a range of phenotypic properties (staining reactions, colonial morphology, and carbohydrate fermentation patterns) and, where appropriate, by using commercial identification kits. In this way it was possible to identify each isolate to the genus or species level. Isolates were stored at −80°C until needed.
DNA extraction and manipulation.
Molecular analysis was undertaken on the tissue samples only. DNA was extracted by standard techniques (24), with the addition of a freeze-thaw step that was repeated three times (2 min in liquid nitrogen and 2 min at 65°C). DNA was extracted directly from the tissue sample and also from individual microbial species cultured from the same wound.
PCR amplification.
The V3 variable region of the 16S rRNA gene was amplified by PCR with primers 341f and 518r (Escherichia coli numbering) as described by Muyzer et al. (32). An improved 40-nucleotide GC clamp was attached to the 5′ end of the forward primer, 5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-3′ (15). The previous use of such universal primers is well documented in the literature for amplification of a broad range of bacteria, including their application in a clinical setting (28, 45, 50). The expected product size for the PCR amplification with these primers was a single band 193 bp in length. The PCR mixture (50 μl) contained 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 25 pmol of each primer, 0.2 mM concentrations of each deoxynucleoside triphosphate (Roche, Lewes, United Kingdom), 1 μg of DNA, 1.5 mM MgCl2 (Promega, Southampton, United Kingdom), and 1.25 U of Taq DNA polymerase (Promega). Touchdown PCR was performed with an initial denaturation step of 95°C for 5 min; followed by 20 cycles of 94°C for 1 min, 65°C (−0.5°C cycle−1) for 45 s, and 72°C for 1 min; followed by 10 cycles of 94°C for 1 min, 55°C for 45 s, and 72°C for 1 min; followed finally by a final elongation step of 72°C for 10 min. Water controls in place of target DNA were carried out for all PCRs. All PCRs were carried out under a laminar flow hood by using filter-sealed tips, ultrapure water, and sterile, clean-room-produced tips and PCR tubes. Molecular weight markers (1 Kb Plus; Invitrogen, Paisley, United Kingdom) were run in the outer lanes of each gel. Gels were stained with 0.5 μg of ethidium bromide (Sigma, Poole, United Kingdom) ml−1 and then visualized by using the Molecular Analyst image analysis system (Bio-Rad Laboratories, Ltd., Hemel Hempstead, United Kingdom).
DGGE.
DGGE was done by using the Bio-Rad D-CODE system. Four bacterial wound isolates (Peptostreptococcus sp., Proteus mirabilis, Pseudomonas aeruginosa, and Micrococcus sp.) chosen on the basis of their widely differing G+C content (27 to 37%, 38 to 45%, 39 to 59%, and 58 to 70%, respectively) (29) were used to optimize the denaturant gradient on a perpendicular gel, and the isolates were then used as marker strains for all subsequent DGGE gels. PCRs for the “in-house” molecular weight markers were done in bulk and completely separately from all other PCRs. The perpendicular gel contained a denaturing gradient of 0 to 100% and was formed according to the manufacturer's instructions. DGGE optimization demonstrated that the optimal range of denaturants for the control strains was between 30 and 60%.
Parallel DGGE was performed essentially as described previously (31, 32). PCR fragments were separated by using 10% (wt/vol) polyacrylamide (acrylamide-bisacrylamide [37.5:1]; Sigma) containing a 30 to 60% linear gradient of denaturants (urea and formamide) increasing in the direction of electrophoresis. Gradients were formed by using a Bio-Rad Gradient Former model 385. PCR samples were applied to gels in aliquots of 20 μl per lane, with 10 μl of DGGE loading buffer and 10 μl of agarose loading buffer. The running buffer used was 1× TAE buffer (40 mM Tris, 20 mM acetic acid, 50 mM EDTA [pH 8.0]). Electrophoresis was performed at 56°C at 70 V for 10 min, followed by 170 V for 3 h and 50 min. Gels were stained with SYBR Green I (Sigma) diluted in 1× TAE buffer (1:10,000) for 15 min and visualized by using the Gel Doc system (Bio-Rad). The PCR products from all cultured isolates from one wound were electrophoresed alongside those amplified directly from tissue, allowing for direct comparison between cultured and amplified products from the same wound.
Excision and sequencing of DGGE fragments.
Bands on the DGGE gel derived from tissue samples that did not align to any bands from bacteria cultured from the same wound were considered unculturable and were excised and sequenced. Bands were cut from the DGGE gels by using sterile scalpel blades and purified by using the “crush-and-soak” method (44). DGGE bands from three cultured wound isolates were also excised and sequenced as controls. The purified DNA was then reamplified by using primers 341f and 534r as described previously. PCR was followed by ligation into the pCR2.1 TOPO vector (Invitrogen) and transformation into Top10-competent Escherichia coli cells (Invitrogen). Blue-white screening of transformants was done on Luria-Bertani agar (44) containing 50 mg of ampicillin (Sigma) ml−1 and top spread with 40 ml of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Sigma) (20 mg ml−1). Clones were screened for inserts of the correct size by M13 amplification. DNA for sequencing was prepared from clones by using Wizard Plus SV Minipreps (Promega). Two clones from each excised band were sequenced, and both strands were sequenced on an automated laser fluorescence sequencer (ABI Prism 377; Applied Biosystems, Warrington, United Kingdom) by using primers M13f and M13r, giving 100% double coverage of the 193-bp 16S ribosomal DNA (rDNA) product for phylogenetic analysis. Sequences obtained were compared to those in the EMBL database (release 71) (47) by using FASTA3 (35, 36) at the European Bioinformatics Institute and with those of the Ribosomal Database Project (RDP) by using SEQUENCE MATCH (10) to identify closely related gene sequences. CHIMERA CHECK (10) at the RDP was used to detect possible chimeric sequence structures, along with manual inspection of the alignments.
Nucleotide sequence accession numbers.
Sequences for selected clones in this study have been submitted to EMBL/GenBank under accession numbers AJ605727 to AJ605739.
RESULTS
Cultural analysis.
Bacterial wound isolates cultured from all 18 wounds (both healing and nonhealing) were identified to genus or species level when possible, and these results are present in Tables 1 and 2. In a few cases in which a bacterial isolate could not be identified due to the limitations of the identification kits used, a description of the cell morphology is given instead. A number of strains were cultured from only the swab or biopsy and not from both sites.
TABLE 1.
Cultured bacterial isolates from swabs and biopsies from individual patients with healing chronic venous leg ulcers and DGGE analysis of the so-called unculturable population from the same wounds
| Patient | Wound surface (swab)a | Wound bed (biopsy)b | DGGE (PCR-cloning/sequencing)c |
|---|---|---|---|
| 12 | Coagulase-negative Staphylococcus sp. and Staphylococcus aureus | Coagulase-negative Staphylococcus sp. | Pseudomonassp. (a) (Pseudomonas putida subgroup) and Salmonellasp. (b) |
| 19 | Coagulase-negative Staphylococcus sp., Micrococcus sp. and Streptococcus sp. | Coagulase-negative Staphylococcus sp. and Micrococcus sp. | Stenotrophomonas maltophilia, (a) Paenibacillus sp., and Pseudomonassp. (b) (Pseudomonas putida subgroup) |
| 24 | Eubacteriumsp. | Staphylococcus aureus, coagulase-negativeStaphylococcus sp., and Peptostreptococcussp. | Staphylococcus sp. (a) (Staphylococcus aureus subgroup) and Pseudomonassp. (b) (Pseudomonas putida subgroup) |
| 26 | Pseudomonassp. and gram-negative nonfermenter | Staphylococcus aureus, coagulase-negativeStaphylococcus sp., and Micrococcussp. | Sphinogomonassp. (a) and Staphylococcus sp. (b) (Staphylococcus aureus subgroup) |
| 28 | Proteus mirabilis, Staphylococcus aureus, coagulase-negativeStaphylococcus sp., Peptostreptococcussp., Bordetellasp., and Stenotrophomonas maltophilia | Proteus mirabilis and Bacteroidessp. | Sphinogomonassp. (a) and Peptoniphilus sp. (b) |
| 48 | Coagulase-negative Staphylococcus sp., Pseudomonas sp., Peptostreptococcussp., Eubacterium sp., and Aerococcussp. | Coagulase-negative Staphylococcus sp., Pseudomonas sp., Streptococcussp., and Proteus mirabilis | Pseudomonas sp. (a) (Pseudomonas aeruginosa subgroup) and Streptococcus dysgalactiae (b) |
| 49 | Coagulase-negative Staphylococcussp., gram-negative nonfermenter, and Aerococcussp. | Pseudomonassp. and Salmonellasp. | Corynebacterium striatum, unidentified β-proteobacteriumd, and-e (c) |
| 53 | Coagulase-negative Staphylococcus sp., Pseudomonas sp., Peptostreptococcus magnus, and Streptococcussp. | Coagulase-negative Staphylococcus sp., Pseudomonas sp., Peptostreptococcus magnus, and Staphylococcus aureus | - |
Underlined species in this column were isolated only from the swab and not from the biopsy.
Underlined species in this column were isolated only from the biopsy and not from the swab.
Underlined species in this column were identified only from PCR cloning-sequencing and not cultured from the wound, i.e., uncultured or unculturable. Where clones could not be identified to a single specific species, the closest related subgroup is given in parentheses. Designations (a), (b), (c), and (d) refer to the bands excised from the DGGE gel in Fig. 1.
The accession no. of the sequence showing closest identity is AF467355.
Identified as the Taq polymerase contaminant, Thermus sp.
TABLE 2.
Cultured bacterial isolates from swabs and biopsies from individual patients with nonhealing chronic venous leg ulcers and DGGE analysis of the so-called unculturable population from the same wounds
| Patient | Wound surface (swab)a | Wound bed (biopsy)b | DGGE (PCR-cloning/sequencing)c |
|---|---|---|---|
| 2 | Staphylococcus aureus, Streptococcus sp., Peptostreptococcus sp., Pseudomonas sp., and Bacteroides fragilis | Staphylococcus aureus, Streptococcus sp., Peptostreptococcus sp., Pseudomonas sp., gram-negative rod (facultative), coagulase-negativeStaphylococcus sp., and Micrococcussp. | Gemellasp. (a) and Peptoniphilus harei (b) |
| 5 | Pseudomonas sp. | Pseudomonas sp. | Pseudomonas sp. (a) (Pseudomonas aeruginosa subgroup) and Staphylococcussp. (b) (Staphylococcus epidermidis group) |
| 6 | Coagulase-negative Staphylococcus sp., Micrococcus sp., Peptostreptococcussp., Eubacteriumsp., and Streptococcussp. | Coagulase-negative Staphylococcus sp., Micrococcus sp., and gram-positive rod (facultative) | Serratia marcescens (a) and Staphylococcus sp. (b) (Staphylococcus epidermidis group) |
| 7 | Coagulase-negative Staphylococcus sp., Pseudomonas sp., Candida sp.d, Micrococcussp., and Eubacteriumsp. | Coagulase-negative Staphylococcus sp., Pseudomonas sp., and Candida sp.d | Pseudomonas sp. (a) (Pseudomonas tolaasir subgroup) and Staphylococcus sp. (b) (Staphylococcus epidermidis group) |
| 14 | Staphylococcus aureus, Micrococcus sp., and coagulase-negativeStaphylococcus sp. | Staphylococcus aureus and Micrococcus sp. | Pseudomonassp. (a) (Pseudomonas putida subgroup) |
| 15 | Staphylococcus aureus, Bordetella sp., Micrococcussp., and Pasteurellasp. | Staphylococcus aureus, Bordetella sp., and Streptococcussp. | Corynebacteriumsp. (a) |
| 17 | Staphylococcus aureus, Micrococcus sp., Serratia sp., and Pseudomonassp. | Staphylococcus aureus, Micrococcus sp., Serratia sp., and Streptococcussp. | Pseudomonas sp. (a) (Pseudomonas tolaasii subgroup), Pseudomonas sp. (b) (Pseudomonas putida subgroup), and Afipiasp. (c) |
| 18 | Pseudomonas sp., Enterobacter sp., Staphylococcus aureus, and gram-positive rod | Pseudomonas sp., Enterobacter sp., Micrococcussp., Enterococcussp., and gram-negative nonfermenter | Staphylococcus sp. (a) (Staphylococcus aureus group) and Pseudomonas sp. (b) (Pseudomonas tolaasii subgroup) |
| 25 | Coagulase-negative Staphylococcus sp., Micrococcussp., Pseudomonassp., and Corynebacterium sp. | Coagulase-negative Staphylococcus sp., Staphylococcus aureus, and Streptococcussp. | Pseudomonas sp. (a) (Pseudomonas putida subgroup), Pseudomonas sp. (b) (Pseudomonas tolaasii subgroup), and Sphingomonassp. (c) |
| 29 | Streptococcus sp., Staphylococcus aureus, and Micrococcussp. | Streptococcus sp., gram-positive rod, Eubacterium sp., Pseudomonassp., Proteussp., and Bacteroidessp. | Pseudomonas sp. (a) (Pseudomonas putida subgroup), Pseudomonas sp. (b) (Pseudomonas putida subgroup), Pseudomonas sp. (c) (Pseudomonas putida subgroup), and Proteus mirabilis (d) |
A high proportion of the wound isolates were found to be Staphylococcus, Pseudomonas, Streptococcus, and Micrococcus species, a number of which were only detected by molecular means. When the results of the swab, tissue, and DGGE (sequence analysis) analyses were pooled, all patients were found to carry Staphylococcus sp.—either Staphylococcus aureus or a coagulase-negative staphylococcus strain. A high level of pseudomonads was also detected; pseudomonads were present in 14 of 18 (80%) patients. When we compared the carriage of Micrococcus and Streptococcus sp. by healers and nonhealers, 2 of 8 (25%) of healers were found to carry Streptococcus sp. in contrast to 6 of 10 (60%) of nonhealers, and for Micrococcus sp. the ratio was even higher: 2 of 8 (25%) healers to 9 of 10 (90%) nonhealers. These organisms are all aerobes or facultative anaerobes, whereas comparison of the carriage of strict anaerobes showed that there was little difference between healers (4 of 8) and nonhealers (4 of 10).
DGGE.
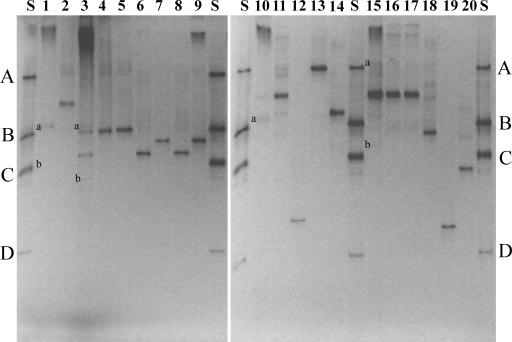
The DGGE profiles of healing and nonhealing wounds amplified directly from chronic wound tissue were compared in Fig. 1. The individual profiles proved to be complex, with the mean number of bands for the healers and nonhealers being 8 and 10, respectively. Although patients had some bands in common, each patient produced a unique banding pattern that was reproducible between successive PCRs (results not shown). Significantly, however, an apparently identical band was found to be present in all of the samples for both healing and nonhealing wounds (highlighted with an arrow in Fig. 1) and was identified from sequence analysis of seven of these bands as being most closely related to Pseudomonas sp. despite the fact that this species was not cultured from the wounds of all patients (i.e., patients 6, 12, 15, 19, 24, and 28). The shortness of the sequence information obtained for these bands was insufficient to allow for species identification, although all but one band 48a (belonging to the Pseudomonas aeruginosa subgroup) had exactly the same identity to up to 13 species in the Pseudomonas putida and Pseudomonas tolaasii subgroups of the γ-proteobacteria. In some cases, the PCR signal for this common band was very weak (i.e., for patients 2, 5, 15, 19, 26, 48, 49, and 53), even though for most of these patients (i.e., patients 2, 5, 26, 48, 49, and 53) high numbers of pseudomonads had been cultured (results not shown). The intensity of the PCR-derived signal for this common band was in general markedly stronger for the nonhealers than for the healers. For a few patients the signal was very faint, although still clearly visible. Also, migration of this fragment in patient 15, was marginally less than for the common bands in all of the other patients. If we assume that the ratio of target human to bacterial DNA does not change significantly between samples and since standard concentrations of target DNA (1 μg) were used in all PCRs, the PCR signal could be regarded as semiquantitative; the strength of signal therefore suggests that the bacterial numbers for this organism may be significantly higher in the nonhealers compared to the healers. From the quantitative cultural analysis of these wounds (results not shown), a large range of Pseudomonas bacterial counts was obtained (8 × 102 to 3.6 × 108 CFU), and these counts revealed no significant differences between healing and nonhealing wounds. Therefore, the PCR results suggest that there may have been an underestimation of bacterial numbers (for Pseudomonas sp. at least) by cultural analysis alone.
FIG. 1.
DGGE analysis showing profiles of healing (n = 8) and nonhealing wounds (n = 10) of 16S rRNA amplified directly from chronic wound tissue. Lanes are identified by patient number. “S” is an in-house four-strain standard comprising Peptostreptococcus sp. (A), Pseudomonas sp. (B), Proteus mirabilis (C), and Micrococcus sp. (D). Bands amplified from the tissues are labeled a to d and correlate to bands that were excised and sequenced. The arrow indicates an apparently identical band found in all tissue samples and identified from sequence analysis to be Pseudomonas sp.
PCR-DGGE and sequencing.
DGGE analysis of 16S rRNA gene fragments from chronic tissues were compared to those amplified from cultured isolates (from both swabs and tissues) from the same wounds. Figure 2 shows the tissue profiles from four patients compared to the profiles obtained from the cultured isolates from the same patients. In these four patients, as well as in the rest of the total 18 patients, the tissue profiles had bands present that were not represented by the bands in the profiles obtained from the cultured isolates from these wounds. These bands are marked a, b, c, and d in Fig. 1 and 2. The 39 bands were excised (from Fig. 1), cloned, and sequenced, allowing the DNA from uncultured bacteria to be identified; the results are presented in Tables 1 and 2.
FIG. 2.
DGGE analysis of 16S rRNA gene fragments from chronic tissues of four patients (all healers) compared to those amplified from cultured isolates from the same wounds. Patients: 12, lanes 1 to 2; 48, lanes 3 to 9; 14, lanes 10 to 14; 26, lanes 15 to 20. Lanes: 1, tissue 12; 2, Staphylococcus aureus; 3, tissue 48; 4, Pseudomonas sp.; 5, Pseudomonas sp.; 6, Aerococcus sp.; 7, Staphylococcus sp.; 8, Streptococcus sp.; 9, Staphylococcus sp.; 10, tissue 14; 11, Staphylococcus aureus; 12, Staphylococcus sp.; 13, Peptostreptococcus sp.; 14, Eubacterium sp.; 15, tissue 26; 16, Staphylococcus aureus; 17, Staphylococcus aureus; 18, Staphylococcus sp.; 19, Micrococcus sp.; 20, gram-negative rod. “S” is an in-house four-strain standard comprising Peptostreptococcus sp. (A), Pseudomonas sp. (B), Proteus mirabilis (C), and Micrococcus sp. (D), all of which were isolated from chronic wounds. Although the bands amplified directly from tissue (labeled a or b) are faint in Fig. 2, they correlate directly with the bands labeled a and b in Fig. 1, from which gel these bands were excised and sequenced. Isolates of the same species from the same wound tissue (i.e., the same wound biopsy) had different colony morphologies.
All 39 different DGGE fragments of partial 16S rRNA genes were sequenced. Despite the use of such short sequences (ca. 200 nucleotides), assignation to its closest relatives was still possible. The percent identity of all clones to known database sequences was >95%, with only one sequence having a lower identity (<93%), namely, 48b (Streptococcus dysgalactiae). The latter was a likely chimera between Streptococcus dysgalactiae and Pseudomonas mosselii (genera of both having been cultured from this wound). None of the other sequences showed evidence of chimera formation. Although a range of bacterial species amplified by using DGGE were also cultured from the wounds, a number of amplification products were from uncultured microorganisms. These included common wound isolates such as pseudomonads and staphylococci as well as Corynebacterium, Salmonella, Serratia, and Stenotrophomonas spp. However, although not cultured from the wounds from which they had been amplified, each of these groups of uncultured organisms had been cultured from at least one wound in this patient group. In contrast, Afipia, Gemella, Paenibacillus, and Sphingomonas spp. were shown to be present by molecular means but were not isolated by culture from any wound in the present study and indeed were not known to have been isolated from chronic wounds in general. All three Sphingomonas sequences were closely related. Two additional fragments were identified by DGGE (from patients 49 and 53) as Thermus spp. Despite the fact that the controls were clear, these must have arisen due to contamination of the Taq polymerase with DNA from Thermus aquaticus YT1 from which it is purified, rather than its being a resident of chronic wounds, i.e., an artifact of PCR-cloning technology (22, 46). As an additional control, one of the bands from the four strain in-house standard ladder was also excised and sequenced (Fig. 1, band A) and was found to be most closely related to Pseudomonas (98% identity over 196 nucleotides; S_ab > 0.930), thereby confirming the API genus identification of this strain.
DISCUSSION
In this study we have applied the use of DGGE and PCR sequencing to the analysis of the chronic wound microflora. This allowed us to quickly and efficiently analyze the whole bacterial population of individual wounds by using a single PCR and to run samples alongside one another on the same gel for direct comparison. In this way, it was possible to concurrently obtain considerable information about the species composition of multiple wounds (18 in all), which would have been prohibitively labor-intensive by direct cloning and sequencing. Bands of specific interest, present in total wound DNA but not corresponding to any bands amplified from the cultured isolates from those wounds (and therefore representing uncultured organisms), were then excised and sequenced.
One of the limitations of DGGE is the length of sequence that can be separated effectively using this technique. The PCR products for DGGE analysis must be <500 bp (33) to allow for efficient resolution and analysis, thereby undoubtedly limiting the amount of sequence information that can be obtained. Although the sequences derived from the DGGE-PCR cloning of products of such small size is insufficient to allow great precision in the construction of phylogenetic trees, it does allow for a presumptive identification at least to genus level (54). The primers used in the present study yielded a very small product (<200 bp), and this was insufficient in many cases to give unambiguous species identification. Better discrimination, however, may be possible with primer products of increased length or from another variable region of the 16S rRNA. In general, DGGE will only separate the predominant species present in a community. In addition, comigration can be a problem for retrieving clean sequences from individual bands. It is also clear from other studies with DGGE that 16S rRNA gene sequences affiliated to specific bacterial species can be found in more than one position in DGGE gels. This is due to the presence of multiple rrN operons. Paenibacillus polymyxa produces more than one DGGE band due to slight sequence heterogeneities between operons (34), and as many as nine different positions were found among 10 Corynebacterium-affiliated sequences (45). There is evidence of multiple operons in the in-house DNA ladder used in the present study in Fig. 1 and 2, particularly since both of these gels were overexposed to enhance the detail of tissue-amplified DNAs. Despite these shortcomings, however, DGGE remains a valuable technique, providing substantial information about complex mixed bacterial populations.
PCR has previously been shown to be more sensitive than culture for the detection of bacteria in clinical samples (40, 45). In the present study, DGGE allowed the identification of a number of strains not detected by cultural means, with ca. 40% of the DGGE fragments sequenced representing organisms not cultured from the wound from which they had been amplified. This highlights the fact that a significant proportion of the resident microflora was not amenable to analysis by culture. In keeping with other studies of the human microflora, it was not that these “molecular isolates” represented unculturable species. In fact, the majority of these amplification products represented strains which, although not cultured from the wound in question, had >95% sequence identity to typical wound microflora organisms, and similar strains were cultured from other wounds in this set of patients. Rather, they were uncultured, perhaps as a result of being in a viable but unculturable state. Moreover, most of these “uncultured” bacteria (i.e., those detected by DGGE alone) were isolated from samples containing large numbers of related bacteria. Hence, they may not have been detected during the initial cultural screening due to competition with other more numerous species and overgrowth on the selective/nonselective media by related microflora. Only one sequence showed <93% sequence identity to any database sequences but this was determined to be a chimeric clone, 48b (S. pyogenes).
Four organisms were identified by sequencing which have not traditionally been associated with chronic wounds. Paenibacillus spp. have been isolated from acute wounds (7), whereas Gemella spp. are closely related to Staphylococcus spp., which were the predominant organisms found in all of the wounds. Sphingomonas and Afipia spp. are closely related to each other, both belonging to the α-division of the proteobacteria, and are species that have not, to our knowledge, been isolated from chronic wounds. Sphingomonas sp. is ubiquitous in the natural environment, has been implicated in community-acquired and nosocomial infections, and was isolated in the present study from both healed and nonhealed patient wounds. Afipia felis is a facultatively intracellular pathogen, although the role of other Afipia sp. as pathogens remains speculative (29).
The contrast between healing and nonhealing wounds was only apparent when looking at the carriage of Micrococcus and Streptococcus spp. in the wounds, both having a higher incidence in the nonhealer group than in the healers. However, although these percentages appear to be potentially significant in this relatively small group of patients, in a larger study in which our group evaluated 66 patients by cultural analysis of the wound microfloras (unpublished results), no significant associations were found between specific bacterial groups and the healing or nonhealing phenotype. Streptococcus and Micrococcus spp. are commensal organisms, but they are also opportunistic pathogens. Both species have been implicated in a range of human infections with Streptococcus sp. in particular initiating (well-documented) soft tissue and skin infections (2) and Micrococcus sp. being implicated in infections in certain immunocompromised patients (29).
Comparison of the amplification signal of the DGGE bands in the nonhealing group of patients to those of the healing group, suggested a possible discrepancy with pseudomonad counts by culture. PCR bias has been widely reported as a reason to view the quantitative interpretation of PCR results with caution (1, 38, 49), although a recent study (8) gives some validity to the quantitative nature of 16S rDNA PCR-DGGE profiles of bacterial communities generated by using this same primer pair (32). Moreover, the particular ratios of human to bacterial DNA are unknown in these samples, and it is these that will significantly influence any PCR biases observed (49). Interestingly, however, bacterial communities in human neonates (17) showed a similar anomaly, with a predominance of Ruminococcus sp. over many months being revealed by DGGE, although these species had been shown by previous cultural analysis to be present in only moderate or low numbers. Recently, real-time PCR was used for the rapid quantification of Pseudomonas aeruginosa in acute wound biopsy samples (37). The application of techniques such as real-time or quantitative PCR (20) to wound DNA would be useful to quantify the pseudomonad (and other species) populations in these wounds to a high degree of accuracy and hence elucidate whether they have a particular significance in nonhealing. Moreover, the sequences generated in the present study can be used directly to aid the design of new primers and probes specifically for such applications. The advantage of such molecular analysis of clinical samples is to further study the relevance and association with healing of selected phylotypes in other samples by DNA probing or PCR analysis by targeting phylotype-specific sequences.
Although a number of strategies such as DNase treatment or restriction endonuclease digestion have been tested for decontaminating PCR reagents, it would seem that none has been wholly successful (9, 25). Hence, it still remains a problem when trying to detect low concentrations of bacterial 16S rDNAs with broad-range PCR primers (22). The fact that we amplified two fragments that appeared to arise from native purified Taq polymerase suggests that a pre-PCR treatment would have been appropriate in the present study to prevent universal primers from amplifying contaminating bacterial DNA. Hence, additional strategies, along with those currently used (i.e., clean-room produced plastics, filter tips, dedicated PCR room, and ultrapure DNase- and RNase-treated water), such as DNase treatment of Taq polymerase (and perhaps other PCR reagents), should be recommended for amplification of bacterial DNA from such clinical samples to improve the validity of universal 16S rRNA gene PCR results by prevention of false positives (22).
The use of multiple primers in conjunction with PCR and sequencing to analyze clinical samples has been highlighted previously in the study of dental (14, 26) and gastrointestinal (48) microflora and is also now beginning to be applied to the field of wound microbiology (41). In a previous study of a single chronic venous leg ulcer (23), we found that using multiple primer sets in conjunction with the sequencing of large numbers of clones, a far more detailed picture of the wound microflora was revealed. Indeed, a far greater diversity of organisms was identified in this way.
The results of the present study together with our previous work (23) have demonstrated the greater diversity of the wound microflora as assessed by a molecular approach in comparison to culture alone. We have also shown that, by the use of DGGE profiling, the molecular analysis of the human microflora need not be limited to small numbers of samples as would be the case for the cloning and sequencing approach. By extension, this labor-saving technique offers the possibility of new clinical approaches, for example, monitoring the status of wounds by serial DGGE profiling. Although the additional complexity documented by a molecular approach could be seen to further complicate the study of these chronic wounds, it will only be by the comprehensive microbial analysis of clinical specimens and the thorough investigation of the biases of both cultural and molecular approaches that associations between specific bacterial types and healing will be established or refuted.
Acknowledgments
We gratefully acknowledge funding for the research described in this study from Leeds Oral Surgery Trust, Department of Oral Surgery, Medicine, and Pathology (Ph.D. studentship for C.E.D.) and from Research into Ageing (project number 196; postdoctoral support for K.E.H.).
We thank Gareth Lewis for sequencing.
REFERENCES
- 1.Becker, S., P. Böger, R. Oehlmann, and A. Ernst. 2000. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analysis of microbial communities. Appl. Environ. Microbiol. 66:4945-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisno, A. L., and D. L. Stevens. 1996. Streptococcal infections of skin and soft tissues. N. Engl. J. Med. 334:240-245. [DOI] [PubMed] [Google Scholar]
- 3.Bowden, G. H., and J. M. Hardie. 1971. Anaerobic organisms from the mouth, p. 177-205. In D. A. Shapton and R. G. Board (ed.), Isolation of anaerobes. Academic Press, Ltd., London, England.
- 4.Bowler, P. G., B. I. Duerden, and D. G. Armstrong. 2001. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 14:244-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowler, P. G. 1998. The anaerobic and aerobic microbiology of wounds: a review. Wounds 10:170-178. [Google Scholar]
- 6.Bowler, P. G. 1999. The prevalence and significance of anaerobic bacteria in wounds. M.Ph. thesis. University of Wales College of Medicine, Cardiff, United Kingdom.
- 7.Bowler, P. G., and B. J. Davies. 1999. The microbiology of acute and chronic wounds. Wounds 11:72-79. [Google Scholar]
- 8.Bruggemann, J., J. R. Stephen, Y.-J. Chang, S. J. Macnaughton, G. A. Kowalchuk, E. Kline, and D. C. White. 2000. Competitive PCR-DGGE analysis of bacterial mixtures an internal standard and an appraisal of template enumeration accuracy. J. Microbiol. Methods 40:111-123. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, N. M., P. Adamson, and N. Okravi. 1999. Elimination of bacterial DNA from Taq DNA polymerases by restriction endonuclease digestion. J. Clin. Microbiol. 37:3402-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagher, F., S. Alongi, and A. Smith. 1978. Bacterial studies of leg ulcers. Angiology 29:641-653. [DOI] [PubMed] [Google Scholar]
- 12.Daltrey, D., B. Rhodes, and J. Chattwood. 1981. Investigation into the microbial flora of healing and non-herlaing decubitus ulcers. J. Clin. Pathol. 34:701-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies, C. E., M. J. Wilson, K. E. Hill, P. Stephens, C. M. Hill, K. G. Harding, and D. W. Thomas. 2001. Use of molecular techniques to study microbial diversity in the skin: chronic wounds re-evaluated. Wound Repair Regen. 9:332-340. [DOI] [PubMed] [Google Scholar]
- 14.Dymock, D., A. J. Weightman, C. Sully, and W. G. Wade. 1996. Molecular analysis of microflora associated with dentoalveolaer abscesses. J. Clin. Microbiol. 34:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellis, R. J., P. Morgan, A. J. Weightman, and J. C. Fry. 2003. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy metal contaminated soil. Appl. Environ. Microbiol. 69:3223-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eriksson, G., A.-E. Eklund, and L. Kallings. 1984. The clinical significance of bacterial growth in venous leg ulcers. Scand. J. Infect. Dis. 16:175-180. [DOI] [PubMed] [Google Scholar]
- 17.Favier, C. F., E. E. Vaughan, W. M. De Vos, and A. D. L. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galpin, J., A. Chow, A. Bayer, and L. Guze. 1976. Sepsis associated with decubitus ulcers. Am. J. Med. 61:346-349. [DOI] [PubMed] [Google Scholar]
- 19.Halbert, A. R., M. C. Stacey, J. B. Rohr, and A. Jopp-McKay. 1992. The effect of bacterial colonization on venous ulcer healing. Australas. J. Dermatol. 33:75-80. [DOI] [PubMed] [Google Scholar]
- 20.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 21.Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heininger, A., M. Binder, A. Ellinger, K. Botzenhart, K. Unertl, and G. Döring. 2003. DNase pretreatment of master mix reagents improves the validity of universal 16S rRNA gene PCR results. J. Clin. Microbiol. 41:1763-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill, K. E., C. E. Davies, M. J. Wilson, P. Stephens, K. G. Harding, and D. W. Thomas. 2003. Molecular analysis of the microflora in chronic venous leg ulceration. J. Med. Microbiol. 52:365-369. [DOI] [PubMed] [Google Scholar]
- 24.Keay, S., C.-O. Zhang, B. R. Baldwin, R. B. Alexander, and J. W. Warren. 1998. Polymerase chain reaction amplification of bacterial 16S rRNA genes from cold-cup biopsy forceps. J. Urol. 160:2229-2231. [DOI] [PubMed] [Google Scholar]
- 25.Klaschik, S., L. E. Lehmann, A. Raadts, A. Hoeft, and F. Stuber. 2002. Comparison of different decontamination methods for reagents to detect low concentrations of bacterial 16S DNA by real-time-PCR. Mol. Biotechnol. 22:231-242. [DOI] [PubMed] [Google Scholar]
- 26.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Pro. Nat. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lookingbill, D., S. Miller, and R. Knowles. 1978. Bacteriology of chronic leg ulcers. Arch. Dermatol. 114:1765-1768. [PubMed] [Google Scholar]
- 28.Miller, M. R., C. J. Linton, D. Glancy, M. Hall, and H. Jalal. 1996. Application of 16S rRNA gene PCR to study bowel flora of preterm infants with or without necrotizing enterocolitis. J. Clin. Microbiol. 34:2506-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 30.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 73:127-141. [DOI] [PubMed] [Google Scholar]
- 31.Muyzer, G., S. Hottentrager, A. Teske, and C. Wawer. 1996. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA: a new approach to analyze the genetic diversity of mixed microbial communities, p. 1-23. In A. D. L. Akermanns, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 32.Muyzer, G., E. C. De Waal, and G. A. Uitterlinden. 1993. Profiling of complex populations by denaturing gradient gel electrophoresis analysis of polymerase-chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers, R. M., S. G. Fischer, L. S. Lerman, and T. Maniatis. 1985. Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel-electrophoresis. Nucleic Acids Res. 13:3131-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence analysis. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 37.Pirnay, J.-P., D. De Vos, L., Duinslaeger, P. Reper, C. Vandenvelde, P. Cornelis, and A. Vanderkelen. 2000. Quantification of Pseudomonas aeruginosa in wound biopsy samples: from bacterial culture to rapid “real-time” polymerase chain reaction. Crit. Care 4:255-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramelet, A.-A., and D. Perrenoud. 1999. Bacteriology of leg ulcers. Curr. Prob. Dermatol. 27:20-25. [DOI] [PubMed] [Google Scholar]
- 40.Rantakokko-Jalava, K., S. Nikkari, J. Jalave, E. Eerola, M. Skurnik, O. Meurman, O. Ruuskanen, A. Alanen, E. Kotilainen, P. Toivanen, and P. Kotilainen. 2000. Direct amplification of rRNA genes in diagnosis of bacterial infections. J. Clin. Microbiol. 38:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Redkar, R., J. Kalns, W. Butler, L. Krock, F. McCleskey, A. Salmen, E. Piepmeier, and V. DelVecchio. 2000. Identification of bacteria from a nonhealing diabetic foot wound by 16S rDNA sequencing. Mol. Cell. Probes 14:163-169. [DOI] [PubMed] [Google Scholar]
- 42.Robson, M. C., R. J. Mannari, P. D. Smith, and W. G. Payne. 1999. Maintenance of wound bacterial balance. Am. J. Surg. 178:399-402. [DOI] [PubMed] [Google Scholar]
- 43.Salaman, R. A. 1999. Compression therapy, neutrophil activation, and cutaneous blood flow in lower limb venous disease. M.D. thesis. University of Wales College of Medicine, Cardiff, United Kingdom.
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Schabereiter-Gurtner, C., S. Maca, S. Rölleke, K. Nigl, J. Lukas, A. Hirschl, W. Lubitz, and T. Barisani-Asenbauer. 2001. 16S rDNA-based identification of bacteria from conjunctival swabs by PCR and DGGE fingerprinting. Investig. Ophthalmol. Vis. Sci. 42:1164-1171. [PubMed] [Google Scholar]
- 46.Schmidt, T. M., B. Pace, and N. R. Pace. 1991. Detection of DNA contamination in Taq polymerase. BioTechniques 11:176-177. [PubMed] [Google Scholar]
- 47.Stoesser, G., W. Baker, A. van den Broek, E. Camon, M. Garcia-Pastor, C. Kanz, T. Kulikova, R. Leinonen, Q. Lin, V. Lombard, R. Lopez, N. Redaschi, P. Stoehr, M. A. Tuli, K. Tzouvara, and R. Vaughan. 2002. The EMBL nucleotide sequence database. Nucleic Acids Res. 30:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel species within the human gut. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki, M., and S. J. Giovanonni. 1996. Bias caused by template annealing in the amplification mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Temmermann, R., I. Scheirlinck, G. Huys, and J. Swings. 2003. Culture-independent analysis of probiotic products by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:220-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial diversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe, K., M. Teramoto, H. Futamata, and S. Harayama. 1998. Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl. Environ. Microbiol. 64:4396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, M. J., A. J. Weightman, and W. G. Wade. 1997. Applications of molecular ecology in the characterisation of uncultured microorganisms associated with human disease. Rev. Med. Microbiol. 8:91-101. [Google Scholar]
- 54.Woese, C. R. 1987. Bacterial evolution. Microbial Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zoetendal, E. G., A. von Wright, T. Vilpponen-Salmela, K. Ben-Amor, A. D. L. Akkermans, and W. M. de Vos. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]