Abstract
The CRISPR/Cas9 system is a powerful genome editing tool for the production of genetically modified animals. To produce mutant mice, chimeric single-guide RNA (sgRNA) is cloned in a plasmid vector and a mixture of sgRNA and Cas9 are microinjected into the fertilized eggs. An issue associated with gene manipulation using the CRISPR/Cas9 system is that there can be off-target effects. To simplify the production of mutant mice with low risks of off-target effects caused by the CRISPR/Cas9 system, we demonstrated that genetically modified mice can be efficiently obtained using chemically synthesized CRISPR RNA (crRNA), trans-activating crRNA (tracrRNA), and modified Cas9s, such as the nickase version and FokI-fused catalytically inactive Cas9, by microinjection into fertilized eggs. Using this method, it is no longer necessary to clone sgRNA into a plasmid vector, and this enables high-throughput production of mutant mice.
Keywords: genome editing, CRISPR/Cas9, crRNA, mouse
Introduction
Genetically modified animals that are carrying mutations or inserted exogenous sequences are useful tools for understanding the functions of a gene of interest in vivo. Recent development of genome editing technologies using zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas9) systems enables rapid gene manipulation in many model organisms [2, 10, 12, 14, 17, 23]. DNA nucleases for genome editing introduce double-strand breaks (DSB) into specific target DNA sequences that induce non-homologous end-joining (NHEJ)-mediated insertions and deletions (indels) or homologous recombination (HR)-mediated insertions when an oligonucleotide or targeting vector containing exogenous DNA sequences with homologous arms is co-injected. The CRISPR/Cas9 system, which is currently the first choice among genome editing tools because of its efficiency and ease of use, consists of human codon-optimized RNA-guided Cas9 from Streptococcus pyogenes and a 102-nt single-guide RNA (sgRNA), including the 20-nt sequence of a target genomic locus. Cas9 and sgRNA form a complex that hybridizes to a target genomic sequence with an NGG protospacer-adjacent motif (PAM), which allows Cas9 to introduce the DSB [4].
The CRISPR/Cas9 system originated from an adaptive immune system in bacteria that required Cas DNA nuclease and two small RNAs, CRISPR RNA (crRNA), and trans-activating crRNA (tracrRNA) [5, 13]. At the 5′ end, crRNA contains a 20-nt sequence that recognizes a target DNA sequence. At its 3′ end, crRNA hybridizes to the 5′ part of the tracrRNA sequence (crRNA/tracrRNA loop) and forms a complex with the Cas protein. To simplify the crRNA/tracrRNA for genome editing applications, a chimeric hybrid of crRNA and tracrRNA that mimics the crRNA/tracrRNA loop has been developed, which was named sgRNA [4, 18]. So far, chimeric sgRNA has been widely used for genome editing of the CRISPR/Cas9 system by cloning a target sequence into an sgRNA expression vector.
There are two options to place an sgRNA in fertilized eggs to produce a mutant mouse by the CRISPR/Cas9 system: microinjection of an sgRNA/Cas9 mRNA into the cytoplasm or injection of an expression vector of sgRNA and Cas9 into the pronucleus of zygotes [20, 23]. In the latter case, there is the possibility of integration of the plasmid sequence into the mouse genome. We therefore believe that the former method is safer; however, it requires RNA synthesis by in vitro transcription, since the chemical synthesis of more than 100 bp of RNA is still difficult. On the other hand, the lengths of the crRNA and tracrRNA are within the range of RNA chemical syntheses. It has recently been shown that mutant mice can be produced by microinjection of crRNA, tracrRNA, and Cas9 into fertilized eggs [1].
An issue associated with gene manipulation using the CRISPR/Cas9 system is that there can be off-target effects, because the recognition sequence of Cas9 is smaller than 20 nt. To avoid off-target effects, various Cas9 mutants, such as Cas9 nickase (Cas9 D10A mutant; nCas9) and inactive Cas9 (dCas9) fused with the FokI DNA nuclease domain (FokI-dCas9; fCas9) have been developed, which lead to DSB only when two sgRNA pairs recognize sequences close to the opposite strands of genomic DNA [6, 21, 22]. We have recently reported that nCas9 and fCas9 can be useful for producing mutant mice by microinjection of in vitro-transcribed sgRNAs into zygotes [7]. To simplify the production of mutant mice with low risks of off-target effects by the CRISPR/Cas9 system, we examined whether genetically modified mice could be obtained using Cas9 and chemically synthesized crRNA and tracrRNA by microinjection into fertilized eggs.
Materials and Methods
Plasmids
Plasmids containing human codon-optimized wild type Cas9 (Cas9WT), nCas9, and fCas9 were obtained from Addgene (Plasmid #41815, #41816, and #52970, respectively).
RNAs
The target sequences of crRNA1, 2, 3, and 4 are the same as sgRNA B1, B2, A3, and A4 in a previous report [6]. The full sequences of crRNA1, 2, 3, and 4 and tracrRNA are listed in Supplementary Table 1. crRNA and tracrRNA were chemically synthesized and purified using HPLC by Fasmac (Atsugi, Kanagawa, Japan). mRNAs for Cas9WT, nCas9, and fCas9 were synthesized using an in vitro transcription reaction as described previously [7]. Briefly, for Cas9WT and nCas9 mRNA synthesis, the Cas9WT and nCas9 sequences were amplified from the coding plasmids by PCR using T7-Cas9 primers (Supplementary Table 1) to add the T7 promoter sequence. PCR products were used as a template for the mMESSAGE/mMACHINE T7 RNA synthesis kit (Ambion, Austin, TX, USA). RNAs were purified with a MegaClear kit (Ambion). For fCas9 mRNA synthesis, coding plasmids linearized with AgeI (Wako, Osaka, Japan) were treated as a template for Cas9WT and nCas9 mRNA synthesis.
Microinjection
Fertilized eggs were collected from superovulated F1 hybrid C57BL/6 and DBA/2 (B6D2F1) female mice crossed with B6D2F1 male mice. For the microinjection of the crRNA/tracrRNA/Cas9WT combination, the RNA concentrations were as follows: 155.3 ng/µl of crRNA, 94.7 ng/µl of tracrRNA, and 250 ng/µl of Cas9 mRNA. For denaturation and renaturation treatments, the RNA mixture of crRNA/tracrRNA was incubated at 95°C for 3 min and transferred to room temperature for 10 min, and then Cas9WT was added. For the microinjection of two crRNAs with tracrRNA and nCas9 or fCas9 mRNA, the RNA concentrations were as follows: 94.7 ng/µl of tracrRNA, 155.3 ng/µl of each crRNA, and 250 ng/µl of nCas9 or fCas9 mRNA. RNAs were injected into the cytoplasm of zygotes using FemtoJet (Eppendorf, Westbury, NY, USA). These embryos were cultured in KSOM medium and transferred to pseudopregnant female mice at the two-cell stage. All mice were obtained from the Sankyo Labo Service Corporation (Tokyo, Japan). The animal protocols were approved by the Animal Care and Use Committee of the National Research Institute for Child Health and Development. All experiments were conducted in accordance with these approved animal protocols.
Genotyping and off-target analysis
Genomic DNA was extracted from embryos at 9.5–10.5 days postcoitum (dpc). The genomic regions around the crRNA/tracrRNA target sequences were amplified by PCR using primers for the Bcr or Abl1 region (Supplementary Table 1). For direct sequencing, PCR products were treated with ExoSAP-IT (USB, Affymetrics Japan, Tokyo, Japan) and subjected to sequencing using the forward primers for Bcr and Abl1 loci, respectively. To determine the nucleotide sequence of mutated alleles, the PCR products were cloned into a pGEM-T Easy Vector (Promega Corp., Madison, WI, USA) and sequenced.
The genotypes were determined from the electropherograms. The criteria for identifying mutant and wild-type embryos were adopted from those reported by Hara et al. [7]. Mutants were further classified as monoallelic, biallelic, and mosaic mutants based on the results of cloning and the sequencing analyses. When wild-type and single mutated alleles were observed or a single mutated allele was observed without a wild-type allele, the genotype of the embryos was considered to be monoallelic. In cases where wild-type alleles were not detected and two or more mutant alleles were observed, the genotype was determined to be biallelic. When there were multiple mutated alleles with a wild-type allele, the genotype was considered to be mosaic. Each genotype was further confirmed by cloning of the PCR product into pGEM-T Easy Vector (Promega) and sequencing of 5–10 clones. All statistical analyses of genome editing efficiencies were assessed by χ2 test.
Confirmation of germline transmission of mutated alleles was performed by mating F0 adult male and C57BL/6N female mice. Tail tips of F1 pups were subjected to the genotyping analysis described above.
For the off-target analysis, possible off-target sites in each crRNA were searched by an online web tool (http://genome-engineering.org/), and two or three putative off-target sites with the highest scores were selected. The off-target sites for each crRNA were amplified by PCR with specific primers (Supplementary Table 1) and sequenced as described previously [7].
Results
Generation of mutant mice using crRNA/tracrRNA with Cas9WT mRNA
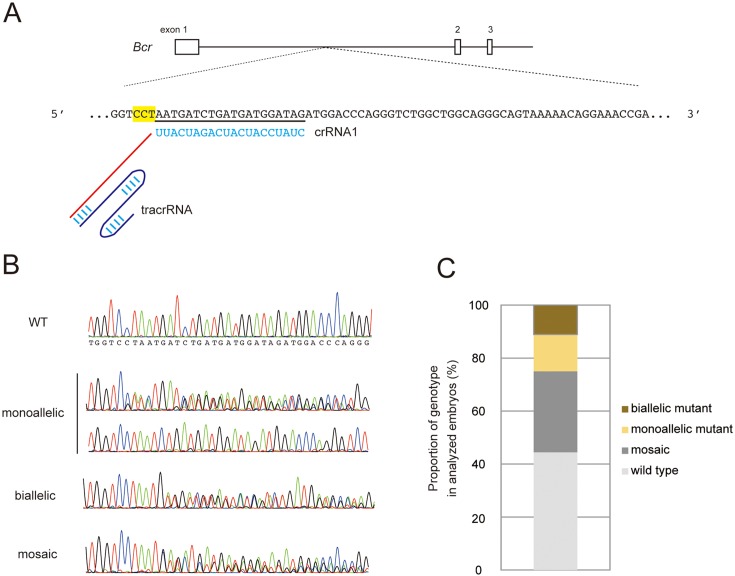
To verify the mutation efficiencies at target loci using crRNA/tracrRNA in mouse zygotes, we designed crRNA1, which targets the intronic region of the mouse Bcr locus as a model (Fig. 1A). The crRNA/tracrRNA were microinjected with Cas9WT mRNA into fertilized eggs. After embryo transfer at the two-cell stage, we obtained 36 embryos from 121 injected zygotes (29.8%) from two independent trials. The genotyping analyses performed by PCR-direct sequencing of the embryos showed that mutations around the crRNA target sequence were detected in 20 out of the 36 embryos (55.6%). We determined these genotypes by cloning and sequencing analyses, and the genotypes of embryos were classified into three groups: monoallelic, biallelic, and mosaic mutants (Fig. 1B). The results showed that there were five monoallelic mutants (25.0%) and four biallelic mutants (20.0%) out of the 20 mutant embryos. The remaining 11 mutated embryos (55.0%) contained two or more mutated alleles and a wild-type allele in each individual and were judged as mosaic mutants (Fig. 1C).
Fig. 1.
Generation of mutant mice using crRNA/tracrRNA in the Bcr locus. (A) The design of the crRNA in the mouse Bcr locus. The genomic structure is indicated at the top. The black bars and boxes show introns and exons, respectively. Blue letters and red lines show specific target sequences and common sequences of crRNA, respectively. The letters with yellow boxes designate PAM sequences. The blue curved lines indicate tracrRNA. Longitudinal blue bars show hydrogen bonds. (B) Representative electropherograms obtained from the sequencing analysis of wild- type, monoallelic, biallelic, and mosaic mutant embryos injected with crRNA1/tracrRNA/Cas9WT. The target sequence of crRNA1 is indicated with a black line. The PAM sequence is indicated with a yellow box. (C) The proportions of the genotypes of embryos obtained by crRNA1/tracrRNA/Cas9WT injection.
To examine whether precise annealing of the crRNA and tracrRNA results in higher mutation efficiencies in zygotes, the same mixture of crRNA/tracrRNA/Cas9WT RNA, but with denaturation and renaturation treatment, was microinjected into fertilized eggs. A total of 40 embryos were obtained from 122 injected zygotes obtained using this strategy. PCR-direct sequencing showed that 18 out of the 40 (45.0%) embryos were mutants, including 27.8% (5/18) monoallelic, 55.6% (10/18) biallelic, and 16.7% (3/18) mosaic mutants (Table 1). The microinjection of the denatured and renatured RNA mix could not improve the efficiency of genome editing compared with the injection of the untreated RNA mixture (untreated vs. denatured; 55.6% vs. 45.0%, P=0.3581), suggesting that the denaturation treatment is dispensable for the generation of mutant mice with the crRNA/tracrRNA mix. We therefore used the untreated crRNA/tracrRNA mixture for the subsequent experiments.
Table 1. A summary of the mutant mice produced using crRNA/tracrRNA.
| Denatured | Trial | Injected/2-cell | Transferred | Genotyped | Mutated | Monoallelic | Biallelic | Mosaic |
|---|---|---|---|---|---|---|---|---|
| untreated | 1 | 58/38 | 36 | 16 | 11 | 2 | 3 | 6 |
| 2 | 63/26 | 26 | 20 | 9 | 3 | 1 | 5 | |
| total | 121/64 | 62 | 36 | 20 (55.6) | 5 (25.0) | 4 (20.0) | 11 (55.0) | |
| denatured | 1 | 51/48 | 36 | 22 | 9 | 3 | 4 | 2 |
| 2 | 71/33 | 32 | 18 | 9 | 2 | 6 | 1 | |
| total | 122/81 | 68 | 40 | 18 (45.0) | 5 (27.8) | 10 (55.6) | 3 (16.7) | |
The numbers in parentheses in the “Mutated” columns represent the percentages calculated from the number of mutants relative to the number of genotyped embryos. The numbers in parentheses in the “Monoallelic”, “Biallelic” and “Mosaic” columns represent the percentages calculated from the number of each type relative to the number of “Mutated”.
Generation of mutant mice using two crRNAs with modified Cas9s
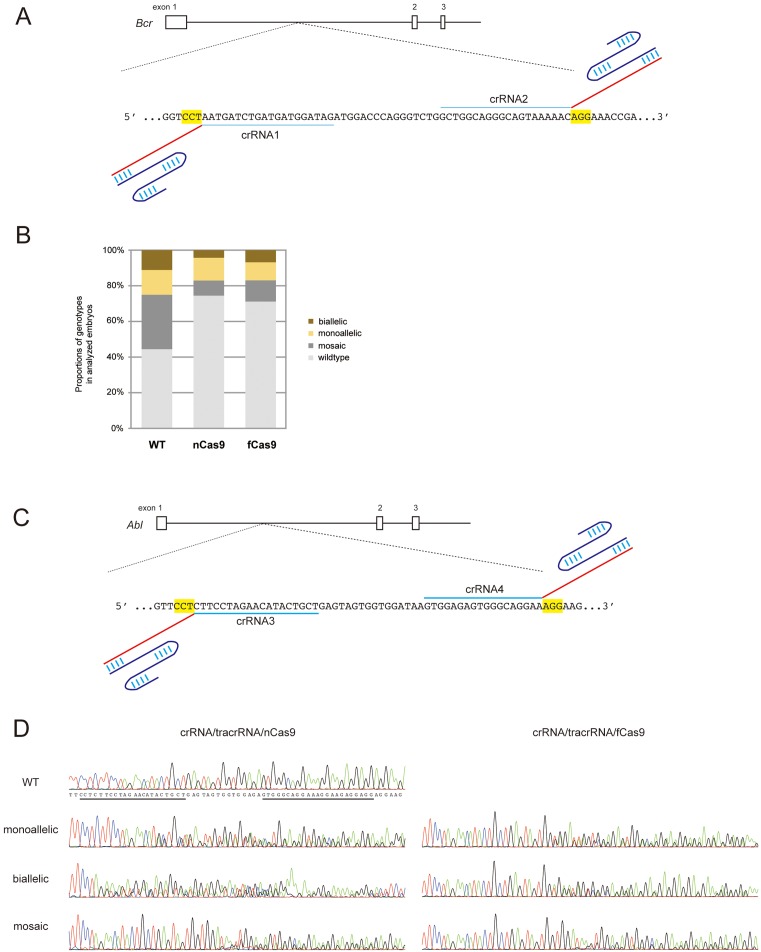
To examine whether the crRNA/tracrRNA approach can be applied as a low off-target method using nCas9 and fCas9, we designed another crRNA (crRNA2) spaced 16 bp apart from crRNA1 in the Bcr locus (Fig. 2A). The RNA mixture containing crRNA1/crRNA2/tracrRNA with nCas9 or fCas9 mRNA was microinjected into zygotes. We obtained a total of 47 and 59 embryos from 136 and 157 fertilized eggs injected with crRNA1/crRNA2/tracrRNA/nCas9 and crRNA1/crRNA2/tracrRNA/fCas9, respectively. The genotyping analysis of the crRNA1/crRNA2/tracrRNA/nCas9-injected embryos showed that 12 out of the 47 embryos (25.5%) were mutated, including 50.0% (6/12) monoallelic, 16.7% (2/12) biallelic, and 33.3% (4/12) mosaic mutant embryos, in a total of three trials. The same analysis of embryos injected with fCas9 with crRNA1/crRNA2/tracrRNA showed that 17 out of 59 embryos (28.8%) were mutated, including 35.3% (6/17) monoallelic, 23.5% (4/17) biallelic, and 41.2% (7/17) mosaic mutants (Fig. 2B and Table 2). χ2 testing of these mutation rates showed that the difference in efficiencies of genome editing between nCas9 and fCas9 with crRNAs/tracrRNA was not significant (P=0.7065), but that these efficiencies were significantly lower than that of Cas9WT with crRNA/tracrRNA (Cas9WT vs. nCas9; P=0.0053, Cas9WT vs. fCas9; P=0.0095, respectively).
Fig. 2.
Generation of mutant mice by co-injection of two crRNAs with Cas9 mutants. (A) The designs of the crRNAs in the mouse Bcr locus. The genomic structure is indicated at the top. The black bars and boxes show introns and exons, respectively. Blue and red lines show specific target sequences and common sequences of crRNA, respectively. The letters with yellow boxes designate PAM sequences. The blue curved lines indicate tracrRNA. Longitudinal blue bars show hydrogen bonds. (B) The proportions of the genotypes of embryos obtained by crRNA1/tracrRNA/Cas9WT (the same data set as shown in Fig. 1C), crRNA1, and 2/tracrRNA/nCas9, and by crRNA1 and 2/tracrRNA/fCas9 injection, respectively. The percentages calculated from the number of mutants relative to the number of analyzed embryos. (C) The designs of the crRNAs for the mouse Abl1 locus. The genomic structure is indicated at the top. The black bars and boxes show introns and exons, respectively. Blue and red lines show specific target sequences and common sequences of crRNAs, respectively. The letters with yellow boxes designate PAM sequences. The blue curved lines indicate tracrRNA. Longitudinal blue bars show hydrogen bonds. (D) Representative electropherograms of embryos injected with crRNA3/crRNA4/tracrRNA and with nCas9 and fCas9, respectively. Underlines indicate the target sequences of each crRNA.
Table 2. A summary of the mutant mice produced using crRNA/tracrRNA with Cas9 mutants.
| Locus | Cas9 | Trial | Injected/2-cell | Transferred | Genotyped | Mutated | Monoallelic | Biallelic | Mosaic |
|---|---|---|---|---|---|---|---|---|---|
| Bcr | nCas9 | 1 | 56/44 | 30 | 24 | 1 | 0 | 0 | 1 |
| 2 | 38/23 | 23 | 8 | 1 | 1 | 0 | 0 | ||
| 3 | 42/33 | 33 | 15 | 10 | 5 | 2 | 3 | ||
| total | 136/100 | 86 | 47 | 12 (25.5) | 6 (50.0) | 2 (16.7) | 4 (33.3) | ||
| fCas9 | 1 | 36/26 | 26 | 21 | 0 | 0 | 0 | 0 | |
| 2 | 43/24 | 24 | 13 | 7 | 1 | 3 | 3 | ||
| 3 | 39/26 | 26 | 11 | 1 | 0 | 0 | 1 | ||
| 4 | 39/30 | 30 | 14 | 9 | 5 | 1 | 3 | ||
| total | 157/106 | 106 | 59 | 17 (28.8) | 6 (35.3) | 4 (23.5) | 7 (41.2) | ||
| Abl1 | nCas9 | 1 | 134/118 | 60 | 39 | 8 (20.5) | 2 (25.0) | 1 (12.5) | 5 (62.5) |
| fCas9 | 1 | 145/115 | 60 | 26 | 10 (38.5) | 2 (20.0) | 6 (60.0) | 2 (20.0) | |
The numbers in parentheses in the “Mutated” columns represent the percentages calculated from the number of mutants relative to the number of genotyped embryos. The numbers in parentheses in the “Monoallelic”, “Biallelic” and “Mosaic” columns represent the percentages calculated from the number of each type relative to the number of “Mutated”.
In order to examine whether genome editing using crRNAs/tracrRNA and nCas9 or fCas9 is applicable for other genomic loci, crRNA pairs (crRNA3 and crRNA4) designed for the mouse Abl1 locus were microinjected with nCas9 or fCas9 mRNA into fertilized zygotes (Fig. 2C). A genotyping analysis showed that 23.1% (9/39) of the embryos injected with nCas9 with crRNA3/crRNA4/tracrRNA were mutated, including 22.2% (2/9) monoallelic, 11.1% (1/9) biallelic, and 66.7% (6/9) mosaic mutant embryos (Fig. 2D). On the other hand, the genotypes of the crRNA3/crRNA4/tracrRNA/fCas9-injected embryos showed that 38.5% (10/26) of the embryos were mutants, including 20.0% (2/10) monoallelic, 60.0% biallelic, and 20.0% mosaic mutant embryos (Table 2). These results suggest that microinjection of two crRNAs and tracrRNA with nCas9 or fCas9 can be useful for mutant mouse production via the microinjection of zygotes.
Next, to examine germline transmission of the mutated alleles to the F1 generation, we crossed F0 mutant mice with wild-type mice. We tested two biallelic mutants and two monoallelic mutants at the Abl1 locus generated by microinjection of crRNA3/crRNA4/tracrRNA with nCas9 (#8 and #11) and fCas9 (#5 and #9), respectively. As a result, the genotyping analysis indicated that all mutated alleles were successfully germline transmitted to F1 pups (Supplementary Table 2). These results suggest that the small indels derived from co-injection of crRNA/tracrRNA with nCas9 or fCas9 can be inherited in the F1 generation.
Off-target analysis of mutant embryos injected with crRNA/tracrRNA with Cas9
Finally, the off-target effects were evaluated in the mutated embryos. We analyzed the putative off-target sites of each crRNA. Possible off-target sites were searched using an online web tool, and three sites with the highest scores for each crRNA were selected. Putative off-target sites for crRNA1 were analyzed by PCR-direct sequencing in 38 mutant embryos, of which 20 and 18 were derived from microinjection of Cas9WT with untreated and denatured/renatured crRNA1/tracrRNA, respectively. The results showed that no off-target effects were detected in the 114 total loci. We also analyzed the mutants produced by the microinjection of crRNA1/crRNA2/tracrRNA/nCas9 and crRNA1/crRNA2/tracrRNA/fCas9, and no off-target mutations were observed. The same analyses were also carried out for crRNA3 and crRNA4 in the Abl1 locus in a total of 19 mutants, including nine nCas9- and ten fCas9-injected embryos, respectively. Possible off-target sites of the two loci for crRNA3 and of the three loci for crRNA4 were examined, which resulted in the identification of no off-target mutations in a total of 95 loci (Supplementary Table 3). Taken together, our results suggest that genome editing using crRNA/tracrRNA does not cause high levels of off-target mutations, and it seems likely that the efficiency of the mutations generated with this method is similar to that of genome editing using sgRNAs.
Discussion
In this study, we showed that crRNA/tracrRNA can be used instead of sgRNA for genome editing with wild-type or modified Cas9s to produce genetically modified mice via microinjection into fertilized eggs. An advantage of utilizing chemically synthesized crRNA/tracrRNA instead of sgRNA is that the synthesis of crRNA/tracrRNA requires much less laboratory work compared to that of sgRNA. Although both crRNA/tracrRNA and sgRNA production require oligonucleotide synthesis, cloning and sequencing are necessary only for sgRNA when an sgRNA coding plasmid itself is used for microinjection. In addition, in vitro transcription and purification are necessary when sgRNA is microinjected as RNA. The chemical synthesis of crRNA/tracrRNA enables the high-throughput production of mutant mice. However, there are several disadvantages of using chemically synthesized crRNA/tracrRNA compared with in vitro transcribed sgRNA: the cost per experiment is higher, the amount of RNAs are limited, and sgRNA can be generated in a week while crRNA/tracrRNA synthesis requires more than one week. In this respect, utilization of sgRNA/Cas9 to produce genetically modified animals requires a shorter period with lower cost than use of crRNA/tracrRNA.
Previously, we reported that mutant mice were produced using Cas9WT, nCas9, and fCas9 with sgRNA, and we compared mutation efficiencies induced by Cas9 variants [7]. In this study, we targeted the same sequences at the Bcr and Abl1 loci. When crRNA1/Cas9WT was used for microinjection, roughly half of the embryos contained mutated alleles. This efficiency is similar to our previous observation when we produced mutant mice using Cas9WT and sgRNA, suggesting that the efficiencies of genome editing are similar between crRNA/tracrRNA and sgRNA. The efficiencies of genome editing at the Bcr locus using nCas9 were similar between crRNA/tracrRNA (27.7%) and sgRNA (33.3%), whereas sgRNA induced a higher level of genome editing (61.7%) than crRNA/tracrRNA (28.8%) when fCas9 was used. At the Abl1 locus, the genome editing efficiency using fCas9 was similar between crRNA/tracrRNA (38.5%) and sgRNA (39.4%, Table 3). In addition, it has been reported that widely used sgRNA exhibits higher mutation efficiencies than that of crRNA in the surveyor assay [9]. A similar tendency was observed in the present study, while Aida et al. (2015) reported that using the crRNA/tracrRNA was more efficient than use of sgRNA with Cas9 protein [1]. There is the possibility that the mutation efficiencies of using sgRNA would be better for some sequences and the efficiencies of using crRNA/tracrRNA would be better for other sequences, or that mutation efficiencies vary with the combination of Cas9 mRNA/protein and sgRNA/crRNA. Direct comparison of the efficiencies of genome editing between the microinjection of Cas9 mRNA and protein with crRNA/tracrRNA would clarify these points.
Table 3. A summary of mutation efficiencies by microinjection of sgRNA and crRNA/tracrRNA with various Cas9s.
Recently, Kotani et al. (2015) showed that the combination of crRNA/tracrRNA/Cas9 protein is more useful than that of crRNA/tracrRNA/Cas9 mRNA when mutations were induced in fertilized zebrafish eggs, possibly because it has immediate enzymatic activity without a translational delay [16]. Based on our analysis, microinjection of crRNA/tracrRNA/Cas9 mRNA into zygotes should be useful for producing mutant mice with small indels, possibly because the cell cycle at the cleavage stage in early mouse embryos is much slower than that of zebrafish. Also, the combination of crRNA/tracrRNA with Cas9 protein may be also useful for producing mutant mice with small indels [1]; however, there are some advantages associated with using Cas9 mRNA instead of Cas9 protein. First, since mRNA can be easily synthesized by an in vitro transcription reaction from plasmids, mRNA would be easier to apply for a variety of methods using modified Cas9, such as Cas9 with altered PAM specificity [15], Cas9 fused with a transactivator [3, 19] or the catalytic domain of epigenetic modification enzymes [8], for example. Second, the cost of mRNA synthesis is lower than that of purchasing Cas9 protein from a commercial source.
The CRISPR/Cas9 system is known to carry the risk of off-target effects. However, several studies have reported that off-target mutations were rarely detected when mutant mice were produced by microinjection of sgRNA with Cas9WT into fertilized eggs [7, 11, 20, 24]. This is consistent with results of our previous study, in which we generated mutant mice using sgRNAs at the Bcr or Abl1 loci [6]. In the current study, no off-target mutations were observed in any case. This suggests that the risk of DSB at off-target sites is similar between the methods using sgRNA and crRNA/tracrRNA. From this point of view, crRNA/tracrRNA may be a good substitute for sgRNA. However, it is better to use crRNA/tracrRNA together with nCas9 or fCas9 instead of Cas9WT when a high level of off-target mutations is predicted by homology searching of the crRNA sequence in a genome database.
In conclusion, our study demonstrated that mutant mice can be produced using a combination of crRNA/tracrRNA/Cas9 mRNA with similar efficiency as the method using sgRNA/Cas9WT. Since it also seems likely that the crRNA/tracrRNA/Cas9 mRNA method does not increase off-target mutations compared with the sgRNA/Cas9WT method, we propose that the crRNA/tracrRNA/Cas9 mRNA method is a good substitute for the sgRNA/Cas9WT method. The crRNA/tracrRNA/Cas9 mRNA method can be applied for low off-target formats using nCas9 or fCas9, enabling the high-throughput production of mutant mice.
Supplementary Material
Acknowledgments
The authors thank all members of the Takada lab for helpful discussions and encouragement. This work was supported in part by JSPS KAKENHI Grant Number 25132713 and in part by a grant from the National Center for Child Health and Development, Grant Number 24–3 to S.T.
References
- 1.Aida T., Chiyo K., Usami T., Ishikubo H., Imahashi R., Wada Y., Tanaka K.F., Sakuma T., Yamamoto T., Tanaka K.2015. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol. 16: 87. doi: 10.1186/s13059-015-0653-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassett A.R., Tibbit C., Ponting C.P., Liu J.L.2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports 4: 220–228. doi: 10.1016/j.celrep.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng A.W., Wang H., Yang H., Shi L., Katz Y., Theunissen T.W., Rangarajan S., Shivalila C.S., Dadon D.B., Jaenisch R.2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23: 1163–1171. doi: 10.1038/cr.2013.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F.2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. doi: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E.2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607. doi: 10.1038/nature09886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guilinger J.P., Thompson D.B., Liu D.R.2014. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 32: 577–582. doi: 10.1038/nbt.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hara S., Tamano M., Yamashita S., Kato T., Saito T., Sakuma T., Yamamoto T., Inui M., Takada S.2015. Generation of mutant mice via the CRISPR/Cas9 system using FokI-dCas9. Sci. Rep. 5: 11221. doi: 10.1038/srep11221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilton I.B., D’Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A.2015. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33: 510–517. doi: 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., Cradick T.J., Marraffini L.A., Bao G., Zhang F.2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31: 827–832. doi: 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K.2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229. doi: 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inui M., Miyado M., Igarashi M., Tamano M., Kubo A., Yamashita S., Asahara H., Fukami M., Takada S.2014. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci. Rep. 4: 5396. doi: 10.1038/srep05396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D.P.2013. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41: e188. doi: 10.1093/nar/gkt780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. doi: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y.G., Cha J., Chandrasegaran S.1996. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 93: 1156–1160. doi: 10.1073/pnas.93.3.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R., Aryee M.J., Joung J.K.2015. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485. doi: 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotani H., Taimatsu K., Ohga R., Ota S., Kawahara A.2015. Efficient multiple genome modifications induced by the crRNAs, tracrRNA and Cas9 protein complex in zebrafish. PLoS ONE 10: e0128319. doi: 10.1371/journal.pone.0128319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T., Huang S., Zhao X., Wright D.A., Carpenter S., Spalding M.H., Weeks D.P., Yang B.2011. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 39: 6315–6325. doi: 10.1093/nar/gkr188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. doi: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M.2013. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31: 833–838. doi: 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mashiko D., Fujihara Y., Satouh Y., Miyata H., Isotani A., Ikawa M.2013. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 3: 3355. doi: 10.1038/srep03355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F.2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. doi: 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K.2014. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 32: 569–576. doi: 10.1038/nbt.2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R.2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. doi: 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yen S.T., Zhang M., Deng J.M., Usman S.J., Smith C.N., Parker-Thornburg J., Swinton P.G., Martin J.F., Behringer R.R.2014. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev. Biol. 393: 3–9. doi: 10.1016/j.ydbio.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.