Abstract
Rabies is a zoonotic disease caused by the Lyssavirus rabies virus (RABV) that can infect most mammals, including humans, where it has a case-fatality rate of almost 100%. Although preventable by vaccination, rabies causes c. 59,000 human fatalities every year worldwide. Thus, there exists an urgent need to establish an effective therapy and/or improve dissemination of vaccines for humans and animals. These outcomes require greater understanding of the mechanisms of RABV pathogenesis to identify new molecular targets for the development of therapeutics and/or live vaccines with high levels of safety. Importantly, a number of studies in recent years have indicated that RABV specifically suppresses host immunity through diverse mechanisms and that this is a key process in pathogenicity. Here, we review current understanding of immune modulation by RABV, with an emphasis on its significance to pathogenicity and the potential exploitation of this knowledge to develop new vaccines and antivirals.
Keywords: immune evasion, interferon, lyssavirus, pathogenesis, rabies
Rabies virus (RABV) is a member of the genus Lyssavirus of the family Rhabdoviridae, order Mononegavirales (MNV, negative-sense single stranded RNA viruses), which causes acute encephalitis (rabies disease) in humans and other mammals with almost invariably lethal outcomes [65, 77]. Although rabies is preventable by vaccination, its incidence remains high (c. 59,000 human deaths/year; 95% confidence intervals: 25,000–159,200) [20], mainly due to the insufficient dissemination of the vaccines for humans and animals, together with the absence of effective therapy for symptomatic rabies. Rabies deaths are largely limited to developing nations in Asia and Africa [77], although deaths in developed nations continue including in “rabies-free” regions where infections by other lyssaviruses have caused fatal rabies-like disease [18]. This highlights the need to develop therapies and/or new live vaccines against RABV/lyssaviruses.
The RABV virion has a “bullet-like” morphology, typical of rhabdoviruses, with an unsegmented negative-sense single-stranded RNA genome of c. 12 kilo-bases, that encodes five structural proteins: nucleoprotein (N protein), phosphoprotein (P protein), matrix (M) protein, glycoprotein (G protein) and large (L) protein [31, 49, 65]. The nucleocapsid “core” consists of the genomic RNA associated with N protein to form a helical N-RNA complex (the template for replication/transcription); the N-RNA interacts with the RNA-dependent RNA polymerase (RdRp) complex, comprising P and L proteins, wherein L is the catalytic subunit and P is a non-catalytic co-factor (Fig. 1) [31, 49, 65]. The molecular function of the latter is unclear, but it appears to involve bridging the RNA associated N protein to the L protein, enabling L to access and transcribe N protein-encapsidated RNA through an unresolved mechanism [31, 57]. M protein binds to the nucleocapsid and mediates its envelopment within the viral envelope. M protein is also thought to bind directly to the cytoplasmic domain of G protein, the extracellular region of which projects from the envelope to participate in binding of virus to the host cell receptors [31, 65].
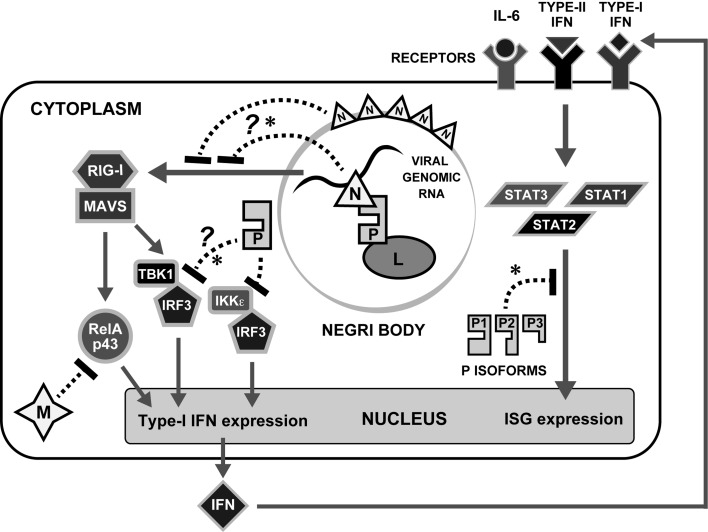
Fig. 1.
IFN antagonism by RABV. Selected molecular processes in the IFN induction and signaling pathways are shown. Following detection of RABV infection by RLRs RIG-I or Mda5 (not shown), signaling platforms are assembled that activate IKKs TBK1 and IKKe, which then phosphorylate the transcription factors IRF3/IRF7; alternate pathways lead to activation of NFkB pathways involving RelAp43. This ultimately activates the expression of type-I IFNs and proinflamatory cytokines. Type I IFN is released from cells and binds to IFNAR to activate classical JAK/STAT signaling pathways through phosphorylation of STAT1 and STAT2 which induce the expression of hundreds of ISGs; IFNs and IL6 family cytokines also activate STAT3. RABV encodes proteins that antagonize the response at multiple stages (see text for details). Host cellular molecules and RABV proteins are indicated by objects with white and black letters, respectively. Mechanisms directly implicated in pathogenicity are indicated by an asterisk.
Typically, RABV in the saliva of an infected animal is transmitted via a bite wound and infects peripheral nerves (either directly or following initial infection of non-neural peripheral cells, such as muscle cells) before spreading to the central nervous system (CNS) through retrograde axonal transport [26, 65]. Pathogenesis is dependent on virus reaching the CNS (reviewed in [26]) where replication and neural network-dependent spread induce severe neurological symptoms including agitation, spasms and paralysis, preceding a lethal outcome [25]. Despite this, it has been long appreciated that pathological changes in the CNS are generally mild, with infected humans/animals lacking significant inflammation or neural cell death [59]. Thus, RABV appears to have evolved neuro-protective capacity involving mechanisms to evade host immunity, thereby replicating in neurons without inducing strong immune responses. This enables RABV to maintain the integrity of the neuronal network, which is important to infection/spread, including post-replication antretrograde transport to deliver new virus to the salivary glands.
A significant body of data now indicates that, despite its apparent molecular simplicity, RABV has evolved diverse strategies for immune evasion as “accessory” functions of the P, N and M proteins (see below and summary in Fig. 1). Although many of these studies have used in vitro approaches focusing on the molecular/cell biology of immune evasion, a number of studies have also evaluated roles in pathogenesis, identifying strong correlations between RABV’s capacity to evade immunity and to cause lethal disease. Here, we outline key studies that have delineated the molecular mechanisms underlying RABV immune evasion and highlight the increasing evidence that these are major factors in pathogenicity, and so represent potential targets for drug/vaccine development.
THE IFN RESPONSE AND ITS INDUCTION BY RABV
The earliest host-cell response to viral infection is activation of the type-I interferon (IFNα/β) system (see Fig. 1), which ultimately leads to the establishment of antiviral responses in infected/neighboring cells and contributes to the shaping of an effective adaptive response [55, 70]. Initiation of the IFN response follows recognition of pathogen associated molecular patterns (PAMPs) by germline-encoded pattern-recognition proteins (PRPs) (e.g. toll-like receptors [TLRs] and retinoic acid-inducible gene I [RIGI]-like receptors [RLRs]) [55, 70]. RNA virus PAMPs include double stranded RNA (dsRNA) and uncapped RNA with a 5′ triphosphate, which are presented by genomic RNA or products of transcription/replication; these variously stimulate the cytoplasmic RLRs RIG-I and melanoma differentiation-associated protein 5 (Mda5) (which principally recognize PAMPs generated within infected cells) and certain of the transmembrane TLRs (which detect extracellularly-derived PAMPs) [6, 22, 55, 70]. RLR signaling proceeds through the adapter protein mitochondrial antiviral-signaling protein (MAVS), ultimately activating the kinases, TANK-binding kinase 1 (TBK1) and I-kappa B kinase ε (IKKε) (Fig. 1); an initially distinct signaling pathway from the double stranded RNA receptor TLR3 also converges at TBK1/IKKε [55]. Subsequent phosphorylation/dimerisation of IFN regulatory factor 3 (IRF3) in most cell types (and IRF7 in certain immune cells or IFN-primed cells) precedes nuclear localization leading to IFNα/β expression. Nuclear factor-kappa B (NFkB) is also activated to form part of the IFN enhanceosome (transcription factor complex), as well as activating the expression of other pro-inflamatory cytokines [55].
Secreted IFNα/β subsequently binds to the type-I IFN receptor (IFNAR) to activate the transcription factors signal transducers and activators of transcription 1 and 2 (STAT1 and 2) (Fig. 1) via phosphorylation at conserved tyrosines [64]. STAT1/2 heterodimerization via reciprocal phosphotyrosine-SH2 interactions precedes nuclear translocation and formation with IRF9 of the IFN-stimulated gene factor 3 (ISGF3), which upregulates several hundred IFN-stimulated gene (ISG) products, including antiviral (e.g. protein kinase R [PKR], Tetherin, myxoma virus resistance [Mx], promyelocytic leukemia [PML]) and immunostimulatory (e.g. major histocompatibility complex [MHC]) proteins, to coordinate a potent antiviral response [55, 63].
Experiments using genetically modified virus and/or infection of dendritic cells to minimize viral mechanisms that inhibit immune signaling (see below and Fig. 1), have indicated that RABV can induce IFN through pathways dependent on RIG-I, Mda5 and MAVS, but independent of TLR signaling, consistent with increased pathogenicity in MAVS-deficient mice [16, 22]. Since pathogenicity is also enhanced in IFNAR−/− mice [13], this indicates that RABV can induce type I-IFNs which are at least partially effective in slowing the onset and/or reducing the severity of symptoms in vivo. Nevertheless, RABV remains a potent killer, indicative of efficient mechanisms to counteract the IFN response.
VIRAL COUNTERMEASURES: IFN ANTAGONISTS
In common with many other viruses, RABV expresses proteins that can directly inhibit IFN signaling pathways. Collectively termed viral IFN antagonists, which encompass a remarkable array of proteins (with hundreds of examples in the literature), variously reported to target all stages of the IFN response through diverse mechanisms [44, 55, 70]. These mechanisms can be broadly categorized as: (i) general inhibition of host gene expression, (ii) sequestration/masking of PAMPs and (iii) sequestration/modification of IFN signaling components or ISG products [70]. More generally, mechanisms may be considered as virus-targeted (where antagonists affect other viral components to minimize PAMP production or exposure to PRPs) or host-targeted (where antagonists, often autonomously, form inhibitory interactions with cellular factors). Many IFN antagonists are multifunctional with roles in the basic viral life cycle as well as accessory roles in IFN antagonism, which often encompass multiple mechanisms targeting different stages of the response [55, 70]. This is particularly important to RNA viruses where limited genome size excludes the expression of dedicated antagonists, resulting in the evolution of accessory IFN-antagonist functions within conserved structural proteins and/or alternative products encoded by conserved genes (e.g. [1, 11, 55, 66, 70, 72]). This has made analysis of the significance of IFN antagonism in infection/pathogenicity challenging due to potential off-target effects in mutagenic studies [76]; however, several recent studies of RABV and other viruses indicate critical roles in disease (see below).
LYSSAVIRUS IFN ANTAGONISTS
RABV P, M and N proteins have IFN antagonist functions that encompass many archetypal features of this class of protein. P protein is the best defined, with in vitro infection and protein expression studies, indicative of several mechanisms, mediated through a large interactome incorporating host innate immune factors, nuclear trafficking receptors and elements of the cytoskeleton (reviewed in [11, 31, 49, 60];, summarized below and in Figs. 1 and 2). Notably, P protein forms many other interactions with host proteins not obviously associated with immunity, indicative of roles for P as a major “hub” at the virus-host interface (e.g. [17, 32, 51, 56]). This is perhaps consistent with a high degree of evolutionary “flexibility” in this non-enzymatic component of the replication machinery, which is encoded by a P gene in all mononegaviruses and is essential to replication, but shows little to no sequence conservation across the MNV order or even between genera of the same family [19].
The first indications of a role for P protein in immune evasion came from the Conzelmann laboratory, through analysis of RABV in which P protein expression was downregulated by translocation of the P gene within the genome [8]. This revealed an inhibitory function toward IRF3/7 phosphorylation by TBK1 (Fig. 1). Although the precise molecular interactions involved in TBK1 inhibition remain unresolved [8, 58], it was recently shown that, P proteins of certain wild-type street strains, but not those of representative fixed laboratory strains, have an additional function to inhibit IKKε, involving physical interaction with this adapter molecule [40].
Subsequently, the Conzelmann and Blondel groups identified an interaction of P protein with STAT1 and STAT2 that inhibits their nuclear translocation and transactivation in response to type-I and type-II IFNs ([9, 71], Fig. 1). This interaction is conserved among lyssaviruses and notably is strongly dependent on IFN-activation (phospholylation of STATs), possibly reflecting a mechanism to ensure that P protein is diverted from replication/transcription function only when required [9, 75]. Recently, P protein was also shown to target STAT3, enabling inhibition of IL6-dependent signaling, indicative of immune antagonistic roles beyond IFNs [33].
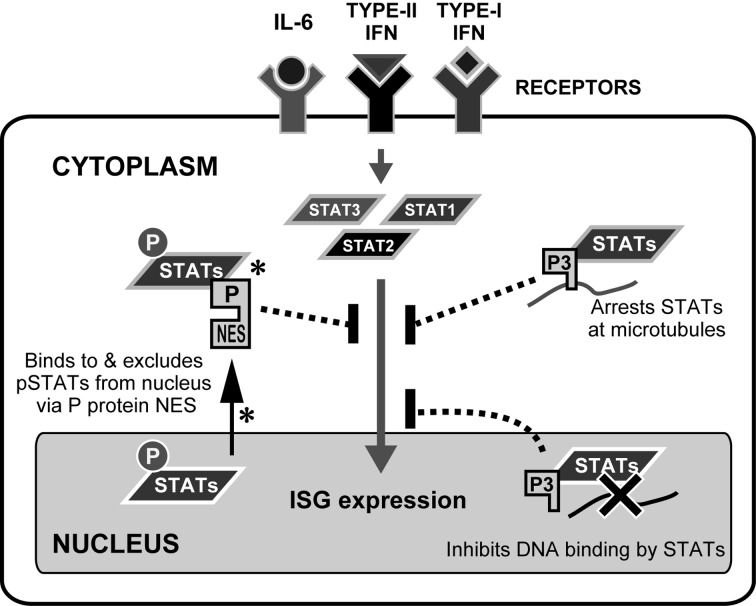
Other than the full length P protein (P or P1), which interacts with L protein in the RdRP complex, RABV P gene expresses four N-terminal truncated isoforms (P2-P5) from four extra in-frame start codons via a ribosomal leaky scanning mechanism [12]. P2-P5 lack the N-terminal L-binding region, suggestive of alternative functions (reviewed in [49]), including specialized roles in IFN antagonism. Indeed, analysis of RABV in which isoform expression is modulated using an internal ribosomal entry site (IRES) has suggested that, while P1 is required for efficient replication, P2 has more important roles in suppressing IFN signaling [35]. Protein expression studies have also identified a number of apparently isoform-specific mechanisms of IFN antagonism, suggestive of a multifaceted strategy (Fig. 2); this might be required to effect an efficient generalized shutdown of the potent antiviral IFN system or to enable targeting of specific elements/pathways of the IFN response that may be more important during different stages of infection and/or in different cell types. P1/P2 exploit the cellular trafficking machinery to undergo active nuclear export via an N-terminal nuclear export sequence (NES), thereby effecting strong nuclear exclusion of P1/P2-associated STATs ([9, 23, 33, 52, 71, 75, 76], Fig. 2). In P3, N-terminal truncation inactivates the NES, activates a nuclear localization sequence (NLS) and induces association with microtubules [7, 47, 50]; together with several additional trafficking sequences [45, 46, 49,50,51,52], this results in P3 distribution to the cytoskeleton and nucleus, enabling cytoskeletal arrest of P3-associated STAT1 and a potential intra-nuclear blockade of STAT1-DNA interaction ([47, 72], Fig. 2). P protein may also target functions of ISG products, since P1 and P3 interact with the PML protein. Although the significance of this interaction is unclear, expression of certain PML isoforms can inhibit infection, suggestive of antiviral functions toward RABV [4, 5].
Fig. 2.
Specific mechanisms of antagonism of IFN signaling by RABV P proteins. The mechanisms involved in inhibition of IFN signaling by RABV P proteins are shown. All of the mechanisms require a physical interaction of P proteins (P1, P2 and P3) with STATs. pSTATs: tyrosine-phosphorylated STATs. Mechanisms directly implicated in pathogenicity are indicated by an asterisk.
M protein immune antagonistic activity involves interaction with a novel isoform of NFkB RelA protein, RelAp43, which was initially identified in a screen for M protein interactors and shown to be a positive regulator of NFkB signaling ([34], Fig. 1). M protein interaction inhibited expression of type I IFN, proinflammatory cytokines and antiviral genes IRF1 and HIAP, consistent with important roles for M-RelAp43 in immune evasion.
Recent data indicate that RABV N protein acts as an IFN antagonist by suppressing IFN induction, although this uses a mechanism distinct from that of P protein, acting upstream of TBK1/IRF3 at the level of RIG-I ([37,38,39], Fig. 1). Notably, this effect was only observed in the context of RABV infection, where the pathogenic RABV strain Nishigahara induced lower IRF3 activation and expression of type-I IFN/chemokines than the attenuated Nishigahara-derivative strain Ni-CE [37]. Mutagenic analysis demonstrated that this is due to substitutions in the Ni-CE N gene affecting two N protein residues [38]. Since single expression of N protein from a plasmid did not inhibit responses to the dsRNA analogue polyI:C or infection by a heterologous virus, this appears to involve a virus-targeted mechanism [37], potentially due to N protein function in encapsidating the RABV genome/replication products and/or forming cage-like structures around cytoplasmic virus factories (Negri bodies) [30, 42], thereby inhibiting PAMP-PRP recognition.
ROLES OF IFN ANTAGONISM IN PATHOGENICITY
Several studies have linked RABV’s capacity to inhibit IFN responses to pathogenicity. Comparative analyses of wild-type street strains and fixed laboratory/attenuated vaccine strains have included the demonstration that the CVS-B2C laboratory strain induces greater inflammatory responses in mice (including elevated type-I IFN-related gene and pro-inflamatory cytokine expression) than the street strain of bat origin (SHBRV), indicative of greater immuno-evasive function in the latter [74]. Similarly, RelAp3 targeting has been reported to be observed for M protein from wild-type lyssaviruses but not from laboratory/vaccine strains [34], with analogous results reported for IKKε targeting by P protein [40]. While these data indicate a correlation between IFN antagonist function and pathogenesis, some other findings argue against such a simple relationship, including the report that the CVS-B2C laboratory strain harbors greater capacity to antagonize IFN signaling than a wild-type dog RABV strain DRV [48], which reverse genetic analysis indicated to be due to differences in the IFN-antagonist-encoding P gene of the respective strains. This is perhaps to be expected, since the attenuated phenotypes of different strains, particularly fixed strains adapted to laboratory animal models and/or cell culture, are likely to have multigenic origin (e.g. [67]); indeed, previous data suggested that G protein is responsible for the attenuated phenotype of CVS-B2C strain via a mechanism distinct from P protein-mediated immune evasion [43]. Similarly, the behavior of street strains derived from host species, such as bats and dogs, may differ significantly when transferred to laboratory animal models. In this respect, it is possible that the P protein-dependent difference observed between DRV and CVS-B2C in murine neuronal cells [48] relates to some extent to the adaptation of the latter to rodents since the parental CVS strain was serially passaged in mouse brains.
Thus, more direct evidence for key roles of IFN-antagonism in disease has come from reverse genetics studies, which have enabled direct comparison of the pathogenicity of viruses that are homogeneous except for altered expression of, or sequences within, specific IFN-antagonists. Suppression of P protein expression in the SAD RABV strain using transcription and translation-based approaches has been shown to cause attenuation in vivo, correlating with reduced capacity to inhibit IFN induction and IFN signaling ([8, 9, 36, 58] and see above). The fact that the P-deficient viruses showed more virulent phenotype in IFNAR-deficient mice is consistent with an important role of IFN antagonism in the viral pathogenesis; however, approaches using altered expression of the entire P protein are complicated by potential “off-target” effects due to P protein’s function in genome transcription/replication. Furthermore, such approaches cannot assess the contribution of particular mechanisms of IFN antagonism. Directed mutagenesis of RABV has enabled such analyses, where deletions within a 10-residue central region of P protein and substitutions in the hydrophobic “W-hole” of the P protein C-terminal domain have been shown to specifically impair antagonism of IFN induction and STAT1/2 signaling, respectively [58, 76]. Importantly, defects in replication/transcription function or growth in IFN-incompetent cells were not apparent for viruses carrying either the deletions or the substitutions, but neurovirulence in mice was reduced in both cases, indicating significant roles for antagonism of both IFN induction and signaling in disease.
Reverse genetics studies using the Nishigahara/Ni-CE viruses (see above) have also revealed roles in virulence for P protein nuclear trafficking and consequent inhibition of STAT1 trafficking/signaling [23, 37,38,39]. Nishigahara is lethal in mice following intracerebral (IC) or intramuscular (IM) inoculation, indicative of neurovirulence and neuroinvasiveness, respectively, while the derivative Ni-CE strain is non-lethal by either route [67, 78]. Although attenuation of Ni-CE is multigenic [67], a significant restoration of pathogenicity in the recombinant CE(NiP) strain, in which the Nishigahara P gene is expressed in the Ni-CE genetic background, identified P protein as a pathogenic determinant by both inoculation routes [23, 67, 78]. This correlated with altered viral sensitivity to IFN in neuronal and muscle cell lines, suggesting that circumvention of IFN responses by P protein is important not only to propagation in neurons, but also in muscle cells en route to peripheral nerves [23, 78].
Analysis of P protein function in neuronal cells indicated that Ni-CE P protein is not defective for suppression of IFN induction or binding to STAT1, but cannot prevent STAT1 nuclear accumulation/signaling due to mutations in the P protein NES that impair nuclear export of P-STAT1 complexes [23]. Notably, in muscle cells, Ni-CE P protein displayed an additional defect in antagonizing IFN induction, suggestive of differences in IFN responses of these cell types [78]. This is consistent with the idea that the diverse IFN-antagonistic mechanisms identified for P protein might have developed to enable immune evasion in different cell-types.
The reverse genetics approach using Nishigahara/Ni-CE chimeric viruses has also highlighted the importance of N protein-dependent inhibition of RIG-I signaling in the pathogenicity: the expression of the Nishigahara N gene in the Ni-CE background [generating the CE(NiN) strain] resulted in increased suppression of viral IFN induction and pathogenicity in mice, with decreased IFN responses and increased viral spread observed in the brain [38, 39].
EVASION OF INFLAMMATORY RESPONSES IN THE CNS
RABV not only impacts on innate immune signaling within infected cells, but also has developed mechanisms to suppress broader adaptive inflammatory responses. In the CNS, despite high levels of viral replication in naturally infected humans and animals, a lack of significant infiltration by inflammatory immune cells is a hallmark of pathogenic infection [59]. Several lines of evidence indicate key roles in pathogenesis [2, 74]. For example, Wang et al. [74] demonstrated that infection of mouse brain with the pathogenic RABV strain SHBRV, only modestly induces expression of chemokines, such as MCP-1 (CCL2), IP-10 (CXCL10) and RANTES (CCL5), that are critical for leukocyte recruitment to sites of inflammation, while the less pathogenic laboratory strain CVS-B2C induced expression more strongly. Cytokines including proinflammatory IL-6, and cytokine receptors, were also up-regulated in CVS-B2C-infected brain. These data indicated a correlation between the ability of RABV to suppress inflammatory responses in the CNS and pathogenicity, suggesting that RABV’s mechanisms to maintain low levels of chemokines and cytokines and/or their cognate receptors in the infected CNS (including the mechanisms described above), suppress inflammation and virus clearance, resulting in a lethal outcome.
Similar approaches have provided insights into the mechanisms by which RABV regulates inflammatory cell infiltration of the CNS, highlighting a role for the blood-brain barrier (BBB). In particular, infection with the attenuated CVS-F3 strain increased BBB permeability, which was accompanied by high-levels of expression of several chemokines including CCL2, CCL5 and CXCL10 in the cerebellum as well as infiltration of CD4+ T cells and CD19+ B cells into this region [53]. In contrast, infection with pathogenic SHBRV maintained BBB integrity and prevented infiltration of immune effectors [61]. These data strongly indicated that enhancement of BBB permeability is important for recruitment of immune effectors into the CNS and subsequent clearance of attenuated RABV, while pathogenic strains harbor mechanisms to maintain the BBB. The key role of BBB integrity in the outcome of viral infection was also supported by the finding that in PLSJL mice, which show moderate resistance to SHBRV infection, reduction of BBB permeability through treatment with a steroid hormone increased the mortality rate of infected mice, while enhancement of BBB permeability by immunization with myelin basic protein resulted in decreased mortality [61]. It was also reported that, in mouse brain infected with CVS-F3 strain, virus-specific antibody produced in situ after recruitment of B cells into the brain plays a critical role in virus clearance [21], highlighting the role of the humoral response in the CNS.
Several studies have begun to delineate the mechanisms by which attenuated RABV infection results in loss of BBB integrity. Phares et al. [54] reported that in CVS-F3-infected mouse, CD4+ T cells, but not CD8+ T cells or B cells, play an important role in enhancement of BBB permeability. The study suggested that IFN-γ released by CD4+ T cells that accumulate in the neurovasculature stimulates neurovascular endothelial cells to produce peroxynitrite (ONOO−), which directly triggers enhancement of BBB permeability. A recent study demonstrated that in mouse brain infected with attenuated CVS-B2C strain, CXCL10 produced by infected neurons promotes infiltration of CXCR3+ CD4+ T cells, which can differentiate into IFN-γ-producing Th1 cells as well as Th17 cells expressing IL-17, an important contributor to disruption of the BBB [10].
The identification of mechanisms by which attenuated RABV infection results in recruitment of immune effectors via enhanced BBB permeability has provided insights into the strategies used by pathogenic RABV to inhibit inflammatory responses in the CNS. Specifically, it is thought that pathogenic RABV minimizes expression of CXCL10 in neurons, thereby preventing infiltration of CXCR3+ CD4+ T cells (Fig. 3A); this hypothesis is based on the observations that infection of mice by pathogenic SHBRV only modestly induces expression of CXCL10 in the mouse brains [74] and that infection of cultured neuroblastoma with Nishigahara induces significantly lower levels of CXCL10 than infection with the attenuated Ni-CE [37]. Thus, reduction of CXCL10 expression in infected neurons may be a general strategy used by pathogenic RABV strains to suppress inflammatory responses in the CNS.
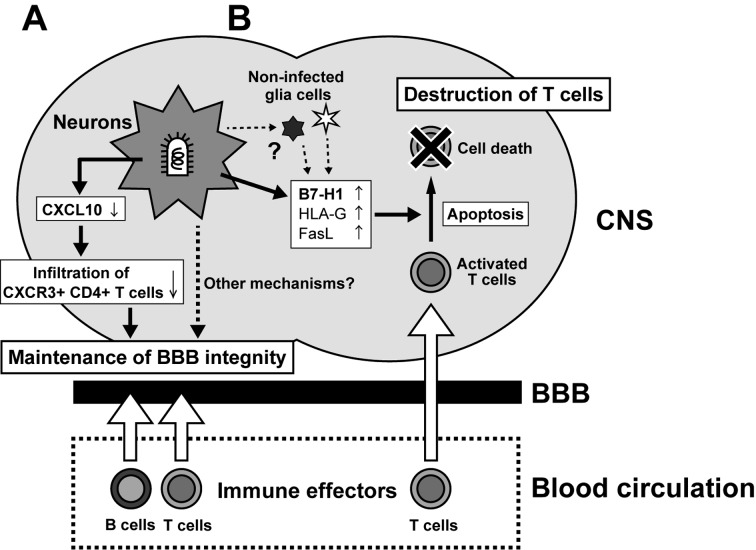
Fig. 3.
Summary of the two mechanisms proposed to effect suppression of neuroinflammation by RABV. (A) Infection of neurons by pathogenic but not attenuated RABV suppresses induction of CXCL10 expression, reducing CXCL10-dependent infiltration of CXCR3+ CD4+ T cells, thereby preventing transition to IL17-producing Th17 cells that promote BBB permeability and immune cell infiltration. (B) RABV infection induces expression of B7-H1 and other immunosuppressive molecules on infected neurons (and potentially non-infected glia), which induce the apoptosis of infiltrating T cells.
Interestingly, studies by other groups have suggested that RABV may have evolved additional, distinct strategies to suppress inflammatory T-cell responses in the CNS. In contrast to the model above, it was reported that infection both by pathogenic CVS strain and by attenuated PV strain led to infiltration of CD3+ T cells into the CNS of infected mice after IM inoculation [3]. However, by seven days post-inocultion, apoptosis of infiltrating T cells and the resultant disappearance of T cells from the CNS were observed in mice infected with CVS, but not with PV.
These findings are particularly intriguing as RABV generally does not infect lymphocytes, raising the question of how pathogenic RABV induces T-cell apoptosis. The most likely scenario appears to relate to the finding that pathogenic RABV infection induces the expression of immunosuppressive molecules, such as B7-H1, HLA-G and FasL, on neurons, which can trigger apoptosis of activated T cells, both in mouse CNS and in cultured neurons [3, 27, 28, 41]. This indicates that pathogenic RABV can hijack host immunosuppressive mechanisms to destroy infiltrating T cells (Fig. 3B). This hypothesis is directly supported by the finding that a pathogenic CVS strain caused significantly milder symptoms in B7-H1 knockout mice than in wild-type mice [28]. Notably, it was also found that the percentages of CD3+ T cells and CD8+ T cells in the total T cell infiltrate in the CNS of B7-H1 knockout mice were higher than those in the CNS of the wild-type mice and that the proportion of apoptotic CD8+ T cells was reduced in the CNS of the B7-H1 knockout mice. These findings indicate that the B7-H1-mediated apoptosis of T cells infiltrating the CNS fulfills a key role not only in suppression of the inflammatory response, but also in the pathogenesis of RABV.
While the findings above are suggestive of a sophisticated strategy for immune evasion by RABV, there is a discrepancy that needs to be resolved: expression of the immunosuppressive B7-H1 and HLA-G, both known ISG products, is induced by IFN [14, 27, 28], but IFN production and signaling is suppressed in neurons infected by pathogenic RABV via the function of N, M and P proteins (see above). However, even pathogenic RABV infection induces modest but detectable level of IFN in the brain and in cultured neurons [37, 74]. Such “leaked” IFN might be sufficient to induce B7-H1 and HLA-G expression and to cause destruction of infiltrating T cells. Although it has been speculated that non-infected glia cells play a role in the destruction of T cells in the CNS [29], this remains to be proven experimentally.
PERSPECTIVES
Current knowledge indicates that RABV effects a potent and multipronged strategy to disable host immunity at many levels, with a growing body of evidence from experiments using genetically modified viruses/hosts indicating critical roles in disease. In IFN-mediated innate immunity, the capacity of P protein to interact with and effect mislocalization of STATs, and that of P and N proteins to inhibit RLR-driven IFN induction, have been demonstrated to impact on pathogenicity in animals. In adaptive immunity/inflammation of the CNS, RABV has a clear capacity to suppress host responses, with two mechanisms suggested in in vivo models, specifically blockage of immune effector infiltration via maintenance of BBB integrity, and induction of T-cell apoptosis (Fig. 3). However, several key elements in RABV immune evasion remain unclear. Firstly, the roles in pathogenicity of a number of IFN-antagonist mechanisms of P protein identified in vitro, such as targeting of PML, STAT3, and microtubules remain unresolved and await research using reverse genetics approaches/animal infection models similar to those that indicated roles in disease for NES-driven P protein nuclear export [23]. Secondly, there are apparently conflicting reports with respect to the roles of IFN signaling/ISG expression in infection (where antiviral and “proviral” roles have been identified), as well as the contribution of altered BBB permeability and T-cell apoptosis in the CNS to RABV suppression of T-cell immunity [28, 62]. Although such observations may relate in part to differences in experimental design/models used, it seems likely that lyssaviruses have evolved diverse mechanisms to effect immune subversion, as indicated by in vitro studies of IFN-antagonism, and that certain mechanisms may predominate in different settings due to factors, such as the viral strains/species examined, cell type infected, stage of infection, infectious dose, etc. For example, a pathogenic strain that only moderately suppresses the expression of IFN and/or CXCL10 in neurons might have evolved mechanisms to induce apoptosis of T cells. Thirdly, in spite of clear evidence that suppression of inflammation in the CNS is important to disease, the molecular mechanisms by which the virus achieves this remain undefined, although involvement of P, M or N protein in suppressing signaling during cytokine/chemokine production seems likely.
Future research using genetically modified viruses/hosts should help to delineate these issues, but what is clear at present is that mutations impacting viral evasion of immunity can potently impact disease, indicating that such mutants are potentially useful for the generation of attenuated vaccine strains. In particular, novel mutations affecting immune evasion may be combined with mutations affecting other genes/mechanisms [15, 24, 38, 67,68,69] to achieve high levels of safety while retaining largely native structure and replication to generate self-adjuventing vaccines; such advances should greatly enhance deliverability, including providing the potential means to achieve single-dose oral vaccination, thereby overcoming current limitations of multiple-dose/needle injection regimes, as well as providing potent protection in diverse host species. Importantly, such vaccines should not simply be “weakened”, but should have the potential to induce stronger immune responses that could generate protection more efficiently. This promise is yet to be realized, however, as a recent study using a P protein-mutated RABV strain unable to prevent IFN induction could induce protection in orally-vaccinated foxes, but gave no apparent improvement in protection compared with the wild-type strain [73]. The effectiveness of such mutations in attenuating strains optimized for immunogenicity, or of alternative strategies targeting STAT antagonism by P protein, or RIG-I-antagonism by N protein, awaits further research.
Finally, another clear corollary of the above considerations is that the mechanisms underlying immune evasion may provide targets for therapeutics for rabies. Several molecular interfaces have been shown to be critical to viral inhibition of immune signaling and virulence (e.g. P protein-STAT1, P-protein-exportin interactions). Others have been defined as important in vitro but not confirmed in vivo (see above), and some involve interactions as yet undefined at the molecular level (e.g. N protein-mediated inhibition of RIG-I). In most cases, however, progress has been made toward elucidating the sites involved by mapping important domains/sequences [5, 38, 76]. The ultimate outcome of such research would be molecular and structural definition of the interfaces, providing knowledge that might guide the development of inhibitors capable of preventing viral evasion of innate immunity. Until the mechanisms underlying viral modulation of BBB permeability and T-cell apoptosis are resolved, potential pathways to therapeutic targeting of these processes are less clear. However, since viral targeting of innate signaling is probably linked to effects on adaptive immunity, the above mentioned therapeutics may have a dual effect on both arms of the response. It is also possible that targeting the host to enhance adaptive responses, rather than targeting viral counter-measures might be useful; indeed, mouse studies indicated that strategies to directly alter BBB permeability can improve outcomes of infection [61], providing proof-of-principal to assess such possibilities towards antiviral therapies.
Acknowledgments
This work was supported by the Grant from the Ministry of Education, Culture, Sports, Science and Technology, Japan, for Joint Research Program of the Research Center for Zoonosis Control, Hokkaido University to N.I. and M.S., JSPS Invitation Fellowship for Research in Japan (Long-term) (L14563) to G.W.M., Australian Research Council Discovery Project (DP150102569) to G.W.M., National Health and Medical Research Council (Australia) Project Grant (#1079211) to G.W.M. and M.S., and Meigunyah Fund Grimwade Fellowship to G.W.M.
REFERENCES
- 1.Audsley M. D., Moseley G. W.2013. Paramyxovirus evasion of innate immunity: Diverse strategies for common targets. World J. Virol. 2: 57–70. doi: 10.5501/wjv.v2.i2.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baloul L., Lafon M.2003. Apoptosis and rabies virus neuroinvasion. Biochimie 85: 777–788. doi: 10.1016/S0300-9084(03)00137-8 [DOI] [PubMed] [Google Scholar]
- 3.Baloul L., Camelo S., Lafon M.2004. Up-regulation of Fas ligand (FasL) in the central nervous system: a mechanism of immune evasion by rabies virus. J. Neurovirol. 10: 372–382. doi: 10.1080/13550280490521122 [DOI] [PubMed] [Google Scholar]
- 4.Blondel D., Kheddache S., Lahaye X., Dianoux L., Chelbi-Alix M. K.2010. Resistance to rabies virus infection conferred by the PMLIV isoform. J. Virol. 84: 10719–10726. doi: 10.1128/JVI.01286-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blondel D., Regad T., Poisson N., Pavie B., Harper F., Pandolfi P. P., De Thé H., Chelbi-Alix M. K.2002. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21: 7957–7970. doi: 10.1038/sj.onc.1205931 [DOI] [PubMed] [Google Scholar]
- 6.Bowie A. G., Unterholzner L.2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8: 911–922. doi: 10.1038/nri2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brice A., Moseley G. W.2013. Viral interactions with microtubules: orchestrators of host cell biology? Future Virol. 8: 229–243. doi: 10.2217/fvl.12.137 [DOI] [Google Scholar]
- 8.Brzózka K., Finke S., Conzelmann K. K.2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 79: 7673–7681. doi: 10.1128/JVI.79.12.7673-7681.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzózka K., Finke S., Conzelmann K. K.2006. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J. Virol. 80: 2675–2683. doi: 10.1128/JVI.80.6.2675-2683.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai Q., She R., Huang Y., Fu Z. F.2015. Expression of neuronal CXCL10 induced by rabies virus infection initiates infiltration of inflammatory cells, production of chemokines and cytokines, and enhancement of blood-brain barrier permeability. J. Virol. 89: 870–876. doi: 10.1128/JVI.02154-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chelbi-Alix M. K., Vidy A., El Bougrini J., Blondel D.2006. Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, Stat1, and PML nuclear bodies. J. Interferon Cytokine Res. 26: 271–280. doi: 10.1089/jir.2006.26.271 [DOI] [PubMed] [Google Scholar]
- 12.Chenik M., Chebli K., Blondel D.1995. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J. Virol. 69: 707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chopy D., Detje C. N., Lafage M., Kalinke U., Lafon M.2011. The type I interferon response bridles rabies virus infection and reduces pathogenicity. J. Neurovirol. 17: 353–367. doi: 10.1007/s13365-011-0041-6 [DOI] [PubMed] [Google Scholar]
- 14.Chopy D., Pothlichet J., Lafage M., Mégret F., Fiette L., Si-Tahar M., Lafon M.2011. Ambivalent role of the innate immune response in rabies virus pathogenesis. J. Virol. 85: 6657–6668. doi: 10.1128/JVI.00302-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietzschold B., Wunner W. H., Wiktor T. J., Lopes A. D., Lafon M., Smith C. L., Koprowski H.1983. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc. Natl. Acad. Sci. U.S.A. 80: 70–74. doi: 10.1073/pnas.80.1.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faul E. J., Wanjalla C. N., Suthar M. S., Gale M., Wirblich C., Schnell M. J.2010. Rabies virus infection induces type I interferon production in an IPS-1 dependent manner while dendritic cell activation relies on IFNAR signaling. PLoS Pathog. 6: e1001016. doi: 10.1371/journal.ppat.1001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouquet B., Nikolic J., Larrous F., Bourhy H., Wirblich C., Lagaudrière-Gesbert C., Blondel D.2015. Focal adhesion kinase is involved in rabies virus infection through its interaction with viral phosphoprotein P. J. Virol. 89: 1640–1651. doi: 10.1128/JVI.02602-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis J. R., Nourse C., Vaska V. L., Calvert S., Northill J. A., McCall B., Mattke A. C.2014. Australian Bat Lyssavirus in a child: the first reported case. Pediatrics 133: e1063–e1067. doi: 10.1542/peds.2013-1782 [DOI] [PubMed] [Google Scholar]
- 19.Gerard F. C., Ribeiro E. A., Jr, Leyrat C., Ivanov I., Blondel D., Longhi S., Ruigrok R. W., Jamin M.2009. Modular organization of rabies virus phosphoprotein. J. Mol. Biol. 388: 978–996. doi: 10.1016/j.jmb.2009.03.061 [DOI] [PubMed] [Google Scholar]
- 20.Hampson K., Coudeville L., Lembo T., Sambo M., Kieffer A., Attlan M., Barrat J., Blanton J. D., Briggs D. J., Cleaveland S., Costa P., Freuling C. M., Hiby E., Knopf L., Leanes F., Meslin F. X., Metlin A., Miranda M. E., Müller T., Nel L. H., Recuenco S., Rupprecht C. E., Schumacher C., Taylor L., Vigilato M. A., Zinsstag J., Dushoff J., Global Alliance for Rabies Control Partners for Rabies Prevention2015. Estimating the global burden of endemic canine rabies. PLoS Negl. Trop. Dis. 9: e0003709. doi: 10.1371/journal.pntd.0003709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper D. C., Phares T. W., Fabis M. J., Roy A.2009. The production of antibody by invading B cells is required for the clearance of rabies virus from the central nervous system. PLoS Negl. Trop. Dis. 3: e535. doi: 10.1371/journal.pntd.0000535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G.2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997. doi: 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- 23.Ito N., Moseley G. W., Blondel D., Shimizu K., Rowe C. L., Ito Y., Masatani T., Nakagawa K., Jans D. A., Sugiyama M.2010. Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J. Virol. 84: 6699–6710. doi: 10.1128/JVI.00011-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito N., Mita T., Shimizu K., Ito Y., Masatani T., Nakagawa K., Yamaoka S., Abe M., Okadera K., Minamoto N., Sugiyama M.2011. Amino acid substitution at position 95 in rabies virus matrix protein affects viral pathogenicity. J. Vet. Med. Sci. 73: 1363–1366. doi: 10.1292/jvms.11-0151 [DOI] [PubMed] [Google Scholar]
- 25.Jackson A. C.2013. Human Disease. pp. 269–98. In: Rabies, 3rd ed. (A. C. Jackson ed.), Academic Press, Oxford. [Google Scholar]
- 26.Jackson A. C., Fu Z. F.2013. Pathogenesis. pp. 299–349. In: Rabies, 3rd ed. (A. C. Jackson ed.), Academic Press, Oxford. [Google Scholar]
- 27.Lafon M.2005. Modulation of the immune response in the nervous system by rabies virus. Curr. Top. Microbiol. Immunol. 289: 239–258. [DOI] [PubMed] [Google Scholar]
- 28.Lafon M.2008. Immune evasion, a critical strategy for rabies virus. Dev. Biol. (Basel) 131: 413–419. [PubMed] [Google Scholar]
- 29.Lafon M.2013. Immunology. pp. 387–408. In: Rabies, 3rd. ed. (Jackson A. C. ed.), Academic Press, Oxford. [Google Scholar]
- 30.Lahaye X., Vidy A., Pomier C., Obiang L., Harper F., Gaudin Y., Blondel D.2009. Functional characterization of Negri bodies (NBs) in rabies virus-infected cells: Evidence that NBs are sites of viral transcription and replication. J. Virol. 83: 7948–7958. doi: 10.1128/JVI.00554-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leyrat C., Ribeiro E. A., Gerard F. C. A., Ivanov I., Ruigrok R. W. H., Jamin M.2011. Structure, interactions with host cell and functions of rhabdovirus phosphoprotein. Future Virol. 6: 465–481. doi: 10.2217/fvl.11.10 [DOI] [Google Scholar]
- 32.Li Y., Dong W., Shi Y., Deng F., Chen X., Wan C., Zhou M., Zhao L., Fu Z. F., Peng G.2016. Rabies virus phosphoprotein interacts with ribosomal protein L9 and affects rabies virus replication. Virology 488: 216–224. doi: 10.1016/j.virol.2015.11.018 [DOI] [PubMed] [Google Scholar]
- 33.Lieu K. G., Brice A., Wiltzer L., Hirst B., Jans D. A., Blondel D., Moseley G. W.2013. The rabies virus interferon antagonist P protein interacts with activated STAT3 and inhibits Gp130 receptor signaling. J. Virol. 87: 8261–8265. doi: 10.1128/JVI.00989-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luco S., Delmas O., Vidalain P. O., Tangy F., Weil R., Bourhy H.2012. RelAp43, a member of the NF-κB family involved in innate immune response against Lyssavirus infection. PLoS Pathog. 8: e1003060. doi: 10.1371/journal.ppat.1003060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marschalek A., Drechsel L., Conzelmann K. K.2012. The importance of being short: the role of rabies virus phosphoprotein isoforms assessed by differential IRES translation initiation. Eur. J. Cell Biol. 91: 17–23. doi: 10.1016/j.ejcb.2011.01.009 [DOI] [PubMed] [Google Scholar]
- 36.Marschalek A., Finke S., Schwemmle M., Mayer D., Heimrich B., Stitz L., Conzelmann K. K.2009. Attenuation of rabies virus replication and virulence by picornavirus internal ribosome entry site elements. J. Virol. 83: 1911–1919. doi: 10.1128/JVI.02055-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masatani T., Ito N., Shimizu K., Ito Y., Nakagawa K., Sawaki Y., Koyama H., Sugiyama M.2010. Rabies virus nucleoprotein functions to evade activation of the RIG-I-mediated antiviral response. J. Virol. 84: 4002–4012. doi: 10.1128/JVI.02220-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masatani T., Ito N., Shimizu K., Ito Y., Nakagawa K., Abe M., Yamaoka S., Sugiyama M.2011. Amino acids at positions 273 and 394 in rabies virus nucleoprotein are important for both evasion of host RIG-I-mediated antiviral response and pathogenicity. Virus Res. 155: 168–174. doi: 10.1016/j.virusres.2010.09.016 [DOI] [PubMed] [Google Scholar]
- 39.Masatani T., Ito N., Ito Y., Nakagawa K., Abe M., Yamaoka S., Okadera K., Sugiyama M.2013. Importance of rabies virus nucleoprotein in viral evasion of interferon response in the brain. Microbiol. Immunol. 57: 511–517. [DOI] [PubMed] [Google Scholar]
- 40.Masatani T., Ozawa M., Yamada K., Ito N., Horie M., Matsuu A., Okuya K., Tsukiyama-Kohara K., Sugiyama M., Nishizono A.2016. Contribution of the interaction between the rabies virus P protein and I-kappa B kinase ε to the inhibition of type I IFN induction signaling. J. Gen. Virol. 97: 316–326. doi: 10.1099/jgv.0.000362 [DOI] [PubMed] [Google Scholar]
- 41.Mégret F., Prehaud C., Lafage M., Moreau P., Rouas-Freiss N., Carosella E. D., Lafon M.2007. Modulation of HLA-G and HLA-E expression in human neuronal cells after rabies virus or herpes virus simplex type 1 infections. Hum. Immunol. 68: 294–302. doi: 10.1016/j.humimm.2006.12.003 [DOI] [PubMed] [Google Scholar]
- 42.Ménager P., Roux P., Mégret F., Bourgeois J. P., Le Sourd A. M., Danckaert A., Lafage M., Préhaud C., Lafon M.2009. Toll-like receptor 3 (TLR3) plays a major role in the formation of rabies virus Negri Bodies. PLoS Pathog. 5: e1000315. doi: 10.1371/journal.ppat.1000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morimoto K., Hooper D. C., Spitsin S., Koprowski H., Dietzschold B.1999. Pathogenicity of different rabies virus variants inversely correlates with apoptosis and rabies virus glycoprotein expression in infected primary neuron cultures. J. Virol. 73: 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moseley G. W.2012. Viral Interferon Antagonism: Making the Leap from the Bench to the Clinic. J. Virol. Antivir. Res. 1: 1–2. doi: 10.4172/2324-8955.1000e103 [DOI] [Google Scholar]
- 45.Moseley G. W., Filmer R. P., DeJesus M. A., Jans D. A.2007. Nucleocytoplasmic distribution of rabies virus P-protein is regulated by phosphorylation adjacent to C-terminal nuclear import and export signals. Biochemistry 46: 12053–12061. doi: 10.1021/bi700521m [DOI] [PubMed] [Google Scholar]
- 46.Moseley G. W., Roth D. M., DeJesus M. A., Leyton D. L., Filmer R. P., Pouton C. W., Jans D. A.2007. Dynein light chain association sequences can facilitate nuclear protein import. Mol. Biol. Cell 18: 3204–3213. doi: 10.1091/mbc.E07-01-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moseley G. W., Lahaye X., Roth D. M., Oksayan S., Filmer R. P., Rowe C. L., Blondel D., Jans D. A.2009. Dual modes of rabies P-protein association with microtubules: a novel strategy to suppress the antiviral response. J. Cell Sci. 122: 3652–3662. doi: 10.1242/jcs.045542 [DOI] [PubMed] [Google Scholar]
- 48.Niu X., Tang L., Tseggai T., Guo Y., Fu Z. F.2013. Wild-type rabies virus phosphoprotein is associated with viral sensitivity to type I interferon treatment. Arch. Virol. 158: 2297–2305. doi: 10.1007/s00705-013-1743-2 [DOI] [PubMed] [Google Scholar]
- 49.Oksayan S., Ito N., Moseley G., Blondel D.2012. Subcellular trafficking in rhabdovirus infection and immune evasion: a novel target for therapeutics. Infect. Disord. Drug Targets 12: 38–58. doi: 10.2174/187152612798994966 [DOI] [PubMed] [Google Scholar]
- 50.Oksayan S., Wiltzer L., Rowe C. L., Blondel D., Jans D. A., Moseley G. W.2012. A novel nuclear trafficking module regulates the nucleocytoplasmic localization of the rabies virus interferon antagonist, P protein. J. Biol. Chem. 287: 28112–28121. doi: 10.1074/jbc.M112.374694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oksayan S., Nikolic J., David C. T., Blondel D., Jans D. A., Moseley G. W.2015. Identification of a role for nucleolin in rabies virus infection. J. Virol. 89: 1939–1943. doi: 10.1128/JVI.03320-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasdeloup D., Poisson N., Raux H., Gaudin Y., Ruigrok R. W., Blondel D.2005. Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology 334: 284–293. doi: 10.1016/j.virol.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 53.Phares T. W., Kean R. B., Mikheeva T., Hooper D. C.2006. Regional differences in blood-brain barrier permeability changes and inflammation in the apathogenic clearance of virus from the central nervous system. J. Immunol. 176: 7666–7675. doi: 10.4049/jimmunol.176.12.7666 [DOI] [PubMed] [Google Scholar]
- 54.Phares T. W., Fabis M. J., Brimer C. M., Kean R. B., Hooper D. C.2007. A peroxynitrite-dependent pathway is responsible for blood-brain barrier permeability changes during a central nervous system inflammatory response: TNF-alpha is neither necessary nor sufficient. J. Immunol. 178: 7334–7343. doi: 10.4049/jimmunol.178.11.7334 [DOI] [PubMed] [Google Scholar]
- 55.Randall R. E., Goodbourn S.2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89: 1–47. doi: 10.1099/vir.0.83391-0 [DOI] [PubMed] [Google Scholar]
- 56.Rawlinson S. M., Moseley G. W.2015. The nucleolar interface of RNA viruses. Cell. Microbiol. 17: 1108–1120. doi: 10.1111/cmi.12465 [DOI] [PubMed] [Google Scholar]
- 57.Ribeiro E. A., Jr, Leyrat C., Gérard F. C. A., Albertini A. A. V., Falk C., Ruigrok R. W. H., Jamin M.2009. Binding of rabies virus polymerase cofactor to recombinant circular nucleoprotein-RNA complexes. J. Mol. Biol. 394: 558–575. doi: 10.1016/j.jmb.2009.09.042 [DOI] [PubMed] [Google Scholar]
- 58.Rieder M., Brzózka K., Pfaller C. K., Cox J. H., Stitz L., Conzelmann K. K.2011. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprotein: inhibition of interferon regulatory factor 3 activation is important for pathogenicity. J. Virol. 85: 842–852. doi: 10.1128/JVI.01427-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rossiter J. P., Jackson A. C.2013. Pathology. pp. 351–386. In: Rabies, 3rd ed. (Jackson A. C. ed.), Academic Press, Oxford. [Google Scholar]
- 60.Rowe C. L., Wagstaff K. M., Oksayan S., Glover D. J., Jans D. A., Moseley G. W.2016. Nuclear Trafficking of the Rabies Virus Interferon Antagonist P-Protein Is Regulated by an Importin-Binding Nuclear Localization Sequence in the C-Terminal Domain. PLoS ONE 11: e0150477. doi: 10.1371/journal.pone.0150477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy A., Hooper D. C.2007. Lethal silver-haired bat rabies virus infection can be prevented by opening the blood-brain barrier. J. Virol. 81: 7993–7998. doi: 10.1128/JVI.00710-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roy A., Phares T. W., Koprowski H., Hooper D. C.2007. Failure to open the blood-brain barrier and deliver immune effectors to central nervous system tissues leads to the lethal outcome of silver-haired bat rabies virus infection. J. Virol. 81: 1110–1118. doi: 10.1128/JVI.01964-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadler A. J., Williams B. R.2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8: 559–568. doi: 10.1038/nri2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindler C., Levy D. E., Decker T.2007. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 282: 20059–20063. doi: 10.1074/jbc.R700016200 [DOI] [PubMed] [Google Scholar]
- 65.Schnell M. J., McGettigan J. P., Wirblich C., Papaneri A.2010. The cell biology of rabies virus: using stealth to reach the brain. Nat. Rev. Microbiol. 8: 51–61. [DOI] [PubMed] [Google Scholar]
- 66.Shaw M. L., Cardenas W. B., Zamarin D., Palese P., Basler C. F.2005. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J. Virol. 79: 6078–6088. doi: 10.1128/JVI.79.10.6078-6088.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shimizu K., Ito N., Mita T., Yamada K., Hosokawa-Muto J., Sugiyama M., Minamoto N.2007. Involvement of nucleoprotein, phosphoprotein, and matrix protein genes of rabies virus in virulence for adult mice. Virus Res. 123: 154–160. doi: 10.1016/j.virusres.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 68.Takayama-Ito M., Ito N., Yamada K., Sugiyama M., Minamoto N.2006. Multiple amino acids in the glycoprotein of rabies virus are responsible for pathogenicity in adult mice. Virus Res. 115: 169–175. doi: 10.1016/j.virusres.2005.08.004 [DOI] [PubMed] [Google Scholar]
- 69.Tuffereau C., Leblois H., Bénéjean J., Coulon P., Lafay F., Flamand A.1989. Arginine or lysine in position 333 of ERA and CVS glycoprotein is necessary for rabies virulence in adult mice. Virology 172: 206–212. doi: 10.1016/0042-6822(89)90122-0 [DOI] [PubMed] [Google Scholar]
- 70.Versteeg G. A., García-Sastre A.2010. Viral tricks to grid-lock the type I interferon system. Curr. Opin. Microbiol. 13: 508–516. doi: 10.1016/j.mib.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vidy A., Chelbi-Alix M., Blondel D.2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J. Virol. 79: 14411–14420. doi: 10.1128/JVI.79.22.14411-14420.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vidy A., El Bougrini J., Chelbi-Alix M. K., Blondel D.2007. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J. Virol. 81: 4255–4263. doi: 10.1128/JVI.01930-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vos A., Conzelmann K. K., Finke S., Müller T., Teifke J., Fooks A. R., Neubert A.2011. Immunogenicity studies in carnivores using a rabies virus construct with a site-directed deletion in the phosphoprotein. Adv. Prev. Med. 2011: 898171. doi: 10.4061/2011/898171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Z. W., Sarmento L., Wang Y., Li X. Q., Dhingra V., Tseggai T., Jiang B., Fu Z. F.2005. Attenuated rabies virus activates, while pathogenic rabies virus evades, the host innate immune responses in the central nervous system. J. Virol. 79: 12554–12565. doi: 10.1128/JVI.79.19.12554-12565.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiltzer L., Larrous F., Oksayan S., Ito N., Marsh G. A., Wang L. F., Blondel D., Bourhy H., Jans D. A., Moseley G. W.2012. Conservation of a unique mechanism of immune evasion across the Lyssavirus genus. J. Virol. 86: 10194–10199. doi: 10.1128/JVI.01249-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wiltzer L., Okada K., Yamaoka S., Larrous F., Kuusisto H. V., Sugiyama M., Blondel D., Bourhy H., Jans D. A., Ito N., Moseley G. W.2014. Interaction of rabies virus P-protein with STAT proteins is critical to lethal rabies disease. J. Infect. Dis. 209: 1744–1753. doi: 10.1093/infdis/jit829 [DOI] [PubMed] [Google Scholar]
- 77.World Health Organization2013. WHO Expert Consultation on Rabies. Second report. World Health Organ. Tech. Rep. Ser. 982: 1–139back cover. [PubMed] [Google Scholar]
- 78.Yamaoka S., Ito N., Ohka S., Kaneda S., Nakamura H., Agari T., Masatani T., Nakagawa K., Okada K., Okadera K., Mitake H., Fujii T., Sugiyama M.2013. Involvement of the rabies virus phosphoprotein gene in neuroinvasiveness. J. Virol. 87: 12327–12338. doi: 10.1128/JVI.02132-13 [DOI] [PMC free article] [PubMed] [Google Scholar]