Abstract
American foulbrood is the most destructive honeybee bacterial disease. The etiological agent, Paenibacillus larvae, has been classified into four genotypes by a repetitive-element PCR (ERIC I-IV) and 21 sequence types by multilocus sequence typing (ST1-21). In this study, we genotyped Japanese P. larvae isolates for the first time and revealed the presence of three genotypes (ERIC I-ST2, ERIC I-ST15 and ERIC II-ST10) in the western region of Aichi prefecture. ERIC I-ST15 and ERIC II-ST10 are globally distributed types, whereas the ERIC I-ST2 isolate was the first isolate of this genotype identified outside the native range of the European honeybee. The ERIC I and II isolates differed in phenotypes including cell morphology, and these may be useful for predicting ERIC types.
Keywords: American foulbrood, ERIC genotypes, MLST, morphology, Paenibacillus larvae
American foulbrood (AFB) is the most severe bacterial disease of the honeybee brood; it is caused by a Gram-positive, spore-forming bacterium, Paenibacillus larvae. Larvae become infected through the ingestion of P. larvae spores. Soon after being ingested, the spores germinate in the midgut lumen, and vegetative bacteria proliferate within the midgut, breach the midgut epithelium and invade the hemocoel [5, 15]. In typical clinical cases, the diseased brood turns creamy brown, then dark brown, becomes ropy and eventually dries out to form hard dark scales. In each diseased brood, P. larvae produces extremely high numbers of spores, which become the infection source in the next host. AFB occurs throughout the world and causes considerable economic losses in the apiculture industry.
P. larvae strains have been classified into four genotypes (ERIC I-IV) by repetitive element PCR (rep-PCR) using enterobacterial repetitive intergenic consensus (ERIC) primers [8, 12]. Among these genotypes, ERIC I and II have frequently been isolated from AFB cases, while ERIC III and IV have not been identified in field isolates in recent years; therefore, ERIC I and II are regarded as two practically important genotypes in AFB [5]. The four ERIC types were previously shown to differ in their phenotypes including colony morphology, metabolism of carbon sources and virulence [8]. For example, ERIC I strains form whitish colonies, whereas colonies of ERIC II and III strains are orange-pigmented. Pigmented and non-pigmented strains have both been reported in the ERIC IV genotype [3, 8].
A multilocus sequence typing (MLST) scheme was recently developed for P. larvae [12]. By using this method, strains of the four ERIC genotypes were further separated into 21 sequence types (STs): ERIC I into 16 STs (ST1-7, 13–21), ERIC II into three STs (ST10-12) and ERIC III and IV into two STs (ST8-9) [12]. An analysis of international isolates by the MLST scheme revealed that some STs are globally distributed while others are more localized, and several STs were only found outside the native range of the European honeybee (Apis mellifera), where honeybees were introduced relatively recently from other countries [12]. These MLST data as well as ERIC typing data have been deposited in the P. larvae MLST database (http://pubmlst.org/plarvae/). The construction of a more comprehensive multinational data set in the database will improve our understanding of AFB.
In the MLST database, five ST10 isolates have been registered as mixed origin isolates from China, Singapore and Japan; however, the actual origin of the isolates currently remains unclear. AFB occurs every year in Japan; however, the detailed characteristics of Japanese P. larvae isolates have not yet been described. As of January 2015, approximately 5,000 A. mellifera colonies were kept at approximately 240 bee farms in Aichi prefecture, Japan. In this area, one to nine AFB cases have been reported every year in the past five years. In the present study, we analyzed seven P. larvae isolates, isolated from AFB cases occurring between 2012 and 2014, in the western region of Aichi prefecture and revealed their genotypic and phenotypic characteristics.
All seven P. larvae isolates were isolated from a foul brood exhibiting the clinical signs of AFB. Brood samples were collected using sterile cotton swabs, inoculated on J-agar [9] and cultured at 37°C in air plus 5% CO2. Bacterial colonies grown on agar and considered to be P. larvae were subcultured, and the genomic DNA of the well-isolated bacteria was extracted by InstaGene Matrix (Bio-Rad Laboratories, Inc., Hercules, CA, U.S.A.) according to the manufacturer’s instructions. Bacterial isolates were identified as P. larvae based on morphological characteristics and a P. larvae-specific PCR assay [11]. The identification was further confirmed by analyzing 16S rRNA gene sequences as described previously [1, 4].
In order to investigate spore formation in vitro and the morphology of the isolates, all isolates were cultured on J-agar without glucose (J´-agar: tryptone 5 g, yeast extract 15 g, K2HPO4 3 g and agar 15 g/l) at 37°C for 14 days in air plus 5% CO2. Bacterial cell morphology was observed by light microscopy after Gram staining. Conventional methods were used to test for the production of catalase. Other biochemical tests were performed using API 20 E, API 20 NE and API 50 CH (bioMérieux, Marcy l’Etoile, France) according to the manufacturer’s instructions. The substrates used in biochemical tests are listed in Table S1 in the supplementary file. In order to identify hemolytic activity, isolates were cultured on Columbia blood agar with 5% horse blood (bioMérieux) and 5% sheep blood agar (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) plates at 37°C for 3 days under air plus 5% CO2 conditions.
The ERIC type of the isolates was determined by rep-PCR with the primers ERIC1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) [6, 8]. PCR was performed in a final reaction volume of 50 µl containing 1× Ex Taq PCR buffer (Mg2+ free), 2.5 mM MgCl2, 0.25 mM of each dNTP, 0.4 µM of each primer, 1.25 U Ex Taq DNA polymerase (Takara Bio, Kusatsu, Japan) and 125 ng template DNA. The reaction conditions consisted of an initial denaturation step at 95°C for 2 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 53°C for 1 min and extension at 72°C for 2.5 min, followed by a final extension at 72°C for 10 min. Five microliters of the amplified product was electrophoresed (100V, 40 min) on a 0.8% agarose gel. The separated products were stained with ethidium bromide and visualized by UV light. The ERIC type was determined by comparing the visualized DNA band patterns with those reported previously [8]. MLST was performed by sequencing seven genes as described previously [12]. Sequencing was carried out with a BigDye Terminator v3.1 cycle sequencing kit using a 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, U.S.A.). The allelic numbers and STs of the isolates were determined by comparing their sequences with those in the P. larvae MLST database (http://pubmlst.org/plarvae/). The genotyping data were deposited in the database.
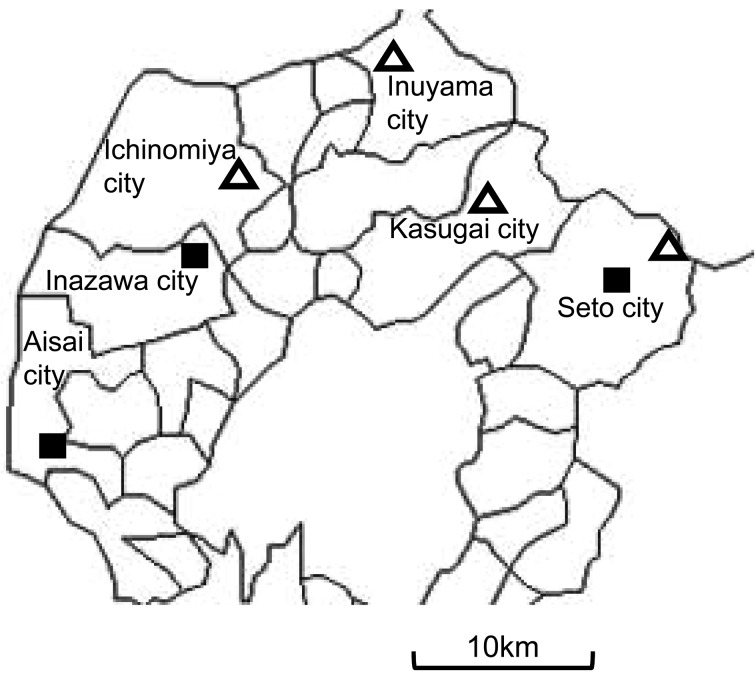
As summarized in Tables 1 and 2, AFB cases found in this area between 2012 and 2014 were not caused by a single-specific strain, but by multiple strains with different phenotypes and genotypes. On J-agar and J´-agar plates, four isolates (N, O, KH and IK) formed orange-pigmented colonies (Fig. 1A). Colonies were more pigmented on J´-agar than on J-agar. These four isolates were all assigned to the same genotype (ERIC II and ST10). On the other hand, colonies of the other three isolates (I, SK and KM) were whitish and somewhat transparent (Fig. 1B), and were assigned to ERIC I. These non-pigmented isolates were further classified into ST15 (I and KM) and ST2 (SK) by MLST. None of the isolates showed hemolysis on the blood agar plates tested (Table 1). These results are consistent with the reported ERIC type/ST correlation and phenotype/genotype correlation [8, 12]. The geographic distribution of the isolates is shown in Fig. 2. Both ERIC types were distributed relatively widely in this area, implying that both types are already widespread in the western region of Aichi prefecture; however, it currently remains unknown when and how the two types spread in this area. No novel genotype was found in this study. All three genotypes have been found in other countries [12]. ERIC I-ST15 in particular is one of the most common and widespread types and has been detected in multiple countries and across five continents. ERIC II-ST10 is also widely distributed and has been found in Europe, North America, Oceania and Asia. Both genotypes were identified not only within the native range of A. mellifera (Europe, Africa and Asia), but also outside this region [12]. In contrast, ERIC I-ST2 strains have so far only been isolated in samples from the native range [12]. Japan is not in the native range of A. mellifera, which was imported into Japan in 1877 in order to increase honey productivity. Therefore, isolate SK is the first example to demonstrate the presence of ST2 P. larvae outside the native host range.
Table 1. P. larvae isolates analyzed in this study.
| Isolate | Source | Site of occurrence | Year of the AFB outbreak | Spores in a diseased brood | Pigmented colonies | Hemolysis | Spore formation in vitroa) | Average days needed to form spores in vitroa) | Genotyping results | |
|---|---|---|---|---|---|---|---|---|---|---|
| ERIC | MLST | |||||||||
| N | Diseased brood | Seto city | 2012 | + | + | − | + | 10 days | II | ST10 |
| O | Diseased brood | Ichinomiya city | 2013 | + | + | − | − | II | ST10 | |
| KH | Diseased brood | Inuyama city | 2014 | + | + | − | − | II | ST10 | |
| IK | Diseased brood | Kasugai city | 2014 | + | + | − | − | II | ST10 | |
| I | Diseased brood | Inazawa city | 2013 | + | − | − | + | 3.6 days | I | ST15 |
| SK | Diseased brood | Seto city | 2014 | + | − | − | + | 4 days | I | ST2 |
| KM | Diseased brood | Aisai city | 2014 | + | − | − | − | I | ST15 | |
a) The formation of spores was examined on J´-agar at 37°C in air plus 5% CO2 for 14 days.
Table 2. Metabolic profiles of P. larvae isolates.
| Isolate | Genotype | Catalasea) | Gelatina) | Fermentationa) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERIC | MLST | D-Trehalose | D-Glucose | D-Fructose | D-Mannose | D-Mannitol | Salicin | Glycerol | D-Ribose | N-Acetyl Glucosamine | D-Tagatose | |||
| N | II | ST10 | − | + | + | ± | + | + | − | − | − | + | + | − |
| O | II | ST10 | − | + | + | ± | + | + | ± | − | − | + | + | − |
| KH | II | ST10 | − | + | + | + | + | + | + | − | − | ± | + | ± |
| IK | II | ST10 | − | + | + | + | + | + | + | − | − | ± | − | ± |
| I | I | ST15 | − | + | + | + | − | − | − | − | − | − | + | − |
| SK | I | ST2 | − | + | + | + | − | − | − | − | − | − | + | − |
| KM | I | ST15 | − | + | + | + | − | − | − | ± | + | ± | + | ± |
a) +, positive; ± , weak positive; −, negative.
Fig. 1.
Morphological characteristics of P. larvae isolates analyzed in this study. (A and B) Colonies grown on J-agar. Pigmented (A) and non-pigmented (B) P. larvae colonies were both isolated from the AFB cases examined in this study. (C and D) Gram staining of P. larvae isolates cultured on J´-agar at 37°C for 3 days. Pigmented ERIC II isolates mainly consisted of short rod-shaped cells (C), while bacterial cells of non-pigmented ERIC I isolates were long rods and occurred in chains (D). Photos of isolates I (ERIC I) and KH (ERIC II) are shown as representatives.
Fig. 2.
Geographic distribution of isolate origins; squares are ERIC I, and triangles are ERIC II.
Although spores were detected in diseased broods in all seven AFB cases, only three of the seven isolates (N, I and SK) formed spores on J´-agar during the examinations (Table 1), and there was no correlation between the ERIC type and spore formation in vitro. However, among the three isolates, ERIC I isolates (I and SK) formed spores earlier than the ERIC II isolate (N) (Table 1). Furthermore, the ERIC I isolates formed much more spores than the ERIC II isolate (data not shown). Interestingly, the bacterial cell morphology of the isolates cultured for 2–3 days on J- and J´-agar differed between ERIC I and II. Cells of ERIC II isolates were short and plump rods (Fig. 1C), whereas those of ERIC I isolates were long rods and occurred in long chains (Fig. 1D).
Biochemical tests showed that all seven isolates metabolized D-trehalose, D-glucose and gelatin, while the metabolic profiles of D-fructose, D-mannose, D-mannitol, salicin, glycerol, D-ribose, N-acetyl glucosamine and D-tagatose varied among the isolates (Table 2). The other biochemical characteristics tested, including catalase activity, were negative in all isolates. Genotype-specific differences were observed in D-fructose and D-mannose. These sugars were only fermented by the ERIC II isolates, and not by the ERIC I isolates (Table 2). These results are consistent with previous findings [2, 13]. Neuendorf et al. found that only genotype AB, which is a genotype determined by another rep-PCR method and correlated with ERIC II, had the ability to metabolize D-fructose and D-mannose [13]. D-fructose was also only fermented by ERIC II strains, and not by ERIC I strains in a recent Italian study [2]. In both these studies, 100% of the investigated ERIC II/genotype AB isolates metabolized D-fructose [2, 13]; therefore, this sugar was reported as a genotype-specific biochemical marker [2]. Our results further support the usefulness of D-fructose as an ERIC II-specific marker.
Genersch et al. [8] also reported genotype-specific differences in the metabolism of salicin and mannitol, namely, ERIC I strains were able to use salicin, but not mannitol, while ERIC II strains metabolized mannitol, but not salicin. In the present study, D-mannitol was only fermented by ERIC II isolates; however, one of the ERIC II isolates did not have the ability to use it. Although salicin was fermented by one ERIC I isolate, the other isolates did not have the ability to metabolize it (Table 2). These results partially differ from the findings of Genersch et al. [8]; however, our results were comparable to a recent study on Italian isolates, in which 86.6% of the ERIC II strains had the ability to use mannitol, and only 5% of the ERIC I strains metabolized salicin [2]. Since the metabolism of salicin may be weak in P. larvae [10], it is not easy to evaluate test results based on color changes of the indicator. Although all studies including this one used API systems (bioMérieux) for biochemical tests, the discrepancies between different studies may be partly due to differences in the reading criteria of the test results. Furthermore, mutations that occurred in the genes responsible for metabolism in the field or during subcultures in laboratories may have caused some of these discrepancies.
ERIC I and II were shown to differ in virulence at both the larval and colony levels [5, 7, 8, 14]. When examined in exposure bioassays, strains of ERIC II were highly virulent against larvae and killed all tested larvae within approximately 7 days. This means that most of the infected larvae die before their brood cells are capped, resulting in the efficient removal of the infected larvae from the colony by hygienic nurse bees and a low level of spore production within the colony. In contrast, ERIC I was less virulent for the individual larva and needed approximately 12 days to kill all infected larvae; therefore, more larvae die after cell capping in ERIC I-strain-infected colonies, leaving nurse bees only a small chance of cleaning out the infected larvae, resulting in a higher level of spore production, the more rapid spread of spores within the colony and faster colony collapse than ERIC II-strain-infected colonies [5, 7, 8, 14]. Since the seven AFB cases in the present study were inspected after all infected colonies had collapsed, we were unable to determine whether the reported differences in disease progression also occurred in these cases.
In the present study, we revealed the existence of ERIC I and II genotypes in Japan and found an ST2 isolate outside the native range of A. mellifera for the first time. We also detected several phenotypic differences between ERIC I and II isolates. Since ERIC III and IV strains have not recently been isolated [5], these phenotypic differences, in particular, differences in cell morphology, along with the previously reported differences in colony pigmentation and fermentation of D-fructose, may become indicators to more easily discriminate between ERIC I and II. Since the numbers and areas of analyzed AFB cases are limited, the population structure of P. larvae in Japan is still unknown. More extensive studies on diverse AFB cases in Japan and the detailed characterization of isolated P. larvae will help us to understand not only the local, but also global epidemiology of this destructive honeybee disease.
SUPPLEMENTARY MATERIAL
REFERENCES
- 1.Arai R., Tominaga K., Wu M., Okura M., Ito K., Okamura N., Onishi H., Osaki M., Sugimura Y., Yoshiyama M., Takamatsu D.2012. Diversity of Melissococcus plutonius from honeybee larvae in Japan and experimental reproduction of European foulbrood with cultured atypical isolates. PLoS ONE 7: e33708. doi: 10.1371/journal.pone.0033708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassi S., Formato G., Milito M., Trevisiol K., Salogni C., Carra E.2015. Phenotypic characterization and ERIC-PCR based genotyping of Paenibacillus larvae isolates recovered from American foulbrood outbreaks in honey bees from Italy. Vet. Q. 35: 27–32. doi: 10.1080/01652176.2014.993095 [DOI] [PubMed] [Google Scholar]
- 3.Dingman D. W.2015. Comparative analysis of Paenibacillus larvae genotypes isolated in Connecticut. Arch. Microbiol. 197: 789–795. doi: 10.1007/s00203-015-1113-4 [DOI] [PubMed] [Google Scholar]
- 4.Dorsch M., Stackebrandt E.1992. Some modifications in the procedure of direct sequencing of PCR amplified 16S rDNA. J. Microbiol. Methods 16: 271–279. doi: 10.1016/0167-7012(92)90017-X [DOI] [Google Scholar]
- 5.Genersch E.2010. American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol. 103Suppl 1: S10–S19. doi: 10.1016/j.jip.2009.06.015 [DOI] [PubMed] [Google Scholar]
- 6.Genersch E., Otten C.2003. The use of repetitive element PCR fingerprinting (rep-PCR) for genetic subtyping of German field isolates of Paenibacillus larvae subsp. larvae. Apidologie 34: 195–206. doi: 10.1051/apido:2003025 [DOI] [Google Scholar]
- 7.Genersch E., Ashiralieva A., Fries I.2005. Strain- and genotype-specific differences in virulence of Paenibacillus larvae subsp. larvae, a bacterial pathogen causing American foulbrood disease in honeybees. Appl. Environ. Microbiol. 71: 7551–7555. doi: 10.1128/AEM.71.11.7551-7555.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genersch E., Forsgren E., Pentikäinen J., Ashiralieva A., Rauch S., Kilwinski J., Fries I.2006. Reclassification of Paenibacillus larvae subsp. pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without subspecies differentiation. Int. J. Syst. Evol. Microbiol. 56: 501–511. doi: 10.1099/ijs.0.63928-0 [DOI] [PubMed] [Google Scholar]
- 9.Gordon R. E., Haynes W. C., Pang C. H.-N.1973. The genus Bacillus. Agriculture Handbook No. 427. U.S. Department of Agriculture. Washington, DC. [Google Scholar]
- 10.Heyndrickx M., Vandemeulebroecke K., Hoste B., Janssen P., Kersters K., De Vos P., Logan N. A., Ali N., Berkeley R. C. W.1996. Reclassification of Paenibacillus (formerly Bacillus) pulvifaciens (Nakamura 1984) Ash et al. 1994, a later subjective synonym of Paenibacillus (formerly Bacillus) larvae (White 1906) Ash et al. 1994, as a subspecies of P. larvae, with emended descriptions of P. larvae as P. larvae subsp. larvae and P. larvae subsp. pulvifaciens. Int. J. Syst. Bacteriol. 46: 270–279. doi: 10.1099/00207713-46-1-270 [DOI] [PubMed] [Google Scholar]
- 11.Govan V. A., Allsopp M. H., Davison S.1999. A PCR detection method for rapid identification of Paenibacillus larvae. Appl. Environ. Microbiol. 65: 2243–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrissey B. J., Helgason T., Poppinga L., Fünfhaus A., Genersch E., Budge G. E.2015. Biogeography of Paenibacillus larvae, the causative agent of American foulbrood, using a new multilocus sequence typing scheme. Environ. Microbiol. 17: 1414–1424. doi: 10.1111/1462-2920.12625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuendorf S., Hedtke K., Tangen G., Genersch E.2004. Biochemical characterization of different genotypes of Paenibacillus larvae subsp. larvae, a honey bee bacterial pathogen. Microbiology 150: 2381–2390. doi: 10.1099/mic.0.27125-0 [DOI] [PubMed] [Google Scholar]
- 14.Rauch S., Ashiralieva A., Hedtke K., Genersch E.2009. Negative correlation between individual-insect-level virulence and colony-level virulence of Paenibacillus larvae, the etiological agent of American foulbrood of honeybees. Appl. Environ. Microbiol. 75: 3344–3347. doi: 10.1128/AEM.02839-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue D., Nordhoff M., Wieler L. H., Genersch E.2008. Fluorescence in situ hybridization (FISH) analysis of the interactions between honeybee larvae and Paenibacillus larvae, the causative agent of American foulbrood of honeybees (Apis mellifera). Environ. Microbiol. 10: 1612–1620. doi: 10.1111/j.1462-2920.2008.01579.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.