Abstract
A 761-bp portion of the tuf gene (encoding the elongation factor Tu) from 28 clinically relevant streptococcal species was obtained by sequencing amplicons generated using broad-range PCR primers. These tuf sequences were used to select Streptococcus-specific PCR primers and to perform phylogenetic analysis. The specificity of the PCR assay was verified using 102 different bacterial species, including the 28 streptococcal species. Genomic DNA purified from all streptococcal species was efficiently detected, whereas there was no amplification with DNA from 72 of the 74 nonstreptococcal bacterial species tested. There was cross-amplification with DNAs from Enterococcus durans and Lactococcus lactis. However, the 15 to 31% nucleotide sequence divergence in the 761-bp tuf portion of these two species compared to any streptococcal tuf sequence provides ample sequence divergence to allow the development of internal probes specific to streptococci. The Streptococcus-specific assay was highly sensitive for all 28 streptococcal species tested (i.e., detection limit of 1 to 10 genome copies per PCR). The tuf sequence data was also used to perform extensive phylogenetic analysis, which was generally in agreement with phylogeny determined on the basis of 16S rRNA gene data. However, the tuf gene provided a better discrimination at the streptococcal species level that should be particularly useful for the identification of very closely related species. In conclusion, tuf appears more suitable than the 16S ribosomal RNA gene for the development of diagnostic assays for the detection and identification of streptococcal species because of its higher level of species-specific genetic divergence.
Streptococci are a heterogeneous group of bacteria, consisting of as many as 48 species, including important human pathogens such as Streptococcus pneumoniae, S. pyogenes, and S. agalactiae (13, 39). S. pneumoniae is considered a common agent of community-acquired pneumonia, otitis media, and endocarditis. S. pyogenes causes a wide array of serious infections, including pharyngitis, soft-tissue infections, scarlet fever, and toxic shock-like syndromes. S. agalactiae is an important cause of serious neonatal infections characterized by sepsis and meningitis. Most other streptococci are members of the normal human floras (39). Their presence in aseptic body sites often indicates subacute bacterial endocarditis.
Current systems for identification of clinically relevant streptococcal species largely depend on an array of culture-based biochemical tests (12). Some simple and rapid presumptive physiological tests or serological tests are available (12, 15, 39). The most clinically significant pathogens among streptococci can be rapidly identified using phenotypic and immunological tests. However, additional phenotypic tests may be required to confirm identification at the species level. In fact, identification of streptococci to the species level may require up to 7 days because these bacteria grow slowly and because identification may rely on a cumbersome classification system that does not always correlate with phylogenetic analysis (13, 22, 46).
Many DNA-based methods have been applied for the identification and detection of clinically important streptococcal species. Hybridization-based assays for the specific detection of S. pneumoniae (8, 14), S. pyogenes (19, 37), S. agalactiae (3), and S. bovis (47) have been developed. However, these probe-based tests are prone to a lack of sensitivity. Consequently, a number of PCR-based approaches having increased analytical sensitivities and allowing detection of streptococci directly from clinical specimens have been developed (1, 6, 10, 17, 21, 25, 34, 35). However, there is no published study on the development of a Streptococcus-specific PCR assay. The use of genus- or group-specific PCR assays coupled with species-specific internal probes should allow researchers to substantially decrease the number of primers used for bacterial identification, thereby simplifying the development of molecular assays for bacteria (2).
Phylogenetic analyses of streptococci conducted using several conserved genes, including those coding for 16S rRNA, heat shock proteins, glucose pyrophosphorylase, and superoxide dismutase, have been reported (4, 13, 23, 33, 43). All of these phylogenetic studies demonstrated the usefulness of genetic approaches to improve the accuracy of streptococcal species identification.
Genus-specific PCR-based assays, targeting the tuf gene encoding elongation factor Tu, for the specific detection of enterococci (26) and staphylococci (31) have been previously described by our group. Similarly, newly generated tuf streptococcal sequences were used in the present study for PCR detection and extensive phylogenetic analysis of streptococci.
MATERIALS AND METHODS
Microorganisms.
Reference strains representing 58 gram-positive species (including 28 streptococcal species) and 44 gram-negative species were used in this study (Table 1). All of these strains were obtained from the American Type Culture Collection (Manassas, Va.) or from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). Clinical isolates of streptococci (n = 153) obtained from the microbiology laboratory of the Centre Hospitalier Universitaire de Québec (CHUQ), Pavillon CHUL (Sainte-Foy, Québec, Canada), were also used (Table 2). The identification of all streptococcal strains was confirmed by conventional biochemical and/or immunological testing.
TABLE 1.
Reference bacterial strains used to validate the Streptococcus-specific assay
| Strain |
|---|
| Gram-positive bacteria (58 species) |
| Abiotrophia defectiva ATCC 49176 |
| E. avium ATCC 14025 |
| E. casseliflavus ATCC 25788 |
| E. dispar ATCC 51266 |
| E. durans ATCC 19432 |
| E. faecalis ATCC 19433 |
| E. faecium ATCC 19434 |
| E. gallinarum ATCC 49573 |
| E. hirae ATCC 8043 |
| E. mundtii ATCC 43186 |
| E. raffinosus ATCC 49427 |
| E. solitarius ATCC 49428 |
| Gemella haemolysans ATCC 10379 |
| Granulicatella adiacens ATCC 49175 |
| Lactobacillus acidophilus ATCC 4356 |
| L. lactis subsp. lactis ATCC 11454 |
| Leifsonia aquatica ATCC 14665 |
| Listeria ivanovii ATCC 19119 |
| Listeria monocytogenes ATCC 15313 |
| Listeria seeligeri ATCC 35967 |
| Micrococcus luteus ATCC 9341 |
| Staphylococcus aureus ATCC 25923 |
| Staphylococcus capitis subsp. capitis ATCC 27840 |
| Staphylococcus epidermidis ATCC 14990 |
| Staphylococcus haemolyticus ATCC 29970 |
| Staphylococcus hominis subsp. hominis ATCC 27844 |
| Staphylococcus lugdunensis ATCC 43809 |
| Staphylococcus saprophyticus ATCC 15305 |
| Staphylococcus simulans ATCC 27848 |
| Staphylococcus warneri ATCC 27836 |
| S. acidominimus ATCC 51726 |
| S. agalactiae ATCC 13813 |
| S. anginosus ATCC 33397 |
| S. bovis ATCC 33317 |
| S. constellatus subsp. constellatus ATCC 27823 |
| S. criceti ATCC 19642 |
| S. cristatus ATCC 51100 |
| S. downei ATCC 33748 |
| S. dysgalactiae ATCC 43078 |
| S. equi subsp. equi ATCC 9528 |
| S. ferus ATCC 33477 |
| S. gordonii ATCC 10558 |
| S. intermedius ATCC 27335 |
| S. macacae ATCC 35911 |
| S. mitis ATCC 49456 |
| S. mutans ATCC 25175 |
| S. oralis ATCC 35037 |
| S. parasanguinis ATCC 15912 |
| S. parauberis DMS 6631 |
| S. pneumoniae ATCC 27336 |
| S. pyogenes ATCC 19615 |
| S. ratti ATCC 19645 |
| S. salivarius ATCC 7073 |
| S. sanguinis ATCC 10556 |
| S. sobrinus ATCC 27352 |
| S. suis ATCC 43765 |
| S. uberis ATCC 19436 |
| S. vestibularis ATCC 49124 |
| Gram-negative bacteria (44 species) |
| Acinetobacter baumannii ATCC 19606 |
| Acinetobacter haemolyticus ATCC 17906 |
| Bordetella pertussis ATCC 9797 |
| Burkholderia cepacia ATCC 25416 |
| Citrobacter koseri ATCC 27028 |
| Citrobacter freundii ATCC 8090 |
| Enterobacter aerogenes ATCC 13048 |
| Enterobacter cloacae ATCC 13047 |
| Pantoea agglomerans ATCC 27155 |
| Escherichia coli ATCC 25922 |
| Haemophilus ducreyi ATCC 33940 |
| Haemophilus haemolyticus ATCC 33390 |
| Haemophilus influenzae ATCC 9007 |
| Haemophilus parahaemolyticus ATCC 10014 |
| Haemophilus parainfluenzae ATCC 7901 |
| Hafnia alvei ATCC 13337 |
| Klebsiella oxytoca ATCC 13182 |
| Klebsiella pneumoniae subsp. pneumoniae ATCC 13883 |
| Moraxella atlantae ATCC 29525 |
| Moraxella catarrhalis ATCC 25240 |
| Moraxella osloensis ATCC 19976 |
| Morganella morganii subsp. morganii ATCC 25830 |
| Neisseria caviae ATCC 14659 |
| Neisseria elongata subsp. elongata ATCC 25295 |
| Neisseria gonorrhoeae ATCC 35201 |
| Neisseria meningitidis ATCC 13077 |
| Neisseria mucosa ATCC 19696 |
| Pasteurella aerogenes ATCC 27883 |
| Proteus hauseri ATCC 13315 |
| Proteus mirabilis ATCC 25933 |
| Providencia alcalifaciens ATCC 9886 |
| Providencia rettgeri ATCC 9250 |
| Providencia rustigianii ATCC 33673 |
| Providencia stuartii ATCC 29914 |
| Pseudomonas aeruginosa ATCC 27853 |
| Pseudomonas fluorescens ATCC 13525 |
| Pseudomonas stutzeri ATCC 17588 |
| Salmonella enterica serovar Typhimurium ATCC 14028 |
| Serratia marcescens ATCC 8100 |
| Shigella flexneri ATCC 12022 |
| Shigella sonnei ATCC 29930 |
| Stenotrophomonas maltophilia ATCC 13843 |
| Suttonella indologenes ATCC 25869 |
| Yersinia enterocolitica ATCC 9610 |
TABLE 2.
Validation of the PCR assay with DNA from clinical isolates of streptococci
| Species | No. of strains testeda | No. of PCR- positive strains |
|---|---|---|
| S. agalactiae | 21 | 21 |
| S. anginosus | 3 | 3 |
| S. bovis | 3 | 2 |
| Group C streptococci | 3 | 3 |
| Group G streptococci | 9 | 9 |
| S. mitis | 13 | 13 |
| S. mutans | 7 | 7 |
| S. pneumoniae | 24 | 24 |
| S. pyogenes | 21 | 21 |
| S. salivarius | 21 | 21 |
| S. sanguinis | 13 | 13 |
| Streptococcus spp. | 4 | 4 |
| S. viridans group streptococci | 11 | 11 |
| Total | 153 | 152 |
All strains were obtained from the microbiology laboratory of the CHUQ, Pavillon CHUL.
DNA sequencing.
Purified genomic DNA was prepared using a G NOME DNA extraction kit (Qbiogene Inc., Carlsbad, Calif.) (26). An 865-bp portion of tuf was amplified from 28 selected streptococcal species as previously described (31). Direct sequencing of these 865-bp amplicons provided edited 761-bp tuf sequences for all species. When required, a 1,471-bp portion of the 16S rRNA genes was amplified using universal primers (30). Direct sequencing of these 16S ribosomal RNA gene (rDNA) amplicons allowed verification of the accuracy of public database sequences and/or confirmation of the identification of streptococcal species. After electrophoresis, the gel was stained with methylene blue and PCR products having the predicted size were recovered using a QIAquick gel extraction kit (Qiagen Inc., Mississauga, Ontario, Canada) (31). The purified PCR products were then sequenced using a BigDye Ready Reaction cycle sequencing kit with a 377 sequencer (Applied Biosystems, Foster City, Calif.). To exclude the possibility of sequencing errors attributable to misincorporations by Taq DNA polymerase, each strand was sequenced twice using PCR products obtained from two independent rounds of PCR.
Oligonucleotides.
The partial tuf gene sequences obtained in this study as well as those available from public databases were analyzed using GCG Wisconsin software (version 10.3; Accelrys Inc., San Diego, Calif.). PCR primers were analyzed using Oligo primer analysis software (version 5.0; Molecular Biology Insights, Cascade, Col.). Oligonucleotides were synthesized with a model 394 DNA/RNA synthesizer (Applied Biosystems).
Streptococcus-specific PCR.
For all bacterial species tested, PCR amplifications using the Streptococcus-specific primers were performed from 1 μl of a genomic DNA preparation at 1 ng/μl which was transferred directly to a 19-μl PCR mixture containing 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 2.5 mM MgCl2, 0.4 μM concentrations of each of the Streptococcus-specific primers (Str1 [5′-GTACAGTTGCTTCAGGACGTATC-3′] and Str2 [5′-ACGTTCGATTTCATCACGTTG-3′]), 200 μM (each) deoxynucleoside triphosphate (Amersham Biosciences, Piscataway, N.J.), 3.3 μg of bovine serum albumin (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) per μl, and 0.5 U of Taq DNA polymerase (Promega, Madison, Wis.) combined with the TaqStart antibody (BD Biosciences Clontech, Palo Alto, Calif.). Thermal cycling for PCR amplification and agarose gel analysis of the amplified products were performed as previously described (32). The analytical sensitivity (i.e., the minimal number of genome copies detected per PCR) of the PCR assay was determined using serial twofold dilutions of quantitated genomic DNA purified from bacterial strains representing 28 streptococcal species (Table 1). Strict precautions to prevent carry-over of amplified DNA were used (28). Pre- and post-PCR manipulations were conducted in separate areas. Aerosol-resistant pipette tips were used to handle all reagents and samples. Control reactions to which no DNA was added were routinely performed to verify the absence of DNA carry-over.
Phylogenetic analysis.
Multiple sequence alignments were performed using PILEUP (GCG Wisconsin package, version 10.3) and/or CLUSTAL W software (version 1.83) (44) and checked manually with a GCG SeqLab editor to verify the quality of the alignments. The SeqLab editor was also used to identify regions containing gaps, indels, or ambiguities to be excluded for phylogenetic analysis. This edition process yielded a 761-bp tuf sequence suitable for phylogenetic analysis and a 1,260-bp sequence for 16S rDNA analysis. Distance phylogenetic trees were generated using a neighbor-joining or heuristic method with MEGA2 software (version 2.1) (27). Evolutionary distance values were calculated by using Kimura's two-parameter substitution model (18). Bootstrap values were obtained for 1,000 randomly generated trees. This number of replicates was sufficient to produce stable tree topologies. Maximum parsimony analyses were performed using the heuristic method of PAUP software (version 4.0b10; Sinauer Associates Inc., Sunderland, Mass.) with general search parameters (41). A sequence of Enterococcus faecalis V583 was used as an outgroup, because this species is phylogenetically close to streptococci.
Nucleotide sequence accession numbers.
GenBank accession numbers for the 761-bp tuf sequences determined in this study are as follows: AY266992 for S. acidominimus; AY266993, AY266994, AY266995, AY266996, and AF276256 for S. agalactiae; AF276257 for S. anginosus; AY266997 and AF276258 for S. bovis; AF276259 for S. constellatus subsp. constellatus; AF276260 for S. criceti; AF276261 for S. cristatus; AF276262 for S. downei; AF276263, AY582541, and AY582542 for S. dysgalactiae; AF276264 for S. equi subsp. equi; AF276265 for S. ferus; AF276266 and AY267005 for S. gordonii; AF276267 for S. intermedius; AF276268 for S. macacae; AF276269 and AY582543 for S. mitis; AF274741 for S. mutans; AF276270 and AY582544 for S. oralis; AF276271 for S. parasanguinis; AY267004 for S. parauberis; AF274742, AY267000, AY267001, AY267002, and AY267003 for S. pneumoniae; AF274743 for S. pyogenes; AF276272 for S. ratti; AF276273 for S. salivarius; AF276274 for S. sanguinis; AF276275 for S. sobrinus; AF274744 for S. suis; AF276276 for S. uberis; and AF276277 for S. vestibularis. GenBank accession numbers for the 1,260-bp 16S rDNA sequences are as follows: AY584476 for S. cristatus; AY584477 for S. parauberis; AY584478 for S. dysgalactiae; and AY584479 for S. ferus.
RESULTS
Sequencing of the streptococcal tuf gene.
We amplified and sequenced a portion of the tuf gene for all 28 streptococcal species tested using universal primers developed previously (31). The edited 761-bp tuf sequence was highly conserved among streptococci (86.1 to 99.1% identity at the nucleotide level) (Table 3). There were three pairs of streptococcal species having more than 98.0% identity (S. mitis versus S. pneumoniae [98.7% identity]; S. pyogenes versus S. dysgalactiae [98.6%]; and S. salivarius versus S. vestibularis [99.1%]).
TABLE 3.
DNA sequence identities between the streptococcal tuf sequencesa
| Species no.b | Species name | % Identity with species no.b:
|
|||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | ||
| 1 | S. acidominimus ATCC 51726 | 91.6 | 90.0 | 90.3 | 90.0 | 90.1 | 89.5 | 89.5 | 89.6 | 89.1 | 92.2 | 90.1 | 89.9 | 87.9 | 89.8 | 88.6 | 89.1 | 90.4 | 88.3 | 88.9 | 89.2 | 90.5 | 90.5 | 89.2 | 89.4 | 91.2 | 90.1 | 90.7 | 82.4 | 76.3 | 77.5 | 78.6 | |
| 2 | S. agalactiae ATCC 13813 | 90.2 | 91.6 | 92.4 | 91.3 | 89.4 | 93.4 | 89.8 | 93.0 | 92.1 | 91.3 | 92.1 | 91.2 | 88.2 | 92.2 | 89.0 | 92.6 | 93.0 | 93.2 | 91.9 | 92.8 | 90.1 | 93.0 | 91.6 | 88.6 | 91.6 | 93.0 | 92.5 | 81.7 | 76.9 | 77.9 | 77.9 | |
| 3 | S. anginosus ATCC 33397 | 90.2 | 88.2 | 92.8 | 98.0 | 90.0 | 90.5 | 89.8 | 90.4 | 88.8 | 90.1 | 91.3 | 97.2 | 90.0 | 91.7 | 90.4 | 91.5 | 91.3 | 89.0 | 91.6 | 90.5 | 91.2 | 91.5 | 91.2 | 89.4 | 89.5 | 91.2 | 91.2 | 81.7 | 76.1 | 78.8 | 77.4 | |
| 4 | S. bovis ATCC 33317 | 88.9 | 89.5 | 92.2 | 92.1 | 90.9 | 92.0 | 90.0 | 90.5 | 89.8 | 90.4 | 91.5 | 92.0 | 89.2 | 92.0 | 90.8 | 90.8 | 91.6 | 88.6 | 91.1 | 90.1 | 91.9 | 92.0 | 91.1 | 90.0 | 91.1 | 91.1 | 91.5 | 82.8 | 75.0 | 77.8 | 77.0 | |
| 5 | S. constellatus ATCC 27823 | 89.5 | 86.9 | 96.7 | 90.2 | 89.4 | 90.8 | 89.1 | 89.9 | 88.7 | 89.8 | 90.8 | 97.8 | 90.1 | 91.2 | 90.1 | 90.9 | 91.3 | 89.2 | 91.1 | 89.5 | 90.5 | 91.5 | 90.7 | 88.4 | 89.0 | 90.5 | 91.2 | 81.6 | 76.0 | 78.7 | 77.8 | |
| 6 | S. criceti ATCC 19642 | 88.9 | 88.2 | 90.2 | 90.9 | 89.5 | 89.4 | 96.7 | 87.8 | 86.9 | 90.8 | 89.0 | 89.1 | 87.6 | 88.4 | 89.6 | 87.8 | 89.5 | 86.2 | 87.9 | 87.1 | 91.2 | 90.8 | 89.5 | 96.2 | 88.6 | 87.9 | 90.8 | 81.1 | 73.5 | 77.7 | 76.5 | |
| 7 | S. cristatus ATCC 51100 | 86.9 | 93.5 | 88.2 | 87.6 | 86.9 | 87.6 | 89.2 | 92.5 | 90.7 | 91.2 | 95.0 | 90.7 | 87.5 | 95.0 | 88.3 | 95.5 | 94.4 | 90.7 | 94.6 | 92.4 | 90.3 | 93.2 | 95.9 | 88.7 | 92.6 | 92.5 | 92.9 | 83.0 | 75.8 | 79.6 | 77.3 | |
| 8 | S. downei ATCC 33748 | 89.5 | 90.2 | 92.2 | 90.9 | 90.2 | 98.0 | 89.5 | 87.4 | 87.5 | 91.6 | 88.0 | 88.8 | 87.1 | 88.0 | 89.2 | 87.9 | 89.2 | 86.6 | 87.8 | 86.6 | 90.3 | 90.1 | 89.1 | 95.0 | 88.7 | 87.5 | 90.1 | 80.0 | 73.1 | 77.0 | 75.6 | |
| 9 | S. dysgalactiae ATCC 43078 | 86.9 | 92.2 | 85.6 | 85.0 | 83.0 | 84.3 | 92.8 | 85.6 | 93.4 | 90.0 | 91.9 | 89.6 | 86.2 | 93.4 | 87.8 | 93.7 | 92.5 | 93.7 | 92.9 | 98.6 | 88.6 | 92.2 | 92.4 | 87.0 | 91.3 | 95.0 | 92.1 | 81.6 | 75.7 | 78.6 | 76.9 | |
| 10 | S. equi ATCC 9528 | 88.9 | 92.2 | 85.0 | 86.9 | 83.0 | 84.3 | 92.2 | 86.3 | 90.2 | 89.2 | 90.8 | 88.4 | 86.6 | 90.3 | 87.9 | 90.8 | 90.9 | 90.5 | 90.4 | 92.8 | 88.6 | 90.3 | 90.1 | 86.6 | 90.3 | 90.3 | 90.1 | 80.2 | 74.2 | 77.5 | 75.6 | |
| 11 | S. ferus ATCC 33477 | 90.9 | 86.9 | 87.6 | 84.3 | 86.9 | 88.2 | 85.6 | 90.2 | 84.3 | 84.3 | 90.3 | 89.9 | 88.3 | 91.1 | 90.4 | 90.5 | 91.9 | 90.0 | 90.5 | 89.4 | 91.7 | 91.7 | 90.5 | 89.9 | 91.7 | 90.9 | 91.2 | 80.6 | 75.6 | 79.9 | 77.4 | |
| 12 | S. gordonii ATCC 10558 | 89.5 | 90.9 | 88.2 | 88.9 | 86.9 | 90.2 | 94.1 | 90.9 | 88.9 | 89.5 | 85.0 | 90.7 | 87.8 | 95.1 | 87.8 | 94.4 | 95.1 | 89.8 | 95.0 | 91.7 | 90.7 | 92.5 | 97.2 | 87.9 | 92.4 | 91.3 | 92.2 | 82.9 | 76.2 | 78.6 | 77.5 | |
| 13 | S. intermedius ATCC 27335 | 89.5 | 87.6 | 98.0 | 90.9 | 96.7 | 89.5 | 88.9 | 91.5 | 85.0 | 84.3 | 88.2 | 87.6 | 90.3 | 91.3 | 90.4 | 91.3 | 91.1 | 89.2 | 91.7 | 89.2 | 90.8 | 90.9 | 90.8 | 88.7 | 88.8 | 90.8 | 91.1 | 81.3 | 76.1 | 79.1 | 77.7 | |
| 14 | S. macacae ATCC 35911 | 88.9 | 87.6 | 90.2 | 89.5 | 92.2 | 87.6 | 87.6 | 88.2 | 83.7 | 85.6 | 84.3 | 87.6 | 90.2 | 86.7 | 90.1 | 86.3 | 88.6 | 86.9 | 86.7 | 88.5 | 90.8 | 87.6 | 87.0 | 88.3 | 87.1 | 86.5 | 87.8 | 79.0 | 73.9 | 77.3 | 75.3 | |
| 15 | S. mitis ATCC 49456 | 88.9 | 88.9 | 88.2 | 85.6 | 85.6 | 85.0 | 89.5 | 86.9 | 92.8 | 85.6 | 84.3 | 92.2 | 87.6 | 85.6 | 87.8 | 97.1 | 93.3 | 91.3 | 98.7 | 92.8 | 89.9 | 93.0 | 95.5 | 87.9 | 93.0 | 93.0 | 92.9 | 83.7 | 76.9 | 78.2 | 77.3 | |
| 16 | S. mutans ATCC 25175 | 85.0 | 85.0 | 90.9 | 93.5 | 88.9 | 88.2 | 84.3 | 88.2 | 81.7 | 83.0 | 84.3 | 84.3 | 90.2 | 88.2 | 83.7 | 86.9 | 88.7 | 88.0 | 86.9 | 87.3 | 91.2 | 88.3 | 87.8 | 89.5 | 87.5 | 87.3 | 88.2 | 79.6 | 75.7 | 79.4 | 76.7 | |
| 17 | S. oralis ATCC 35037 | 88.9 | 92.8 | 88.2 | 85.6 | 85.6 | 85.6 | 93.5 | 87.6 | 96.7 | 89.5 | 85.6 | 89.5 | 87.6 | 86.3 | 96.1 | 82.4 | 93.7 | 91.5 | 97.4 | 93.0 | 89.4 | 93.3 | 94.5 | 87.3 | 93.2 | 93.4 | 93.2 | 82.9 | 76.5 | 79.1 | 77.7 | |
| 18 | S. parasanguinis ATCC 15912 | 88.2 | 92.2 | 86.9 | 87.6 | 86.9 | 89.5 | 95.4 | 90.2 | 90.2 | 89.5 | 86.3 | 94.8 | 87.6 | 86.9 | 89.5 | 84.3 | 92.2 | 90.9 | 93.4 | 92.2 | 91.5 | 93.8 | 95.0 | 88.7 | 92.9 | 92.2 | 93.3 | 82.3 | 76.3 | 80.8 | 78.3 | |
| 19 | S. parauberis DSM 6631 | 83.7 | 91.5 | 82.4 | 82.4 | 82.4 | 81.0 | 90.9 | 82.4 | 96.7 | 87.6 | 82.4 | 86.3 | 83.0 | 84.3 | 90.2 | 80.4 | 93.5 | 88.2 | 91.1 | 93.6 | 88.4 | 90.4 | 89.9 | 86.1 | 89.9 | 92.6 | 90.1 | 80.7 | 76.2 | 78.8 | 76.3 | |
| 20 | S. pneumoniae ATCC 27336 | 90.2 | 89.5 | 89.5 | 86.3 | 86.9 | 85.6 | 90.2 | 87.6 | 93.5 | 86.3 | 85.6 | 92.8 | 88.9 | 86.9 | 98.7 | 83.0 | 96.1 | 90.2 | 90.9 | 92.2 | 89.8 | 92.6 | 95.4 | 87.5 | 93.0 | 92.9 | 92.5 | 83.6 | 77.0 | 78.6 | 77.5 | |
| 21 | S. pyogenes ATCC 19615 | 86.9 | 92.2 | 85.6 | 85.0 | 83.0 | 84.3 | 92.8 | 85.6 | 100.0 | 90.2 | 84.3 | 88.9 | 85.0 | 83.7 | 92.8 | 81.7 | 96.7 | 90.2 | 96.7 | 93.5 | 88.0 | 91.6 | 92.1 | 86.6 | 91.1 | 94.4 | 91.5 | 81.3 | 75.6 | 78.4 | 76.7 | |
| 22 | S. ratti ATCC 19645 | 90.2 | 88.2 | 93.5 | 94.1 | 91.5 | 89.5 | 88.9 | 89.5 | 85.0 | 86.3 | 85.6 | 90.9 | 91.5 | 93.5 | 86.3 | 90.2 | 86.3 | 88.2 | 82.4 | 87.6 | 85.0 | 90.7 | 90.8 | 91.1 | 89.6 | 88.3 | 89.9 | 82.8 | 74.6 | 78.6 | 76.3 | |
| 23 | S. salivarius ATCC 7073 | 88.2 | 92.2 | 87.6 | 88.2 | 87.6 | 86.3 | 89.5 | 86.9 | 86.9 | 85.6 | 83.0 | 88.9 | 86.9 | 87.6 | 87.6 | 83.7 | 90.2 | 92.8 | 86.9 | 87.6 | 86.9 | 88.2 | 92.4 | 90.1 | 93.2 | 92.1 | 99.1 | 81.3 | 76.1 | 79.0 | 77.9 | |
| 24 | S. sanguinis ATCC 10556 | 87.6 | 90.2 | 88.9 | 88.2 | 86.3 | 88.2 | 94.8 | 90.2 | 89.5 | 88.9 | 85.6 | 98.0 | 88.2 | 86.3 | 92.8 | 83.7 | 90.2 | 92.8 | 86.9 | 93.5 | 89.5 | 88.9 | 87.6 | 89.0 | 92.0 | 91.3 | 92.1 | 83.4 | 75.8 | 79.0 | 77.4 | |
| 25 | S. sobrinus ATCC 33478 | 89.5 | 87.6 | 94.1 | 90.9 | 92.2 | 92.2 | 85.6 | 94.1 | 83.0 | 85.6 | 86.9 | 86.9 | 92.8 | 92.2 | 85.0 | 89.5 | 85.6 | 86.3 | 81.0 | 86.3 | 83.0 | 91.5 | 85.6 | 86.3 | 88.4 | 87.1 | 90.3 | 80.9 | 74.1 | 77.3 | 75.7 | |
| 26 | S. suis ATCC 43765 | 89.5 | 90.2 | 84.3 | 83.7 | 83.0 | 83.0 | 88.9 | 85.0 | 89.5 | 87.6 | 86.3 | 86.9 | 83.7 | 83.7 | 88.2 | 79.7 | 92.2 | 89.5 | 88.9 | 88.2 | 89.5 | 84.3 | 88.2 | 86.3 | 83.7 | 91.5 | 93.2 | 81.5 | 75.7 | 77.9 | 77.9 | |
| 27 | S. uberis ATCC 19436 | 88.2 | 92.2 | 86.9 | 85.0 | 84.3 | 85.0 | 91.5 | 86.9 | 97.4 | 88.9 | 86.9 | 87.6 | 86.3 | 85.0 | 92.8 | 83.0 | 96.7 | 90.2 | 94.1 | 93.5 | 97.4 | 85.0 | 88.2 | 88.2 | 84.3 | 89.5 | 92.0 | 81.2 | 75.6 | 79.2 | 76.9 | |
| 28 | S. vestibularis ATCC 49124 | 88.2 | 90.2 | 86.3 | 85.6 | 86.3 | 85.6 | 88.9 | 86.3 | 86.9 | 85.6 | 82.4 | 88.2 | 85.6 | 88.2 | 87.6 | 82.4 | 90.2 | 92.2 | 86.3 | 87.6 | 86.9 | 86.3 | 97.4 | 86.9 | 85.6 | 87.6 | 88.2 | 81.5 | 76.0 | 79.0 | 77.8 | |
| 29 | L. lactis ATCC 19435 | 84.3 | 80.4 | 81.7 | 82.4 | 80.4 | 82.4 | 81.0 | 81.0 | 82.4 | 79.1 | 81.0 | 82.4 | 81.0 | 83.7 | 82.4 | 81.0 | 82.4 | 81.0 | 79.1 | 83.7 | 82.4 | 85.0 | 81.0 | 82.4 | 80.4 | 77.8 | 83.0 | 82.4 | 75.4 | 77.3 | 76.3 | |
| 30 | E. durans ATCC 19432c | 71.9 | 69.9 | 67.3 | 64.7 | 66.0 | 64.7 | 66.7 | 65.4 | 69.3 | 67.3 | 68.6 | 67.3 | 66.7 | 64.7 | 68.0 | 64.7 | 69.9 | 68.0 | 67.3 | 69.3 | 69.3 | 65.4 | 68.0 | 66.0 | 66.7 | 69.3 | 69.3 | 68.0 | 68.6 | 77.7 | 88.3 | |
| 31 | E. durans ATCC 19432 tufBc | 70.6 | 71.9 | 72.5 | 70.6 | 71.9 | 72.5 | 75.8 | 72.5 | 74.5 | 71.9 | 70.6 | 73.2 | 72.5 | 69.3 | 71.9 | 72.5 | 75.2 | 77.8 | 73.9 | 72.5 | 74.5 | 71.9 | 73.2 | 71.9 | 71.9 | 71.2 | 74.5 | 73.9 | 73.9 | 69.3 | 78.4 | |
| 32 | E. faecalis V853 | 76.5 | 74.5 | 69.3 | 68.6 | 69.9 | 70.6 | 70.6 | 71.2 | 71.9 | 71.9 | 73.2 | 70.6 | 68.6 | 69.9 | 69.9 | 68.0 | 71.9 | 71.9 | 71.2 | 70.6 | 71.9 | 67.3 | 73.9 | 69.9 | 68.6 | 74.5 | 72.5 | 73.9 | 71.9 | 86.9 | 74.5 | |
The streptococcal sequences were also compared with those obtained from E. durans and L. lactis. The numbers in the upper right triangle are the percentages of identity for a 761-bp segment from the tuf genes (positions 340 to 1100 of the complete gene of S. pneumoniae R6; accession number AE008504), while those in the lower left triangle represent the percentages of identity for the 153-bp sequences (positions 706 to 858 of the complete gene of S. pneumoniae R6; accession number AE008504) flanked by the two Streptococcus-specific PCR primers. The identity scores were obtained by use of the GAP program from the GCG Wisconsin package.
Species numbers are arbitrarily assigned to clarify species comparisons.
Two divergent copies of the tuf gene are found in E. durans (24).
Nucleotide sequence analysis revealed that streptococcal tuf genes are generally more variable than those coding for 16S rRNA. Sequence identities among the 28 different streptococcal species determined for the 761-bp portion of tuf ranged from 86.1 to 99.1% (Table 3). On the other hand, a comparative nucleotide sequence analysis of a 1,260-bp portion of the 16S rRNA genes for these same 28 streptococcal species revealed a higher level of sequence identities (89.4 to 99.8%). More specifically, interspecies sequence identities within the three streptococcal groups consisting of the most closely related species were as follows: (i) mitis group (including S. mitis, S. gordonii, S. pneumoniae, S. oralis, S. sanguinis, S. parasanguinis, and S. cristatus), 93.3 to 98.7% for tuf and 97.1 to 99.7% for 16S rDNA; (ii) anginosus group (including S. anginosus, S. constellatus, and S. intermedius), 97.2 to 98.0% for tuf and 96.3 to 99.8% for 16S rDNA; and (iii) pyogenes group (including S. pyogenes, S. agalactiae, S. dysgalactiae, S. equi, S. uberis, and S. parauberis), 90.3 to 98.6% for tuf and 94.5 to 97.1% for 16S rDNA. Therefore, tuf sequences of streptococci generally offer more discrimination power than 16S rDNA sequences and should allow identification at the species level of even the most closely related streptococcal species.
A multiple alignment of the tuf sequences from streptococcal species as well as those from staphylococci, enterococci, and lactococci revealed regions conserved among streptococci but distinct from those of other bacteria. The regions for which the sequence mismatches for the nonstreptococcal bacterial species were mainly located at the 3′ end were chosen as targets for the Streptococcus-specific PCR primers (Str1 and Str2). This strategy allowed effective discriminatory PCR, since mismatches at the 3′ end of primers are the most detrimental to PCR. The selected Streptococcus-specific primers generated 197-bp amplicons. Direct sequencing of these amplicons provided sequence information for the 153-bp sequence between the two Streptococcus-specific primers. Comparisons of these 153-bp sequences obtained for the 28 streptococcal species tested revealed sequence identities ranging from 79.7 to 100% (Table 3). For these sequences, there were two pairs of streptococcal species showing more than 98% identity (S. mitis versus S. pneumoniae [98.7%] and S. pyogenes versus S. dysgalactiae [100%]).
The tuf sequences either determined in our laboratory or available in public databases for different strains of the same streptococcal species allowed the analysis of intraspecies sequence variations. As shown in Fig. 1, the species for which two or more strains have been sequenced are S. pneumoniae (n = 8), S. agalactiae (n = 7), S. pyogenes (n = 6), S. gordonii (n = 3), S. dysgalactiae (n = 3), S. mitis (n = 2), S. oralis (n = 2), S. bovis (n = 2), S. mutans (n = 2), and S. uberis (n = 2). We compared the region corresponding to the 153-bp sequence for all of these available sequences and found that the level of intraspecies sequence variation ranged from 0 to 2.6%, depending on the species. More specifically, intraspecies variations were (i) 0% for S. agalactiae, S. dysgalactiae, S. mutans, and S. uberis, (ii) 0 to 0.7% for S. pneumoniae, (iii) 0.7% for S. mitis, (iv) 0 to 1.3% for S. pyogenes, (v) 0.7 to 2.6% for S. gordonii, (vi) 2% for S. oralis, and (vii) 2.6% for S. bovis.
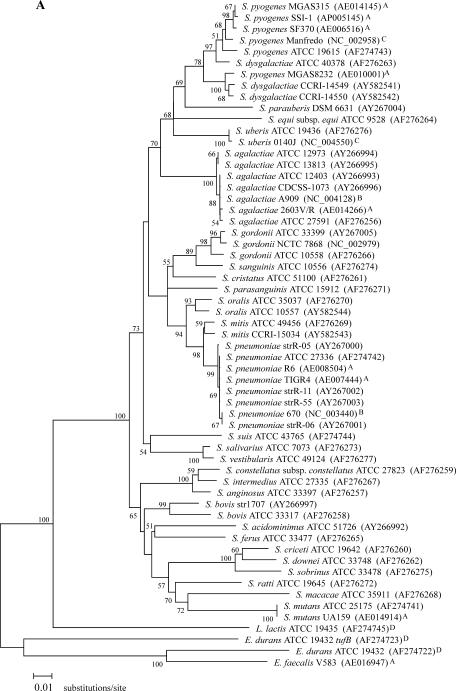
FIG. 1.
Phylogenetic relationships among 28 streptococcal species. (A) Phylogenetic tree based on a 761-bp portion of tuf. (B) Phylogenetic tree based on a 1,260-bp portion of 16S rDNA. The trees were generated using the MEGA2 heuristic method, and evolutionary distance values were calculated by Kimura's two-parameter substitution model. The value on each branch represents the percentage of bootstrap replications supporting the branch. A total of 1,000 bootstrap replications were calculated. Bootstrap values lower than 50% are not shown. GenBank accession numbers are given in parentheses. The tuf and 16S rDNA portions correspond to nucleotide positions 340 to 1,100 of the complete tuf gene of S. pneumoniae R6 (AE008504) and 93 to 1,382 of the complete 16S rRNA gene of S. pneumoniae R6 (AE008546). All sequences used for these phylogenetic analysis were obtained either from this study or from the following sources: GenBank (http://www.ncbi.nlm.nih.gov) (A), TIGR ongoing genome projects (http://www.tigr.org) (B), Sanger ongoing genome projects (http://www.sanger.ac.uk) (C), and our group (for previously determined sequences) (24) (D); sequences from these four sources are indicated with A, B, C, and D, respectively.
Streptococcus-specific PCR assay.
The PCR assay using primers Str1 and Str2 amplified efficiently genomic DNA from all 28 streptococcal species tested. The detection limits ranged from 1 to 10 genome copies per PCR. The specificity of the PCR assay was verified by performing 40-cycle PCR amplifications using a battery of gram-negative (44 species from 23 genera) and gram-positive (58 species from 10 genera) bacteria, including the 28 streptococcal species (Table 1). No DNAs from any nonstreptococcal bacterial species were amplified by the assay except for those from Lactococcus lactis and Enterococcus durans. Analysis of tuf sequences from L. lactis and E. durans revealed that there was no mismatch at the 3′ end (first 6 to 8 nucleotides) of each of the PCR primers and that there were 0 to 2 mismatches elsewhere in the primer binding sites. The 153-bp amplicon sequences generated for these two bacterial species showed nucleotide identities ranging from 69.3 to 85.0% compared to the corresponding sequences for the 28 streptococcal species tested (Table 3). This relatively high level of sequence divergence between lactococci, enterococci, and streptococci suggests that it should be easy to develop genus-specific and species-specific internal probes for streptococci. Finally, testing DNAs from a collection of 153 streptococcal isolates from the microbiology laboratory of the CHUQ (Pavillon CHUL) showed a uniform amplification signal for all clinical strains except for one S. bovis strain which was not detected (Table 2). Phenotypic identification confirmed that the nondetectable S. bovis strain was of biotype II, which is more genetically heterogeneous than the other S. bovis biotypes (13, 43, 45).
Phylogenetic analysis.
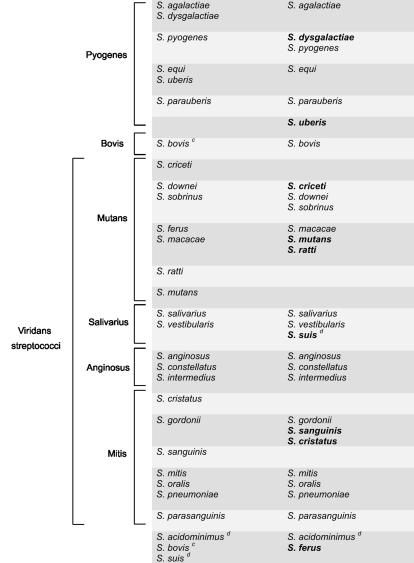
The entire Streptococcus group is monophyletic as determined on the basis of tuf and 16S rDNA phylogenies and forms a phylum distinct from L. lactis (Fig. 1). tuf-based phylogenetic relationships between the 28 streptococcal species revealed clusters that are generally in agreement with those observed with 16S rDNA phylogeny (Fig. 1 and Table 4). Incidentally, the 16S rDNA tree, which we constructed mostly using available database sequences for the species selected for the present study, was similar in terms of branching to the 16S rDNA tree recently reported by Facklam (13) for 55 streptococcal species. Comparison of tuf and 16S rDNA phylogenetic trees revealed a different ancestor for the pyogenes group (Fig. 1). As determined on the basis of tuf phylogeny, that streptococcal group is linked with the mitis and salivarius groups whereas 16S rDNA results show that the pyogenes group branches separately from the other streptococcal groups. For the other major deep branches, the bootstrap values were too low with both tuf- and 16S rDNA-based phylogenetic trees to permit reliable interpretations (Fig. 1). Phylogenetic analysis of available database sequences for sodA, groEL, atpD (data not shown), and rnpB (42) showed a similar lack of resolution for basal branches of the streptococcal tree. However, branches between closely related taxa are well supported and can be used to establish their grouping (Fig. 1 and Table 4).
TABLE 4.
Phylogenetic clusters observed with tuf and 16S rRNA genes within phenotypic streptococcal species groups
Phylogenetic clusters with 16S rDNA were devised based on trees constructed with sequences available in public databases. Ambiguities and gaps were excluded for phylogenetic analysis.
Species in bold type branched differently based on tuf phylogeny compared to 16S rDNA phylogeny.
Different strains from this species branched with different clusters.
These species have not been assigned to a phenotypic streptococcal species group.
There were some streptococcal species branching differently or forming different cluster groups in the tuf-based trees compared to those derived from 16S rDNA (Fig. 1 and Table 4). These species include S. dysgalactiae, S. suis, S. ferus, S. criceti, S. mutans, S. ratti, S. mitis, S. sanguinis, and S. uberis. Phylogenetic analyses using either tuf or 16S rRNA genes also revealed that three important streptococcal phenotypic groups (i.e., mitis, pyogenes, and mutans) are each composed of up to five phylogenetic clusters. On the other hand, the streptococcal groups anginosus and salivarius were found to be monophyletic on the basis of these two phylogenetic analyses (Table 4).
Distance analysis using neighbor-joining or the MEGA2 heuristic method produced similar trees except for minor differences present in the topology of the basal branches for which bootstrap support is weak. Distance and parsimony phylogenetic analysis of tuf yielded trees showing similar end-branching structures (data not shown). However, many basal branches are still poorly resolved.
DISCUSSION
In previous studies, the usefulness of tuf sequences for the identification of enterococci (26) and staphylococci (31) was demonstrated. In the present study, we used broad-range PCR primers to amplify and sequence a portion of tuf genes from 28 clinically relevant streptococcal species. Determination of regions conserved in streptococci but distinct in other bacteria allowed the development of a Streptococcus-specific PCR assay. Sequence analysis of the streptococcal amplicons revealed interspecies variations that are promising for the development of species-specific internal probes for identification of streptococci.
The use of genus- or group-specific PCR assays can substantially decrease the number of primers used for bacterial identification. Indeed, identification of the most frequently encountered species at the genus level is often sufficient to permit selection of an appropriately targeted antibiotic (2). Furthermore, the use of post-PCR hybridization with species-specific internal probes bound onto a solid support (e.g., oligonucleotide arrays) would allow identification at the species level. Fluorescent probes (e.g., TaqMan probes) may also be used for the development of species-specific real-time PCR assays for clinically important streptococci. We chose the tuf gene as a genetic target to develop a PCR-based assay for the detection of streptococci, because it has both conserved and variable regions suitable for the design of genus- and species-specific probes, respectively (26, 31). Our assay efficiently detected all 28 streptococcal species tested, with an analytical sensitivity of 1 to 10 genome copies per PCR. It also detected DNA purified from two phylogenetically closely related species (i.e., E. durans and L. lactis). However, the 15-to-31% nucleotide sequence divergence in the tuf gene of these two species compared to corresponding sequences for the 28 streptococcal species provides much flexibility for the development of internal probes specific to streptococci.
Based on 16S rDNA sequence analysis, the genus Lactococcus is phylogenetically very closely related to the genus Streptococcus (40). It is therefore not surprising that L. lactis DNA was detected by the Streptococcus-specific PCR assay. Indeed, analysis of the primer binding sites for this species revealed that there was no mismatch in the Str1 primer and only one at the ninth nucleotide of the 3′ end for the Str2 primer. Phylogenetic analysis of tuf sequences from different lactococcal species confirmed that they form a distinct phylum closely related to streptococci (data not shown).
Analysis of tuf sequences from a variety of streptococci and lactococci revealed that only one copy of tuf is present in their genome. The nonspecific amplification of E. durans by the Streptococcus-specific PCR assay can be explained by the finding that E. durans and some closely related enterococcal species carry two divergent tuf genes, one of which shares a common ancestor with the tuf gene of streptococci and lactococci (24). Indeed, it is the horizontally transferred tufB gene of E. durans that was amplified by the Streptococcus-specific PCR-based assay. However, no other enterococcal tufB genes were amplified by this assay.
The 153 clinical isolates representing a variety of streptococci which were obtained from the Quebec City region were all detected by our assay except for one S. bovis strain which was not amplified. The two other clinical strains of S. bovis as well as the reference S. bovis strain ATCC 33317 were amplified efficiently. The nondetectable S. bovis strain was of biotype II. S. bovis of this biotype has been shown to be more genetically polymorphic and frequently associated with animal hosts (13, 43, 45). Sequence data for this S. bovis strain revealed the presence of a single mismatch at the 3′ end (first nucleotide) of the Streptococcus-specific primer Str1 which probably explains the absence of PCR amplification.
S. oralis and S. mitis are phylogenetically very closely related to S. pneumoniae (22). These three species form well-supported distinct groups in tuf phylogeny but not with 16S rDNA phylogeny, where S. mitis is present in two different branches (Fig. 1). The findings with 16S rDNA phylogeny regarding S. mitis are consistent with previous studies showing that the genes coding for the streptococcal pneumolysin (ply), autolysin (lytA), and superoxide dismutase (sodA) are more polymorphic in S. mitis (23, 46). The genetic heterogeneity of S. mitis may be associated with the horizontal transfer of genes between streptococci (9, 20, 29). The present study also revealed a very low intraspecies tuf sequence divergence in S. pneumoniae (0 to 0.3% for 8 strains). The homogeneity of tuf sequences observed in S. pneumoniae could be explained by the clonal spread of a limited number of variants of this bacterial pathogen (9, 11).
Sequence analysis of the 761-bp portion of tuf revealed that sequence variations between S. oralis, S. mitis, and S. pneumoniae ranged from 1.3 to 2.9% (Table 3) compared to 0.3 to 0.6% for the 1,260-bp portion of 16S rDNA (data not shown). Hence, the higher level of sequence variations in tuf compared to 16S rDNA for these very closely related species provides more potential for the development of primers and probes allowing them to be distinguished. These three species can also be distinguished by using less-conserved genes like ddl (coding for d-alanine-d-alanine ligase), lytA, and sodA (16, 17, 23, 38).
According to our tuf sequence data for the 153-bp amplicon sequences, species-specific sequence variations are present for all streptococcal species except for S. pyogenes and S. dysgalactiae, for which the tuf amplicon sequences are identical. However, other regions of tuf may be appropriate to distinguish these two species, as suggested by the 1.4% divergence in their sequences for the 761-bp tuf portion. Surprisingly, the 16S rDNA sequences for S. pyogenes and S. dysgalactiae revealed a significantly higher level of divergence (i.e., 3.2%). Consequently, S. dysgalactiae branched together with S. agalactiae according to 16S rDNA phylogeny while it appeared to be more related to S. pyogenes on the basis of tuf phylogeny (Table 4 and Fig. 1). Phylogenetic studies performed with sodA, groEL, atpD (data not shown), and rnpB (42) support this relationship between S. dysgalactiae and S. pyogenes.
The comparison of tuf and 16S rDNA phylogenetic trees revealed differences in branching topologies and clustering for some streptococcal species. For S. criceti, S. cristatus, S. mutans, S. ratti, S. sanguinis, and S. uberis, the clustering differences for these species could be explained by the lower resolution of the 16S rDNA phylogenetic tree. In the case of S. dysgalactiae, S ferus, and S. suis, apparent discrepancies between the phylogenetic trees are not statistically well supported. It is possible that these differences are associated with different evolutionary rates for tuf and 16S rDNA. It can also be linked to the heterogeneity of 16S rDNA operons in bacteria. Indeed, it has been reported that phylogenetic studies are severely limited by 16S rDNA heterogeneity and that analysis of distinct rRNA operons within the same microbial strain may lead to different results (5, 7, 36). Finally, for S. suis and S. mitis, differential branching could be the result of previously reported greater genetic variation within these two species (4, 13, 23, 43, 46).
Phylogenetic analysis using either tuf or 16S rDNA sequences revealed that three clinically important streptococcal phenotypic species groups (i.e., mitis, pyogenes, and mutans) are composed of three to five phylogenetic clusters and that the groups anginosus and salivarius were monophyletic. This suggests that the mitis, pyogenes, and mutans groups are much more genetically heterogeneous than the anginosus and salivarius groups. It also indicates that the standard streptococcal species grouping, which is based on physiological and biochemical characteristics determined by conventional methods, does not correlate well with the level of genetic diversity within each group.
Phylogenetic analysis of multiple strains of the same streptococcal species revealed that the level of intraspecies tuf sequence variations depends on the streptococcal species. We analyzed the 761-bp tuf sequences from a total of six to eight strains for each of the three most clinically important streptococcal species (i.e., S. pyogenes, S. pneumoniae, and S. agalactiae). This analysis clearly demonstrated that S. pyogenes has more polymorphisms (intraspecies variation of 0.1 to 3.5%) than S. pneumoniae and S. agalactiae (intraspecies variation of 0 to 0.3% and 0 to 0.4%, respectively). By contrast, analysis of 16S rDNA sequences suggests that S. pyogenes has slightly fewer intraspecies sequence variations than S. agalactiae or S. pneumoniae (0 to 0.2% versus 0 to 0.5%). These observations indicate that evolutionary rates for these two genes involved in different components of the protein synthesis machinery may differ for streptococcal species.
In conclusion, we have performed an extensive sequence analysis of streptococci showing that tuf generally offers a better discrimination power than 16S rDNA to distinguish streptococcal species. tuf and 16S rDNA phylogenetic trees were generally in agreement, although different clustering of some closely related streptococcal species was observed. However, these phylogenetic clusters revealed that classical streptococcal phenotypic groups may comprise different genetic subgroups. We have used tuf sequences to develop a PCR-based approach for the detection of streptococci. Future developments will seek to combine this genus-specific assay with detection of species-specific tuf sequence polymorphisms by using internal hybridization probes to provide a molecular diagnostic tool for rapid and accurate diagnosis of streptococcal infections.
Acknowledgments
This study was supported by Infectio Diagnostic (I.D.I.) Inc. (Sainte-Foy, Québec, Canada) and by grant PA-15586 from the Canadian Institutes of Health Research. Marc Ouellette is a holder of a Canada Research Chair in Antimicrobial Resistance.
We thank Louise Côté, director of the microbiology laboratory of CHUQ (Pavillon CHUL) for access to the laboratory and for providing clinical isolates of streptococci. We also thank Gisèle Chassé and Ève Bérubé for their contribution in microbial species culture and identification. Sequence data for S. agalactiae A909, S. gordonii NCTC 7868, and S. pneumoniae 670 were obtained from The Institute for Genomic Research website at http://www.tigr.org. Sequencing of S. agalactiae A909 was accomplished with support from the National Institute of Allergy and Infectious Diseases (NIAID). Sequencing of S. pneumoniae 670 was accomplished with support of the National Institute of Allergy and Infectious Diseases and the University of Alabama. Sequencing of S. gordonii NCTC 7868 was accomplished with support from National Institute of Dental and Craniofacial Research. Sequence data for S. pyogenes Manfredo and S. uberis 0140J were produced by the sequencing groups at the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/sp/ and ftp://ftp.sanger.ac.uk/pub/pathogens/su/.
REFERENCES
- 1.Bergeron, M. G., D. Ke, C. Ménard, F. J. Picard, M. Gagnon, M. Bernier, M. Ouellette, P. H. Roy, S. Marcoux, and W. D. Fraser. 2000. Rapid detection of Group B streptococci in pregnant women at delivery. N. Engl. J. Med. 343:175-179. [DOI] [PubMed] [Google Scholar]
- 2.Bergeron, M. G., and M. Ouellette. 1998. Preventing antibiotic resistance through rapid genotypic identification of bacteria and of their antibiotic resistance genes in the clinical microbiology laboratory. J. Clin. Microbiol. 36:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourbeau, P. P., B. J. Heiter, and M. Figdore. 1997. Use of Gen-Probe AccuProbe Group B streptococcus test to detect group B streptococci in broth cultures of vaginal-anal specimens from pregnant women: comparison with traditional culture method. J. Clin. Microbiol. 35:144-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatellier, S., J. Harel, Y. Zhang, M. Gottschalk, R. Higgins, L. A. Devriese, and R. Brousseau. 1998. Phylogenetic diversity of Streptococcus suis strains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int. J. Syst. Bacteriol. 48:581-589. [DOI] [PubMed] [Google Scholar]
- 5.Clayton, R. A., G. Sutton, P. S. Hinkle, Jr., C. Bult, and C. Fields. 1995. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol. 45:595-599. [DOI] [PubMed] [Google Scholar]
- 6.Dagan, R., O. Shriker, I. Hazan, E. Leibovitz, D. Greenberg, F. Schlaeffer, and R. Levy. 1998. Prospective study to determine clinical relevance of detection of pneumococcal DNA in sera of children by PCR. J. Clin. Microbiol. 36:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahllof, I., H. Baillie, and S. Kjelleberg. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denys, G. A., and R. B. Carey. 1992. Identification of Streptococcus pneumoniae with a DNA probe. J. Clin. Microbiol. 30:2725-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowson, C. G., T. J. Coffey, and B. G. Spratt. 1994. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to β-lactam antibiotics. Trends Microbiol. 2:361-366. [DOI] [PubMed] [Google Scholar]
- 10.du Plessis, M., A. M. Smith, and K. P. Klugman. 1999. Application of pbp1A PCR in identification of penicillin-resistant Streptococcus pneumoniae. J. Clin. Microbiol. 37:628-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 12.Evangelista, A. T., A. L. Truant, and P. P. Bourbeau. 2002. Rapid systems and instruments for the identification of bacteria, p. 22-49. In A. L. Truant (ed.), Manual of commercial methods in clinical microbiology. ASM Press, Washington, D.C.
- 13.Facklam, R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenoll, A., J. V. Martinez-Suarez, R. Munoz, J. Casal, and J. L. Garcia. 1990. Identification of atypical strains of Streptococcus pneumoniae by specific DNA probe. Eur. J. Clin. Microbiol. Infect. Dis. 9:396-401. [DOI] [PubMed] [Google Scholar]
- 15.Freney, J., S. Bland, J. Etienne, M. Desmonceaux, J. M. Boeufgras, and J. Fleurette. 1992. Description and evaluation of the semiautomated 4-hour rapid ID 32 Strep method for identification of streptococci and members of related genera. J. Clin. Microbiol. 30:2657-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnier, F., G. Gerbaud, P. Courvalin, and M. Galimand. 1997. Identification of clinically relevant viridans group streptococci to the species level by PCR. J. Clin. Microbiol. 35:2337-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillespie, S. H., C. Ullman, M. D. Smith, and V. Emery. 1994. Detection of Streptococcus pneumoniae in sputum samples by PCR. J. Clin. Microbiol. 32:1308-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graur, D., and W.-H. Li. 2000. Fundamentals of molecular evolution. Sinauer Associates, Inc., Sunderland, Mass.
- 19.Heiter, B. J., and P. P. Bourbeau. 1993. Comparison of the Gen-Probe Group A Streptococcus Direct Test with culture and a rapid streptococcal antigen detection assay for diagnosis of streptococcal pharyngitis. J. Clin. Microbiol. 31:2070-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaacman, D. J., Y. Zhang, J. Rydquist-White, R. M. Wadowsky, J. C. Post, and G. D. Ehrlich. 1995. Identification of a patient with Streptococcus pneumoniae bacteremia and meningitis by the polymerase chain reaction (PCR). Mol. Cell. Probes 9:157-160. [DOI] [PubMed] [Google Scholar]
- 22.Kawamura, Y., X.-G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 23.Kawamura, Y., R. A. Whiley, S.-E. Shu, T. Ezaki, and J. M. Hardie. 1999. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology 145:2605-2613. [DOI] [PubMed] [Google Scholar]
- 24.Ke, D., M. Boissinot, A. Huletsky, F. J. Picard, J. Frenette, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Evidence for horizontal gene transfer in evolution of elongation factor Tu in enterococci. J. Bacteriol. 182:6913-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ke, D., C. Ménard, F. J. Picard, M. Boissinot, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin. Chem. 46:324-331. [PubMed] [Google Scholar]
- 26.Ke, D., F. J. Picard, F. Martineau, C. Ménard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 1999. Development of a PCR assay for rapid detection of enterococci. J. Clin. Microbiol. 37:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 28.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 29.Lachenauer, C. S., R. Creti, J. L. Michel, and L. C. Madoff. 2000. Mosaicism in the alpha-like protein genes of group B streptococci. Proc. Natl. Acad. Sci. USA 97:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane, J. D. 1991. 16S/23S rRNA sequencing, p. 115-203. In E. Stackbrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley & Sons, New York, N.Y.
- 31.Martineau, F., F. J. Picard, D. Ke, S. Paradis, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2001. Development of a PCR assay for identification of staphylococci at genus and species levels. J. Clin. Microbiol. 39:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martineau, F., F. J. Picard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 1998. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus aureus. J. Clin. Microbiol. 36:618-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mollerach, M., and E. Garcia. 2000. The galU gene of Streptococcus pneumoniae that codes for a UDP-glucose pyrophosphorylase is highly polymorphic and suitable for molecular typing and phylogenetic studies. Gene 260:77-86. [DOI] [PubMed] [Google Scholar]
- 34.Oho, T., Y. Yamashita, Y. Shimazaki, M. Kushiyama, and T. Koga. 2000. Simple and rapid detection of Streptococcus mutans and Streptococcus sobrinus in human saliva by polymerase chain reaction. Oral Microbiol. Immunol. 15:258-262. [DOI] [PubMed] [Google Scholar]
- 35.Ono, T., K. Hirota, K. Nemoto, E. J. Fernandez, F. Ota, and K. Fukui. 1994. Detection of Streptococcus mutans by PCR amplification of spaP gene. J. Med. Microbiol. 41:231-235. [DOI] [PubMed] [Google Scholar]
- 36.Pettersson, B., G. Bolske, F. Thiaucourt, M. Uhlen, and K. E. Johansson. 1998. Molecular evolution of Mycoplasma capricolum subsp. capripneumoniae strains, based on polymorphisms in the 16S rRNA genes. J. Bacteriol. 180:2350-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pokorski, S. J., E. A. Vetter, P. C. Wollan, and F. R. Cockerill III. 1994. Comparison of Gen-Probe Group A Streptococcus Direct Test with culture for diagnosing streptococcal pharyngitis. J. Clin. Microbiol. 32:1440-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poyart, C., C. Quesne, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruoff, K. L., R. A. Whiley, and D. Beighton. 1998. Streptococcus, p. 283-296. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 40.Schleifer, K. H., and R. Kilpper-Balz. 1987. Molecular and chemotaxonomic approaches to the classification of streptococci, enterococci, and lactococci: a review. Syst. Appl. Microbiol. 10:1-19. [Google Scholar]
- 41.Swofford, D. 1998. Phylogenetic analysis using parsimony, version 4.0, beta documentation. Laboratory of Molecular Systematics, Smithsonian Institution, Washington, D.C.
- 42.Täpp, J., M. Thollesson, and B. Herrmann. 2003. Phylogenetic relationships and genotyping of the genus Streptococcus by sequence determination of the RNase P RNA gene, rnpB. Int. J. Syst. Evol. Microbiol. 53:1861-1871. [DOI] [PubMed] [Google Scholar]
- 43.Teng, L.-J., P.-R. Hsueh, J.-C. Tsai, P.-W. Chen, J.-C. Hsu, H.-C. Lai, C.-N. Lee, and S.-W. Ho. 2002. groESL sequence determination, phylogenetic analysis, and species differentiation for viridans group streptococci. J. Clin. Microbiol. 40:3172-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, S. M., M. A. Deighton, J. A. Capstick, and N. Gerraty. 1999. Epidemiological typing of bovine streptococci by pulsed-field gel electrophoresis. Epidemiol. Infect. 123:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whatmore, A. M., A. Efstratiou, A. P. Pickerill, K. Broughton, G. Woodard, D. Sturgeon, R. George, and C. G. Dowson. 2000. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect. Immun. 68:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitehead, T. R., and M. A. Cotta. 1993. Development of a DNA probe for Streptococcus bovis by using a cloned amylase gene. J. Clin. Microbiol. 31:2387-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]