Abstract
The PGMY-PCR for human papillomavirus (HPV) was evaluated, in parallel with nested PCR (nPCR), in samples with noted Hybrid Capture II (HCII) and MY-PCR results. PGMY-PCR detected HPV DNA in 2.5% of HCII-negative-MY-PCR-negative samples and in 71.7% of HCII-positive-MY-PCR-negative samples; also, it detected the MY-PCR-negative-nPCR-negative types HPV-42, HPV-44, HPV-51, HPV-87, and HPV-89.
Human papillomavirus (HPV) is the main etiological agent of cervical cancer (26). The approximately 45 HPV types that infect the genital mucosa are classified low-risk HPV (LR HPV) or high-risk HPV (HR HPV) on the basis of their association with premalignant and malignant lesions (2). Reliable identification of HPV may be relevant for clinical management of cervical lesions and cancer.
At present, the most widely used HPV tests are the hybridization assay Hybrid Capture II (HCII; Digene), capable of detecting 5 LR and 13 HR HPV types (22, 24), and the one-step PCR with general primers MY09 and MY11 (MY-PCR) (10, 15, 19). An additional approach, nested PCR (nPCR) with primers MY09/11 and GP5+/6+ (5, 11), is based on two-step amplifications. Two-step PCR amplifications are generally not recommended for routine diagnosis of HPV infection, being more cumbersome and more susceptible to contamination. However, they have been shown to be an extremely sensitive and reliable means of HPV detection (5, 12, 21); thus, when performed in appropriate conditions to prevent contamination (14, 16), two-step PCR may be considered a high-sensitivity standard to which methods that are new and still under evaluation may be compared. After general (one- or two-step) PCR, the HPV type is identified by different methods (15, 20, 23); in particular, direct cycle sequencing (DCS) is considered the “gold standard” for accurate HPV identification and genotyping (6, 7).
Recently, to improve HPV detection, the MY09/11 primers were redesigned as general, no-degenerate PGMY09/11 primers (9). The one-step PGMY-PCR has been reported to improve the analytical sensitivity, specificity, and reproducibility of MY-PCR (3).
In the present study, the new PGMY-PCR for HPV DNA detection was evaluated, in paired comparison with nPCR, by the use of a series of cervical samples previously examined by HCII and MY-PCR assays. DCS-based HPV genotyping was performed after all PCR assays.
Cervical scrapings were selected from 1,100 specimens that had come to the Laboratory of Virology, Department of Hygiene and Microbiology (University of Palermo, Palermo, Italy), between January 2000 and December 2002. Diagnosis of HPV infection was performed by HCII and MY-PCR assays. A total of 307 samples were selected as follows.
For group I, 64 samples positive by HCII and MY-PCR were selected. These included 44 samples that were LR or HR positive and MY-PCR positive for one of genotypes HPV-6, -11, -16, -18, -31, -33, -39, -45, -52, -53, -54, -56, -58, -61, -62, -66, -68, -70, -81, -82, -83, or LVX160 (two samples for each type) and 20 samples that were positive by HCII for LR and HR genotypes, 18 of which were MY-PCR positive for a single HPV (HPV-6, -16, or -18) and 2 of which were MY-PCR positive for mixed types (HPV-6 and -16 and HPV-11 and -16). Only PGMY-PCR was applied to these samples.
For group II, 200 samples negative by MY-PCR and HCII were randomly selected. Both PGMY-PCR and nPCR were applied to these samples.
For group III, 106 samples that were HCII positive (43 LR HPV, 58 HR HPV, and 5 LR and HR HPV positive) and MY-PCR negative, as confirmed by repeat testing, were selected. Both PGMY-PCR and nPCR were applied to these samples.
Cervical cells, obtained with a spatula and an endocervical cytobrush, were placed into 20 ml of PreservCyt Solution (Cytyc), washed, spun down, and split in three aliquots: the first pellet, resuspended in 1 ml of specimen transport medium (DIGENE), was used for HCII; the second was used for DNA extraction as previously described (1); and the third pellet was stored at −70°C for future use.
The HCII assay was performed according to the instructions in the manufacturer's package insert. The results were expressed as relative light units with respect to a cutoff: relative light units/cutoff ≥ 1.0 indicated the presence of HPV DNA.
Amplifications were carried out in a Mastercycler (Eppendorf, Germany), and the products were analyzed in 8% polyacrylamide gel. The sensitivity assays were performed by amplification of HPV-positive HeLa cells from 103 (20,000 to 50,000 HPV-18 copies) to 10−2 (2 to 5 copies) and SiHa cells from 104 (10,000 to 20,000 HPV-16 copies) to 10−1 (1 to 2 copies). Negative controls were blank control and the HPV-negative Wi-38 cell line. Special care was taken to control contaminations (14, 16). The standard MY-PCR (15) was performed using an ultrasensitive amplification profile (9); the amplification product was approximately 450 bp. The nPCR was performed with nested GP5+/6+ primers (4): 2 μl of the MY-PCR product was amplified with 50 pmol of primers, in the same reaction mixture as described before, except that the MgCl2 concentration was 3.5 mM; the program was 10 min at 95°C followed by 40 cycles of 60 s at 94°C, 60 s at 42°C, 45 s at 72°C, and 30 s at 72°C. The amplification product was approximately 140 bp. The PGMY-PCR (3, 9) was performed with 50 μl of 50 mM KCl-10 mM Tris-HCl (pH 8.4)-2 mM MgCl2-200 μM of each dNTP-10 pmol each of nonbiotinylated PGMY09-PGMY11-1.5 U of AmpliTaq Gold. A total of 5 μl of lysate was amplified as in the MY-PCR; the amplification product was 450 bp. The sequencing analysis was performed using 5 ng of MY- and PGMY-PCR product and 3 ng of nPCR product as previously described (8). The MY- and PGMY-PCR program was 25 cycles of 30 s at 96°C, 45 s at 50°C, and 1 min at 60°C; the nPCR program was 25 cycles of 30 s at 96°C, 45 s at 45°C, and 1 min at 60°C. Genotypes were considered LR or HR according to the L1 HPV phylogenetic tree (17).
The HPV detection rate was analyzed by the Z test (statistical significance, P ≤ 0.05). The agreement between PGMY-PCR and nPCR was measured by Cohen's kappa statistic (κ > 0.75, substantial agreement; κ = 0.4 to 0.75, fair to good agreement; κ < 0.40, poor agreement) or by two separate agreement indexes, ppos and pneg, representing a generalization of Cohen's κ without chance correction.
In the amplification of HeLa and SiHa cells, PGMY-PCR detected 10 to 20 copies of HPV-16 and -18, MY-PCR detected 100 to 200 copies, and nPCR detected 1 to 2 copies.
In the 64 samples with HCII- and MY-PCR-positive results (group I), PGMY-PCR detected all the 22 different HPV genotypes identified by HCII and MY-PCR. In the subgroup of 20 specimens HCII positive for mixed types, where MY-PCR found two cases of double infections, PGMY-PCR identified four cases.
In the 200 specimens negative by HCII and MY-PCR (group II) and analyzed by both PGMY-PCR and nPCR (Table 1), PGMY-PCR yielded 2.5% and nPCR yielded 12.0% HPV DNA-positive results (P = 0.0004). Although the concordance between PGMY-PCR and nPCR was high (88.5%), the statistically significant agreement attained was poor (κ = 0.173; P = 0.001) due to symmetrically unbalanced marginal frequencies in HPV DNA positive and negative results. Concordance between PGMY-PCR and nPCR was then expressed by separate indexes of agreement, showing a concordance of identification of negative samples (pneg = 0.938) and poor agreement for detection of positive cases (ppos = 0.20). In this group, PGMY-PCR detected some common HPV genotypes (HPV-6, -11, -16, -18, -33, and -68) also identified by nPCR and was the only assay to detect the LR type HPV-89.
TABLE 1.
Observed and expected counts of joint distribution of the 200 samples with HPV-negative results in HCII and MY-PCR by PGMY-PCR and nPCR
| nPCR result | PGMY-PCR sample resulta
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Negative
|
Positive
|
Total
|
|||||||
| Count | Expected count | % Of total | Count | Expected count | % Of Total | Count | Expected count | % Of total | |
| Negative | 174 | 171.6 | 87.0 | 2 | 4.4 | 1.0 | 176 | 171.0 | 88.0 |
| Positive | 21 | 23.4 | 10.5 | 3 | 0.6 | 1.5 | 24 | 24.0 | 12.0 |
| Total | 195 | 195.0 | 97.5 | 5 | 5.0 | 2.5 | 200 | 200.0 | 100.0 |
HPV DNA detection rates by PGMY-PCR and nPCR are shown in boldface. Measures of agreement:
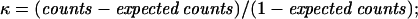
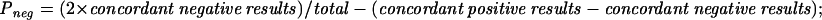
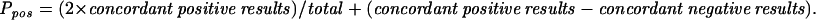
In the 106 specimens positive by HCII and negative by MY-PCR (group III), analyzed by both PGMY-PCR and nPCR (Table 2), PGMY-PCR results were positive in 71.7% and nPCR results were positive in 56.6% of the samples (P = 0.0003); high concordance (84.9%) and good agreement (κ = 0.68; P < 0.00001) of HPV DNA detection was evident. In 60 samples of this group, both PGMY-PCR and nPCR yielded positive results. Agreement of HPV type identification results between HCII and the two PCR assays was evident in 58 samples, in which HPV types that tested as LR by HCII were identified by both PCR assays as HPV-6 or -11 and HPV types that tested as HR were identified as HPV-16,-18, -31, -33, -39, -53, -61, -68, -81, or -83; discrepancy was found in 2 samples that tested as LR or HR HPV positive by HCII and in which PGMY-PCR and nPCR identified HPV-6 alone. For 16 samples of this group, PGMY-PCR gave positive results and nPCR gave negative results. Agreement of HPV type identification between HCII and PGMY-PCR was present in 13 cases, in which HPV types identified as LR by HCII were identified as HPV-44 or -87 and HR types were identified as HPV-16 or -51; discrepancy was found in three samples LR or HR positive by HCII where PGMY-PCR found HPV-42 alone.
TABLE 2.
Observed and expected counts of joint distribution of the 106 samples with an HPV-positive result in HCII and an HPV-negative result in MY-PCR by PGMY-PCR and nPCR
| nPCR result | PGMY-PCR sample resulta
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Negative
|
Positive
|
Total
|
|||||||
| Count | Expected count | % Of total | Count | Expected count | % Of total | Count | Expected count | % Of total | |
| Negative | 30 | 13.0 | 28.3 | 16 | 33.0 | 15.1 | 46 | 46.0 | 43.4 |
| Positive | 0 | 17.0 | 0.0 | 60 | 43.0 | 56.6 | 60 | 60.0 | 56.6 |
| Total | 30 | 30.0 | 28.3 | 76 | 76.0 | 71.7 | 106 | 106.0 | 100.0 |
HPV DNA detection rates by PGMY-PCR and nPCR are shown in boldface. Measure of agreement:
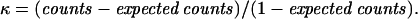
The present study compared the new PGMY-PCR for HPV DNA to the well-established methods HCII and MY-PCR as well as to a two-step, nPCR system (8). Actually, nPCR is the most sensitive HPV DNA assay (5, 12), but it is impractical for high-throughput HPV detection. In this analysis, the DCS-based genotyping approach permitted a wider range of identification of HPV types amplified by PGMY-PCR than the line-blot hybridization of the original assay format (9, 13).
PGMY-PCR was proven efficient in the amplification of 22 different HPV types; of these, HPV LVX160 has never been previously detected in PGMY-PCR analysis based on the line-blot system, which does not include a specific probe for this type. In this DCS-based analysis of PGMY-PCR, it did not improve the identification of multiple genotypes compared to MY-PCR. Generally, DCS methods using a general amplification primer as a sequencing primer have been shown to be of limited use in cases of multiple infections (24); this aspect of PGMY-PCR could be better evaluated by newly developed DCS approaches (7, 25), suited for selective detection and genotyping of HPV in multiple-variant-infected samples.
In sensitivity assays of HeLa and SiHa cells, PGMY-PCR showed a 10-fold increase in sensitivity over MY-PCR but not over nPCR, confirmed as the most sensitive method. Also, in clinical specimens, PGMY-PCR was shown to be slightly more sensitive than MY-PCR and less sensitive than nPCR. The higher sensitivity of PGMY-PCR was also the most likely reason for the 2.5% HPV detection rates in HCII-negative samples, since all types found are included in the HCII probe sets, and HPV-89, which is not included, is detectable by cross-hybridization (18).
Results of PGMY-PCR for HCII-positive and MY-PCR-negative samples pointed out some advantages of PGMY-PCR over MY-PCR and nPCR. First, the PGMY primers were more robust, leading to greater consistency of amplification and one-step detection of HPV DNA, as evident in samples with PGMY-PCR-positive results which tested MY-PCR negative (these specimens were confirmed as HPV positive by nPCR). Second, a broader range of HPV types was detected, as shown by PGMY-PCR-positive results for samples negative not only by MY-PCR but also by nPCR and found positive for genotypes HPV-42, -44, -51, and -87. Of note, genotypes HPV-44 and -87 are newly identified by PGMY-PCR in the present analysis. And third, improved analytical sensitivity was seen for the HR type HPV-16, also reported in other studies as detected with difficulty by MY-PCR (13).
In conclusion, compared to MY-PCR, PGMY-PCR seems only slightly more sensitive but remarkably more efficient and also shows an enlarged HPV detection range. Compared to HCII, PGMY-PCR shows the same slightly increased sensitivity as MY-PCR. Compared to nPCR, PGMY-PCR represents a tradeoff of lesser sensitivity for a greater number of HPV types detected.
The highly sensitive and robust PGMY-PCR assay may be considered a versatile HPV amplification system, suitable to be used for clinical purposes, particularly when HPV characterization more specific than the group risk identification provided by the HCII test is required.
Acknowledgments
This work was supported by the Ministry of Instruction and University Research (fund MIUR [60%], 2000, and fund PRIN [40%], 2001).
REFERENCES
- 1.Ammatuna, P., L. Giovannelli, D. Giambelluca, S. Mancuso, E. Rubino, P. Colletti, G. Mazzola, P. Belfiore, and R. Lima. 2000. Presence of human papillomavirus and Epstein-Barr virus in the cervix of women infected with the human immunodeficiency virus. J. Med. Virol. 62:410-415. [DOI] [PubMed] [Google Scholar]
- 2.Burd, E. M. 2003. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 16:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coutlée, F., P. Gravitt, J. Kornegay, C. Hankins, H. Richardson, N. Lapointe, H. Voyer, The Canadian Women's HIV Study Group, and E. Franco. 2002. Use of PGMY primers in L1 consensus PCR improves detection of human papillomavirus DNA in genital samples. J. Clin. Microbiol. 40:902-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Roda Husman, A. M., J. M. M. Walboomers, A. J. van den Brule, C. J. L. M. Meijer, and P. J. F. Snijders. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 76:1057-1062. [DOI] [PubMed] [Google Scholar]
- 5.Evander, M., K. Edlund, E. Bodén, Å. Gustafsson, M. Jonsson, R. Karlsson, E. Rylander, and G. Wadell. 1992. Comparison of a one-step and a two-step polymerase chain reaction with degenerate general primers in a population-based study of human papillomavirus infection in young Swedish women. J. Clin. Microbiol. 30:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feoli-Fonseca, J. C., L. L. Oligny, P. Brochu, P. Simard, S. Falconi, and W. Yotov. 2001. Human papillomavirus (HPV) study of 691 pathological specimens from Quebec by PCR-direct sequencing approach. J. Med. Virol. 63:284-292. [DOI] [PubMed] [Google Scholar]
- 7.Gharizadeh, B., M. Ghaderi, D. Donnelly, B. Amini, K. Wallin, and P. Nyrén. 2003. Multiple-primer DNA sequencing method. Electrophoresis 24:1145-1151. [DOI] [PubMed] [Google Scholar]
- 8.Giovannelli, L., G. Campisi, A. Lama, O. Giambalvo, J. Osborn, V. Margiotta, and P. Ammatuna. 2002. Human papillomavirus DNA in oral mucosal lesions. J. Infect. Dis. 185:833-836. [DOI] [PubMed] [Google Scholar]
- 9.Gravitt, P. E., C. L. Peyton, T. Q. Alessi, C. M. Wheeler, F. Coutlée, A. Hildesheim, M. H. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of human papillomaviruses. J. Clin. Microbiol. 38:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harnish, D. G., L. M. Belland, E. E. Scheid, and T. E. Rohan. 1999. Evaluation of human papillomavirus-consensus primers for HPV detection by the polymerase chain reaction. Mol. Cell. Probes 13:9-21. [DOI] [PubMed] [Google Scholar]
- 11.Husniak, K., M. Grce, L. Magdic, and K. Pavelic. 2000. Comparison of five different polymerase chain reaction methods for detection of human papillomavirus in cervical cell specimens. J. Virol. Methods 88:125-134. [DOI] [PubMed] [Google Scholar]
- 12.Kay, P., K. Meehan, and A. L. Williamson. 2002. The use of nested polymerase chain reaction and restriction fragment length polymorphism for the detection and typing of mucosal human papillomaviruses in samples containing low copy numbers of viral DNA. J. Virol. Methods 105:159-170. [DOI] [PubMed] [Google Scholar]
- 13.Kornegay, J. R., A. P. Shepard, C. Hankins, E. Franco, N. Lapointe, H. Richardson, The Canadian Women's HIV Study Group, and F. Coutlée. 2001. Nonisotopic detection of human papillomavirus DNA in clinical specimens using a consensus PCR and a generic probe mix in an enzyme-linked immunosorbent assay format. J. Clin. Microbiol. 39:3530-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwok, and R. Higuchi. 1989. Avoiding false positivities with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 15.Manos, M. M., Y. Ting, D. K. Wright, A. J. Lewis, T. R. Broker, and S. M. Wolinsky. 1989. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells 7:209-214. [Google Scholar]
- 16.Mant, C., B. Kell, J. M. Best, and J. Cason. 1997. Polymerase chain reaction protocols for the detection of DNA from mucosal human papillomavirus types -6, -11, -16, -18, -31 and -33. J. Virol. Methods 66:169-178. [DOI] [PubMed] [Google Scholar]
- 17.Myers, G., F. Sverdrup, and C. Baker (ed.). 1997. Human papillomavirus 1997: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.M.
- 18.Poljak, M., I. J. Marin, K. Seme, and A. Vince. 2002. Hybrid Capture II HPV test detects at least 15 human papillomavirus genotypes not included in its current high-risk probe cocktail. J. Clin. Virol. 25:S89-S97. [DOI] [PubMed] [Google Scholar]
- 19.Qu, W., G. Jiang, Y. Cruz, C. J. Chang, G. Y. F. Ho, R. S. Klein, and R. D. Burk. 1997. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J. Clin. Microbiol. 35:1304-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rady, P. L., R. Chin, I. Arany, T. K. Hughes, and S. Tyring. 1993. Direct sequencing of consensus primer generated PCR fragments of human papillomavirus. J. Virol. Methods 43:335-350. [DOI] [PubMed] [Google Scholar]
- 21.Strauss, S., J. Z. Jordens, U. Desselberger, and J. J. Gray. 2000. Single-tube real-time nested polymerase chain reaction for detecting human papillomavirus DNA. Diagn. Mol. Pathol. 9:151-157. [DOI] [PubMed] [Google Scholar]
- 22.Terry, G., L. Ho, P. Londesborough, J. Cuzick, I. Mielzynska-Lohnas, and A. Lorincz. 2001. Detection of high-risk HPV types by the Hybrid Capture 2 test. J. Med. Virol. 65:155-162. [PubMed] [Google Scholar]
- 23.van den Brule, A. J. C., R. Pol, N. Fransen-Daalmeijer, L. M. Schouls, C. J. L. M. Meijer, and P. J. F. Snijders. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40:779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vernon, S. D., E. R. Unger, and D. Williams. 2000. Comparison of human papillomavirus detection and typing by cycle sequencing, line blotting, and hybrid capture. J. Clin. Microbiol. 38:651-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinayagamoorthy, T., K. Mulatz, and R. Hodkinson. 2003. Nucleotide sequence-based multitarget identification. J. Clin. Microbiol. 41:3284-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.zur Hausen, H., L. Gissman, and J. R. Schlehofer. 1984. Viruses in the etiology of human genital cancers. Prog. Med. Virol. 30:170-186. [PubMed] [Google Scholar]


