Abstract
Human adenovirus type 4 is one of the major serotypes isolated from patients with adenoviral conjunctivitis. In 2001 we encountered nosocomial infections with epidemic conjunctivitis in the ophthalmology ward of one hospital in Sapporo, which is in the northern part of Japan. Adenoviruses were isolated from the patients with this nosocomial infection and identified as adenovirus type 4 (AdV-4) by a neutralization test with serotype-specific antiserum. When the cleavage patterns of the isolates were compared with the full viral genome with BamHI and SmaI, the cleavage patterns of the isolates were shown to be different from those of AdV-4p and other previously known AdV-4 variants. The nucleotide sequences of the fiber gene of the isolates showed the highest homologies (94.3%) with AdV-4 among the nucleotide sequences available from GenBank and formed a monophyletic cluster along with the prototype strain of AdV-4. The isolates, however, were located in a different lineage from those of AdV-4p and the AdV-4 variant from the sporadic infections. We conclude that the nosocomial infection that appeared in 2001 was caused by a new genome type of AdV-4, which was designated AdV-4c.
Human adenoviruses (AdVs) in the genus Mastadenovirus of the family Adenoviridae form a group of viruses that include 51 serotypes (5, 22). They have been classified into six species, human AdV-A to -F, according to their biologic, immunologic, and oncogenic properties (4, 29). The human AdVs have been shown to cause enteric and upper and lower respiratory tract infections, hemorrhagic cystitis, and ocular infections.
A diverse tissue tropism exists within human AdVs (18). Upper respiratory infections have mainly been caused by AdV-1, -2, -3, -4, -5, and -6, and enteric infections have mainly been caused by AdV-40 and -41. Adenoviral conjunctivitis is the most common viral infection of the human eye in many parts of the world. It is caused by AdVs, mainly types 3, 4, 8, 19, and 37. AdV-8, -19, and -37 are members of human AdV-D, and AdV-3 is a member of human AdV-B. AdV-4 is the only member of human AdV-E and often causes outbreaks of epidemic conjunctivitis (2) and respiratory infections (8, 17). AdV-4 ocular infections were quite rare up to the 1960s, but in the latter half of the 1970s, AdV-4 caused worldwide outbreaks of epidemic conjunctivitis in, among other locations, Rome in 1974, Bristol in 1978, and Chicago in 1981 (12, 14, 27).
Some AdV-4 infections caused nosocomial infections of epidemic conjunctivitis. In Japan, no AdV-4 strains were isolated from patients with adenoviral conjunctivitis from 1974 through 1977, but in 1979 and 1980, 33 AdV-4s were isolated from patients with conjunctivitis (2). By DNA restriction analysis, Wadell et al. showed that there were at least two genome types of AdV-4, the AdV-4 prototype (AdV-4p) and AdV-4a, and made it clear that since 1965, AdV-4a has caused many outbreaks of adenoviral conjunctivitis throughout the world, including China, the United States, and Holland (13, 29). In Japan, AdV-4a also became the major cause of adenoviral conjunctivitis from 1980. Data from the Infectious Agents Surveillance Reports of Japan revealed that AdV-4 epidemics occurred in 1984 and 1992. However, the incidence of AdV-4 conjunctival infection has been decreasing in Japan (15, 16), and no large epidemics of conjunctivitis caused by AdV-4 have been observed since 1993 (3, 9, 10).
In September 2001, nosocomial infection with epidemic conjunctivitis was caused by AdV-4 originating in the ophthalmology ward of a university hospital in Sapporo, Japan. In this study, we investigated the molecular biological characteristics of the isolates with the PCR-restriction fragment length polymorphism (PCR-RFLP) method and the DNA restriction method for genome typing and phylogenetic analysis, along with the AdV-4 isolates taken from patients with epidemic conjunctivitis in Sapporo during the past 17 years.
MATERIALS AND METHODS
Virus strains.
Thirty-six AdV-4 isolates from conjunctival swabs from patients with acute conjunctivitis were used in this study. Of the 36 isolates, 2 were isolated in 1984, 12 in 1985, 2 in 1991, 2 in 1996, 6 in 1998, and 12 in 2001. Among the 12 strains isolated in 2001, 8 were from nosocomial infections, and the others were from sporadic infections in clinics in Sapporo, Japan. The six strains isolated in 1998 were provided by K. Fujita of the Sapporo City Institute of Public Health. All isolates were propagated in HeLa, A549, or Hep-2 cells and identified as AdV-4 by the neutralization method with type-specific antisera against adenovirus, which was purchased from Denka Seiken Co., Ltd. (Tokyo, Japan). The human AdV-4 prototype strain RI-67 was purchased from the American Type Culture Collection (Manassas, Va.).
Genome typing.
Viral DNA was extracted from the infected cells in a 75-cm2 plastic flask with 3 ml of Hirt lysis solution (10 mM Tris, 1 mM EDTA, 0.6% sodium dodecyl sulfate, pH 8) (28). Proteinase K was added at a final concentration of 50 μg/ml, and the samples were incubated at 37°C for 1 h. Cellular DNA was precipitated with 1 M NaCl overnight at 4°C. After phenol-chloroform extraction, the supernatant was treated with RNases A and T1 (final concentrations of 25 mg/ml and 80 U/ml, respectively; Sigma, St. Louis, Mo.) and phenol-chloroform extraction. Viral DNA was precipitated with isopropanol and suspended in 50 μl of TE buffer (1 mM Tris-HCl, 0.1 mM EDTA, pH 8.0).
Aliquots of 1 μg of viral genomic DNA were digested with 5 U of restriction endonucleases BamHI, EcoRI, SmaI, and XhoI (Takara Shuzo, Kyoto, Japan). The digested viral DNA was loaded onto 1% agarose gels containing 0.1 μg of ethidium bromide per ml. The DNA bands were photographed with a UV transilluminator and a Polaroid camera, and the patterns of fragments were analyzed by comparison with those of previously reported AdV-4 variants and the prototype.
PCR and phylogenetic analysis.
Viral DNA was extracted from 100 μl of virus solution with a Sumitest EX-R&D kit (Genome Science Laboratories Co., Ltd., Fukushima, Japan), according to the manufacturer's instructions. The extracted DNA was suspended in 100 μl of TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). PCR was carried out in 50 μl of reaction mixture containing 5 μl of extracted viral DNA, 5 μl of 10× Taq buffer, 200 μM each dATP, dGTP, dCTP, and dTTP, 0.5 μM each primer, AdnU-S′ (nucleotides 20743 to 20762 of GenBank accession no. J01917, 5′-TTCCCCATGGCNCACAACAC-3′), and AdnU-A (nucleotides 21698 to 21679 of GenBank accession no. J01917, 5′-GCCTCGATGACGCCGCGGTG-3′), and 2.5 U of Taq DNA polymerase (Roche Diagnostic Systems, Pleasanton, Calif.). With this pair of primers, we could amplify the 3′ end of the 956-bp hexon gene (19). After initial denaturation at 94°C for 5 min, 40 cycles of denaturing at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min; final extension at 72°C for 7 min was performed with a Gene Amp PCR System 9600 (Applied Biosystems, Foster City, Calif.). The 956-bp PCR product from the hexon gene was separated in 1% agarose gel and purified with a QIAquick gel extraction kit (Qiagen, Valencia, Calif.).
The nucleotide sequences of the partial hexon gene of the PCR products were determined with a 373A DNA auto sequencer (Applied Biosystems) with fluorescent dideoxy chain terminators (Applied Biosystems) with AdnU-S′ and AdnU-A as primers. An internal forward primer, AdEHxs (nucleotides 2348 to 2367, 5′-GTGGCCCAGTGCAACATGAC-3′), and a reverse primer, AdEHxa (nucleotides 2421 to 2402, 5′-CCCTGGTAGCCGATGTTGTA-3′), were also used for sequencing. The positions of the primers used were numbered according to the nucleotide sequence of the hexon gene of the AdV-4 RI-67 strain (GenBank accession number AF065062).
For nucleotide sequence analysis of the fiber gene, the viral DNA of the isolate was extracted with the Smitest EX-R&D kit (Genome Science Laboratories Co., Ltd., Fukushima, Japan) as described above. PCR was carried out in 50-μl volumes containing 5 μl of extracted viral DNA, 5 μl of 10× Taq buffer, 200 μM each deoxynucleoside triphosphates, 0.5 μM each of a pair of primers, the AdEf1 forward primer (nucleotides 1 to 20, 5′-TGCAAACTTCCTCCACACTC-3′), the AdEf2 reverse primer (nucleotides 1375 to 1356, 5′-TGGGTTGGCATGCAGGGTGG-3′), and 2.5 U of Taq DNA polymerase (Roche Diagnostic Systems, Pleasanton, Calif.). The positions of the primers used were numbered according to the nucleotide sequence of the fiber gene of the AdV-4 RI-67 strain (GenBank accession number X76547). After initial denaturation at 94°C for 5 min, 40 cycles of denaturing at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min, with a final extension at 72°C for 7 min, was performed with a Gene Amp PCR system. The products were separated in a 1% agarose gel and purified with a QIAquick gel extraction kit (Qiagen, Valencia, Calif.). The nucleotide sequence was determined with a 373A DNA auto sequencer (Applied Biosystems) with fluorescent dideoxy chain terminators (Applied Biosystems) with the AdEf1 and AdEf2 primers. An internal forward primer, AdEs2 (nucleotides 475 to 494, 5′-AGCTTTTGGCTCAGGTTTAG-3′) and AdEs3 (nucleotides 915 to 934, 5′-TGGCCACTGTATCAGTTTTG-3′), and reverse primer AdEr2 (nucleotides 988 to 969, 5′-TTGAGCACTGCTTACTGTGC-3′) or AdEr3 (nucleotides 572 to 549 5′-GGGTAATCTTTATATTCCCTTTAT-3′) were also used for sequencing. The position of the each primer was numbered according to the nucleotide sequence of the fiber gene of the AdV-4 RI-67 strain (GenBank accession number X76547).
A total of 36 sequences from the isolates were analyzed, along with previously known adenovirus prototypes, with SINCA software (Fujitsu Limited, Tokyo, Japan). For the analysis of hexon, we used 916-bp sequences of the 956-bp PCR products. For the fiber, we compared the nucleotide sequence of the entire open reading frame of the fiber gene. We estimated the evolutionary distances with the Kimura two-parameter method (11) and constructed unrooted phylogenetic trees with the neighbor-joining method (21). Bootstrap analyses were performed by 1,000 resamplings of the data sets. Bootstrap values of ≥70 were considered statistically significant for the grouping (6).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the nucleotide sequences determined in this study are AB098598 to AB098607.
RESULTS
Restriction analysis of full viral genome.
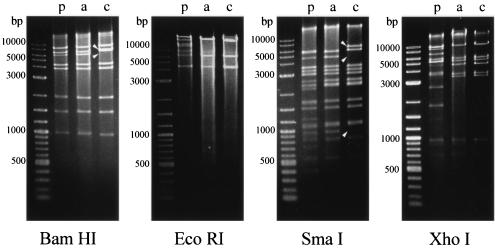
The full viral genome from the isolates was analyzed with four restriction endonucleases: BamHI, EcoRI, SmaI, and XhoI. According to their restriction patterns, the 36 isolates were divided into two groups. One group included the isolates from 1985 to 1998, and the other included 12 isolates obtained in 2001, of which 4 isolates were from sporadic infections and 8 were from nosocomial infections. In the first group, four restriction patterns were identical to those of AdV-4a. In the second group, the EcoRI and XhoI restriction patterns were identical to that of AdV-4a. However, the BamHI and SmaI restriction patterns were different from those of AdV-4p and all previously known genome types of AdV-4 (AdV-4p, p1-3, a, a1, b, and ch) (13) (Fig. 1). Pairwise comparison of the pairwise comigrating restriction fragment (PCRF) showed that AdV-4c shared 31 fragments (91.2%) with AdV-4a (total, 34 fragments). Therefore, we temporarily named these isolates the genome type of AdV-4c.
FIG. 1.
Restriction patterns of AdV-4p, AdV-4a, and AdV-4c, the new variant associated with nosocomial infection. Lane M, molecular size markers, 2-log DNA ladder (New England Biolabs, Beverly, Mass.). Lane p, adenovirus type 4 prototype. Lane a, representative isolate (TC-4822/s-84) of AdV-4a. Lane c, representative isolate (TC-33376/s-01n) of AdV-4c. Arrowheads indicate bands present in AdV-4c but absent from AdV-4a or those present in AdV-4a but absent from AdV-4c.
Phylogenetic analysis.
To date, only 11 hexon sequences (including two of human AdV-A [AdV-12 and -18], five of human AdV-B [AdV-3, -7, -16, -21, and -34], two of human AdV-C [AdV-2 and -5], and two of human AdV-F [AdV-40 and -41]) are available from GenBank. No hexon sequences of the prototype strains from human AdV-D and -E are available from GenBank. Therefore, we determined the nucleotide sequences of the prototype strains of 33 serotypes from human AdV-D and -E in a previous study (23a) and made it appear that the phylogenetic analysis of the partial hexon gene was very useful for the genetic investigation of adenovirus.
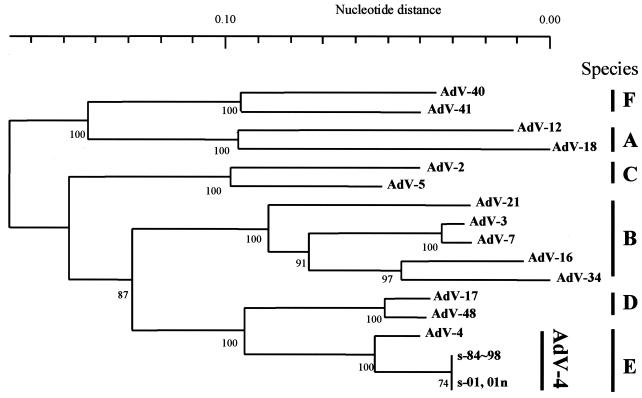
To assess the genetic relationships of the 36 isolates, an alignment of the partial hexon nucleotide sequences from these isolates along with 14 prototype strains was performed with the SINCA genetic software program. It appeared that these isolates and the 14 prototype strains contained 916 bases and that no deletions or insertions had occurred in any of them. To analyze the genetic relationships among these sequences, a phylogenetic tree based on the partial hexon nucleotide sequences was constructed. As shown in Fig. 2, the prototype strains of human AdVs were segregated into six major clusters, A to F. These clusters corresponded well to the six newly designated human adenovirus species A to F (4). The genetic relationships of the clinical isolates and prototype strain were further analyzed by one-by-one comparison. All isolates had an identical nucleotide sequence and showed the highest identity (96.1%) to the AdV-4 prototype strain. The phylogenetic tree showed that all clinical isolates formed a distinct cluster with the AdV-4 prototype strain in cluster E (Fig. 2). In this cluster, however, the isolates had a different lineage from the lineage of the prototype strain. Therefore, we identified all clinical isolates as AdV-4 variants.
FIG. 2.
Phylogenetic analysis of adenovirus prototype and clinical isolates of AdV-4 described in this study. The nucleotide sequence of the partial hexon gene (916 bp) was analyzed by the neighbor-joining method. Numbers at nodes are percentages of 1,000 bootstrap values. The following published sequences were used: AdV-2 (GenBank accession no. J01917), AdV-3 (X76549), AdV-4 (AB099380), AdV-5 (M73260), AdV-7 (AF065065), AdV-12 (X73487), AdV-16 (X74662), AdV-17 (AB099353), AdV-18 (Y17249), AdV-21 (AB053116), AdV-34 (AB052911), AdV-40 (L19443), AdV-41 (X51783), and AdV-48 (AB099377).
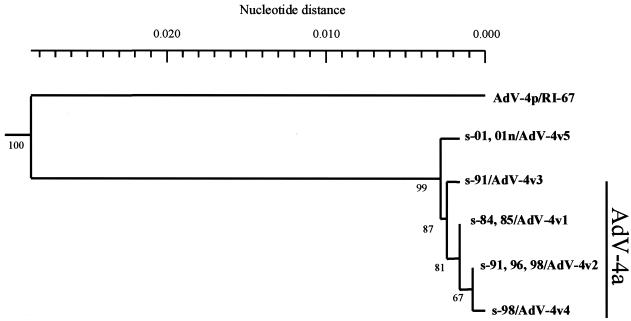
To analyze the genetic relationship of the isolates in more detail, we determined the full length of their fiber nucleotide sequence, because fiber plays an important role when AdVs attach to a target cell. We constructed the phylogenetic tree based on the full length of the fiber nucleotide sequences along with the available nucleotide sequences of AdV-4 fiber from GenBank. All isolates were segregated from AdV-4 prototype strains and formed distinct clusters (Fig. 3). Within a cluster, the isolates were divided into five groups, variants v1 to v5. Group 1, AdV-4v1, contained two isolates from 1984 and 12 isolates from 1985. Group 2, AdV-4v2, contained one isolate from 1991, two isolates from 1996, and five isolates from 1998. Group 3, AdV-4v3, and group 4, AdV-4v4, contained only one isolate from 1991 and one from 1998, respectively. The last group, AdV-4v5, contained four isolates from sporadic infections of conjunctivitis in 2001 and all eight isolates from nosocomial infections in 2001. Therefore, the new genome type, AdV-4c, was included in the AdV-4v5 group.
FIG. 3.
Phylogenetic analysis of isolates in this study and the AdV-4 prototype based on the fiber gene. The fiber gene of the new genome type (s-01, 01n/AdV-4v5) is located on another branch from that of the AdV-4a group. Numbers at nodes are percentages of 1,000 bootstrap values. The AdV-4p fiber gene sequence has been published (GenBank accession no. X76547).
From 1984 to 1985, the dominant variant of isolates in Sapporo was AdV-4v1. The dominant variant was AdV-4v2 in the 1990s and then AdV-4v5 in 2001. AdV-4v1 to AdV-4v4 formed a distinct cluster, with 87% bootstrap support, and their genomes were typed as AdV-4a. AdV-4v5 was located on a different branch from AdV-4v1 to -v4, however (Fig. 3).
DISCUSSION
In this study, we analyzed the nucleotide sequences of AdV-4 strains which caused nosocomial infections of epidemic conjunctivitis in Sapporo in 2001 together with other AdV-4 strains isolated from sporadic cases of patients with adenoviral conjunctivitis over the past 17 years. It was revealed that a new variant strain of AdV-4 strain caused the nosocomial infections.
To analyze the genetic characteristics of AdV-4 strains associated with nosocomial infection and to compare the differences between isolates from nosocomial cases and sporadic ones, we performed genome typing with the full length of the viral DNA. Until now, eight genome types of AdV-4, i.e., AdV-4p, p1-3, a, a1, b, and chimpanzee (ch), have been reported (13), and the major causative agent of the epidemics of adenoviral keratoconjunctivitis in the world was AdV-4a (7, 9, 12, 13, 26, 27). In this study, strains isolated before 2001 were found to be identical to AdV-4a. However, genome typing of the isolates from nosocomial and sporadic infections in 2001 with BamHI and SmaI showed different patterns from those of previously known AdV-4 variants, i.e., AdV-4p, p1-3, a, a1, and b. Therefore, we temporarily named this new genome type of AdV-4 strain AdV-4c. The pairwise comigrating restriction fragment (PCRF) analysis showed that AdV-4 consisted of at least three clusters (cluster 1 [AdV-4p and p1-3], cluster 2 [AdV-4a, a1, and b], and cluster 3 [AdV-4ch]) (13). The percentage of PCRF within a cluster and between clusters was 95 to 99% and 25 to 48%, respectively. In this study, the isolates from nosocomial infections showed high homology with AdV-4a (91.2% of PCRF). Thus, it was considered that AdV-4c was closely related to cluster 2.
So far, the PCR-RFLP method based on the hexon gene has been used to rapidly determine AdV serotypes (1, 19). When nosocomial infection occurred, we performed PCR-RFLP method (19) with the eye swab to rapidly investigate the serotype of the strain that caused the nosocomial infection. However, we could not identify the serotype because the PCR-RFLP patterns were not identical to that of AdV-4p and other all prototype strains (data not shown). We recently developed a molecular diagnotic method for human AdV-D and -E by phylogeny-based classification with a partial hexon sequence (Shimada et al., in press). In this method, we used the same PCR products with the PCR-RFLP method reported by Saitoh-Inagawa et al. (19). We used this classification to investigate the genetic characterizations of the strains that caused the nosocomial infections and why identification by the PCR-RFLP method was unsuccessful.
By nucleotide sequence analysis, the nucleotide sequences of the partial hexon genes of the clinical isolates were not identical to that of the AdV-4 prototype strain. The phylogeny-based classification identified all isolates as AdV-4 variants, including AdV-4a (Fig. 2). The homology rate of AdV-4p and the clinically isolated strain (AdV-4a and -4c) was 96.1%, and several point mutations occurred on the target site of the restriction enzymes with the PCR-RFLP method. Indeed, the PCR-RFLP method was useful for the rapid diagnosis of AdV, but this method has one disadvantage, which is that serotype identification might become impossible when a point mutation occurs on the target site of the restriction enzymes, because the basis of serotype identification with this method is the restriction pattern. For example, some isolates of AdV-7 (25), -8 (24), and -34 (20) have been reported to be unidentifiable by the PCR-RFLP method.
In the phylogeny-based classification with a partial hexon sequence, AdV-4a and -4c represented the same sequence. However, the isolates causing nosocomial infections had a different fiber gene lineage than that of AdV-4a in the phylogenetic tree, based on the full length of the fiber nucleotide sequence (Fig. 3). Fiber plays an important role when adenovirus adheres to the human cell (23). The mutation of the fiber gene might be related to the pathogenesis of this nosocomial infection. Further analysis of the fiber protein of AdV-4c may lead to elucidation of adenoviral adhesion to conjunctival cells.
In conclusion, we revealed that the new genome type of AdV-4 caused the nosocomial infections of epidemic conjunctivitis in Sapporo in 2001. The new genome type of AdV-4 showed a different restriction pattern from that of previously known variants of AdV-4. Therefore, we named the new variants type AdV-4c. We would like to propose to ophthalmologists that they need to carefully observe cases of adenoviral conjunctivitis caused by this new variant, AdV-4c, in order to prevent nosocomial infections.
Acknowledgments
This study was supported by a grant from the Takeda Science Foundation and in part by a grant for Research on Sensory and Communicative Disorders, Ministry of Health, Labor and Welfare, Japan, and by a grant-in-aid for Scientific Research awarded by the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
REFERENCES
- 1.Allard, A., B. Albinsson, and G. Wadell. 2001. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 39:498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, K., M. Kato, H. Ohtsuka, K. Ishii, N. Nakazono, and H. Sawada. 1982. Clinical and aetiological study of adenoviral conjunctivitis, with special reference to adenovirus types 4 and 19 infections. Br. J. Ophthalmol. 66:776-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, K., and Y. Tagawa. 2002. A twenty-one year surveillance of adenoviral conjunctivitis in Sapporo, Japan. Int. Ophthalmol. Clin. 42:49-54. [DOI] [PubMed] [Google Scholar]
- 4.Benkö, M., B. Harrach, and W. C. Russell. 2000. Adenoviridae, p. 227-238. In M. H. V. C. M. F. van Regenmortel, D. H. L. Bishop, E. B. Carstens, M. K. S. M. L. Estes, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and E. B. Wickner (ed.), Virus taxonomy. Academic Press, San Diego, Calif.
- 5.De Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:738-791. [DOI] [PubMed] [Google Scholar]
- 7.Guo, D. F., M. Shinagawa, K. Aoki, H. Sawada, S. Itakura, and G. Sato. 1988. Genome typing of adenovirus strains isolated from conjunctivitis in Japan, Australia, and the Philippines. Microbiol. Immunol. 32:1107-1118. [DOI] [PubMed] [Google Scholar]
- 8.Hilleman, M. R., and J. H. Werner. 1954. Recovery of new agent from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 85:183-188. [DOI] [PubMed] [Google Scholar]
- 9.Itakura, S., K. Aoki, H. Sawada, N. Ishiguro, and M. Shinagawa. 1991. Changes in subgenome types of adenovirus type 4 isolated from patients with ocular disease between 1985 and 1989 in Sapporo, Japan. J. Clin. Microbiol. 29:1740-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanai, H. 2000. Genome analysis with restriction endonucleases recognizing 4- or 5-base pair sequences of adenovirus type. Jpn. J. Ophthalmol. 44:463-466. [DOI] [PubMed] [Google Scholar]
- 11.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 12.Levandowski, R. A., and M. Rubenis. 1981. Nosocomial conjunctivitis caused by adenovirus type 4. J. Infect. Dis. 143:28-31. [DOI] [PubMed] [Google Scholar]
- 13.Li, Q. G., and G. Wadell. 1988. The degree of genetic variability among adenovirus type 4 strains isolated from man and chimpanzee. Arch. Virol. 101:65-77. [DOI] [PubMed] [Google Scholar]
- 14.Muzzi, A., G. Rocchi, B. Lumbroso, G. Tosato, and F. Barbieri. 1975. Acute haemorrhagic conjunctivitis during an epidemic outbreak of adenovirus-type-4 injection. Lancet ii:822-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Institute of Infectious Diseases. 1999. Adenovirus infections. Infect. Agents Surveil. Rep. 20:315. [Google Scholar]
- 16.National Institute of Infectious Diseases. 1995. Viruses isolated from the eye, Japan, 1990-1994. Infect. Agents Surveil. Rep. 16:97-98. [Google Scholar]
- 17.Ren, C. S., N. Nakazono, S. Ishida, S. Fujii, T. Yoshii, S. Yamazaki, K. Ishii, and K. Fujinaga. 1985. Genome type analysis of adenovirus type 4 isolates, recently obtained from clinically different syndromes in some areas in Japan. Jpn J. Med. Sci Biol. 38:195-199. [DOI] [PubMed] [Google Scholar]
- 18.Russel, W. C. 1998. Adenoviruses, p. 281-290. In L. Collier, A. Balows, and M. Sussman (ed.), Microbiology and microbial infections, 9th ed., vol. 1: virology. Arnold, London, England.
- 19.Saitoh-Inagawa, W., A. Oshima, K. Aoki, N. Itoh, K. Isobe, E. Uchio, S. Ohno, H. Nakajima, K. Hata, and H. Ishiko. 1996. Rapid diagnosis of adenoviral conjunctivitis by PCR and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 34:2113-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitoh-Inagawa, W., K. Tanaka, E. Uchio, N. Itoh, S. Ohno, and K. Aoki. 2001. Genome typing of adenovirus type 34 isolated in two cases of conjunctivitis in Sapporo, Japan. J. Clin. Microbiol. 39:4187-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 22.Schnurr, D., and M. E. Dondero. 1993. Two new candidate adenovirus serotypes. Intervirology 36:79-83. [DOI] [PubMed] [Google Scholar]
- 23.Shenk, T. E. 2001. Adenoviridae: the viruses and their replication, p. 2265-2300. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23a.Shimada, Y., T. Ariga, Y. Tagawa, K. Aoki, S. Ohno, and H. Ishiko. 2004. Molecular diagnosis of human adenoviruses D and E by a phylogeny-based classification method using a partial hexon sequence. J. Clin. Microbiol. 42:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi, S., N. Itoh, E. Uchio, K. Tanaka, N. Kitamura, H. Kanai, K. Isobe, K. Aoki, and S. Ohno. 1999. Adenovirus strains of subgenus D associated with nosocomial infection as new etiological agents of epidemic keratoconjunctivitis in Japan. J. Clin. Microbiol. 37:3392-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka, K., N. Itoh, W. Saitoh-Inagawa, E. Uchio, S. Takeuchi, K. Aoki, E. Soriano, M. Nishi, R. B. Junior, C. M. Harsi, L. Tsuzuki-Wang, E. L. Durigon, K. E. Stewien, and S. Ohno. 2000. Genetic characterization of adenovirus strains isolated from patients with acute conjunctivitis in the city of Sao Paulo, Brazil. J. Med. Virol. 61:143-149. [PubMed] [Google Scholar]
- 26.Tsuzuki-Wang, L., K. Aoki, K. Isobe, S. Shiao, K. Toba, N. Kobayashi, Y. Noguchi, and S. Ohno. 1997. Genome analysis of adenovirus type 4 strains isolated from acute conjunctivitis in Japan. Jpn. J. Ophthalmol. 41:308-311. [DOI] [PubMed] [Google Scholar]
- 27.Tullo, A. B., and P. G. Higgins. 1980. An outbreak of adenovirus type 4 conjunctivitis. Br. J. Ophthalmol. 64:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wadell, G., and J. C. de Jong. 1980. Restriction endonucleases in identification of a genome type of adenovirus 19 associated with keratoconjunctivitis. Infect. Immun. 27:292-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wadell, G., M. L. Hammarskjold, G. Winberg, T. M. Varsanyi, and G. Sundell. 1980. Genetic variability of adenoviruses. Ann. N. Y. Acad. Sci. 354:16-42. [DOI] [PubMed] [Google Scholar]