Abstract
A variety of sex determination mechanisms can be observed in evolutionary divergent teleosts. Sex determination is genetic in Atlantic cod (Gadus morhua), however the genomic location or size of its sex-locus is unknown. Here, we characterize the sex-locus of Atlantic cod using whole genome sequence (WGS) data of 227 wild-caught specimens. Analyzing more than 55 million polymorphic loci, we identify 166 loci that are associated with sex. These loci are located in six distinct regions on five different linkage groups (LG) in the genome. The largest of these regions, an approximately 55 Kb region on LG11, contains the majority of genotypes that segregate closely according to a XX-XY system. Genotypes in this region can be used genetically determine sex, whereas those in the other regions are inconsistently sex-linked. The identified region on LG11 and its surrounding genes have no clear sequence homology with genes or regulatory elements associated with sex-determination or differentiation in other species. The functionality of this sex-locus therefore remains unknown. The WGS strategy used here proved adequate for detecting the small regions associated with sex in this species. Our results highlight the evolutionary flexibility in genomic architecture underlying teleost sex-determination and allow practical applications to genetically sex Atlantic cod.
Teleosts are characterized by a remarkable diversity of independently evolved sex-determining mechanisms that range from those using environmental cues to those under strict genetic control1,2,3,4. Different genes control sexual fate in a variety of teleosts and a variety master sex determining (SD) genes have been described, dmY, gsdfY and sox3Y in the medaka genus5,6,7,8, amhr2 in fugu9, amhy in Patagonian pejerry10 and Nile Tilapia11, dmrt1 in half-smooth tongue sole12, gdf6Y in Killifish13 and sdY in rainbow trout14. Of these examples, only sdY represents a case whereby a gene without previously known functionality during sexual development has been recruited as a SD gene, and could be an example of de novo SD evolution4,14. The potentially rapid evolution of SD mechanisms15 and their striking diversity, even in closely related lineages like those of the medaka genus16 and the stickleback family17,18,19 hinders the identification and characterization of such mechanisms in divergent teleost lineages.
Here, we characterize the sex-locus in Atlantic cod (Gadus morhua), an economically and culturally important marine resource in the North Atlantic region. In this species, divergent and sex-dimorphic expression between females and males has been observed for several genes, including copies of dmrt20, sox9 and cyp19a121 and several estrogen receptors (ER)22. Nevertheless, these genes regulate the downstream process of sex differentiation, and there is no direct evidence for their role in sex-determination. Therefore the identity of the sex-locus in Atlantic cod has remained unknown.
Atlantic cod exhibits variable levels of sexual dimorphism, with females becoming longer and heavier than males23. Sex further alters the propensity to mature early in aquaculture, with males maturing earlier than females, leading to reduced male somatic growth, resulting in considerable economical loss24. Several observations provide evidence for a largely genetic rather than environmental sex determination in Atlantic cod. First, equal male to female ratios are found in wild Atlantic cod specimens below 60 cm25, regardless of the highly skewed sex ratios that can be observed during spawning26. Equal sex ratios are also obtained in controlled experimental crosses27. Finally, the generation of all-female populations by gynogenesis or breeding with masculinized females provides not only evidence for genetic sex-determination, but also for male-heterogametic sex-determination, i.e. a XX-XY system27,28.
We analyzed whole genome sequence (WGS) data from 227 Atlantic cod specimens in order to find the putative location of the sex-locus in this species. First, the analysis of a stringently filtered SNP dataset containing genotypes from 48 individuals identifies a single genomic region on linkage group (LG) 11 (sensu Hubert et al.29) that contains the only genotypes that segregate according to a male-heterogametic sex-determination system in this reduced dataset. Subsequently, we confirmed this regions’ association with gender by determining genetic sex in 179 unrelated individuals of known sex. Through analysis of an unfiltered variant dataset of all 227 individuals, we identified five additional genomic regions on alternate linkage groups that can be associated with sex, although this association is inconsistent. The reference genome has been based on data from a male specimen. To exclude large-scale assembly errors as the most likely explanation for these sex-linkage patterns, PacBio read data obtained from a female individual was used to independently confirm assembly accuracy. The identified sex-associated region on LG11 is less than 55 Kb and lacks clear sequence homology compared to genes or regulatory regions previously identified as sex-locus. Moreover, it is not directly associated with genes known to be involved in sex differentiation. The functionality of this region remains therefore obscure.
Results
We obtained average Illumina sequence coverage of 10.9 X per individual (Supplementary Table 1), retaining 1,573,340 SNPs after filtering and 55,160,622 variable sites without filtering. Out of these variants, we selected those polymorphisms with a maximum of two heterozygote genotypes in the homogametic sex, and a maximum of two homozygote genotypes in the heterogametic sex in a subset of 21 males and 27 females. Following our criteria, three and thirteen loci show sex-linked segregation (homozygous females and heterozygous males) using the filtered or unfiltered dataset, respectively (Table 1). Thus, filtering using recommended practices substantially reduces the number of loci that are identified, including several insertion-deletion polymorphisms. All of the identified loci in the subset of individuals are located within a single, relatively small genomic region on linkage group (LG) 11. In contrast, the reverse selection criteria (heterozygous females and homozygous males) do not yield a single polymorphism.
Table 1. Polymorphisms with sex-linked genotypic segregation in 48 Atlantic cod specimens (21 ♂ and 27 ♀).
| LG | Position (bp) | Allele |
Genotype (Ref/Het/Alt1) |
||
|---|---|---|---|---|---|
| Reference | Alternative | Females | Males | ||
| 11 | 118857532 | G | T | 0/2/24 | 1/20/0 |
| 11 | 118868732 | T | A | 0/0/27 | 1/20/0 |
| 11 | 11888434 | T | C | 27/0/0 | 0/21/0 |
| 11 | 118931182 | T | A | 27/0/0 | 1/18/1 |
| 11 | 11897471 | G | GTGT | 0/0/27 | 0/21/0 |
| 11 | 11897513 | C | T | 0/0/27 | 0/20/1 |
| 11 | 11897519 | A | T | 0/0/27 | 0/21/0 |
| 11 | 11897566 | AATCC | A | 0/1/25 | 1/19/0 |
| 11 | 11899188 | A | G | 0/0/27 | 0/20/1 |
| 11 | 11899196 | G | T | 0/0/27 | 0/21/0 |
| 11 | 11899391 | C | CT | 0/0/27 | 0/21/0 |
| 11 | 11899539 | G | T | 0/2/25 | 0/20/0 |
| 11 | 11899548 | TA | T | 0/2/25 | 0/20/0 |
Limited numbers of polymorphisms (3 out of 1,573,340 in the filtered, 13 out of 55,160,622 in the unfiltered dataset) are identified, which have highly homozygous genotypes for females and heterozygous genotypes for males. All polymorphisms are co-located within a 15 Kb region on linkage group (LG) 11.
1Ref = homozygous reference, Het = heterozygous, Alt = homozygous alternative.
2Identified in the filtered SNP dataset.
We calculated individual values of the inbreeding coefficient (F) based on the 13 identified sex-linked polymorphisms. Positive F values indicate largely homozygous and negative F values heterozygous genotypes. By classifying individuals based on these F values, we confirm female or male phenotypic sex in a total of 179 unrelated specimens, misclassifying a single individual. Moreover, the 13 polymorphisms display distinct homo- or heterozygous segregation depending on sex (i.e., the majority of individuals is either fully homozygous or heterozygous, Supplementary Table 1). The single misclassified specimen has heterozygous genotypes for all 13 loci and is therefore clearly a genetic male. The probability of obtaining 13 heterozygous loci in a female given random genotype error or recombination is vanishingly small. We therefore postulate that human error (most likely while recording sex) is responsible for this single misclassification.
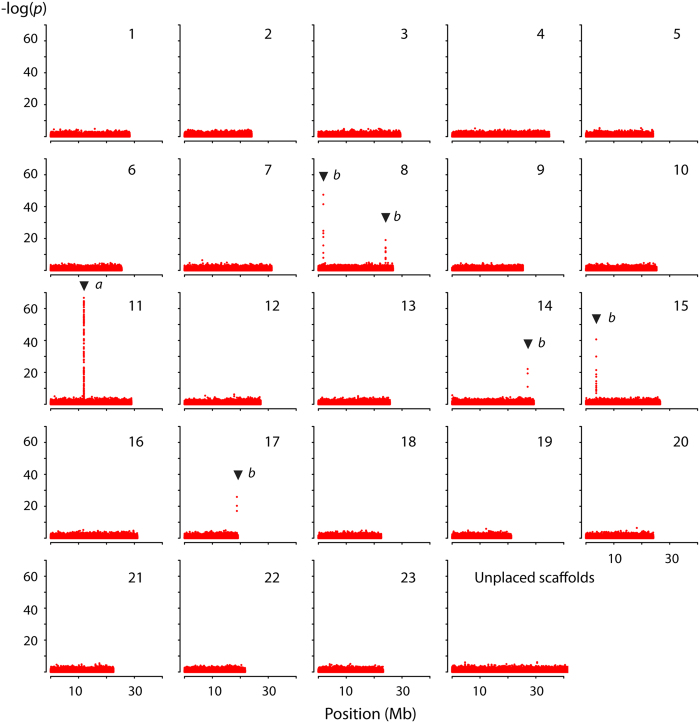
By further investigating sex-linked genotypic segregation in more than 55 million polymorphisms using the unfiltered dataset for 110 males and 116 females (excluding the misclassified individual), we identify six distinct regions with elevated p-values (i.e., six times above the standard deviation (SD) of the mean transformed p-value) on five different LGs using Fishers’ exact test (Fig. 1). LG11 contains the largest region (55 Kb) and the highest number of sex-linked loci (n = 127) with most extreme p-values, whereas the other LGs contain relatively small regions and lower number of less extreme p-values (Table 2). Among all sex-linked loci, those with transformed p-values above 50 (n = 36) show a near exclusive XX-XY segregation of genotypes, although genotypes with inconsistent sex-linkage do occur in that range (Supplementary Table 2). Loci with p-values below 50 exhibit a higher proportion of genotypes that are inconsistently linked to sex-specific segregation (Supplementary Table 2).
Figure 1. Sex linked segregation of genotypes in Atlantic cod.
Over 55 million polymorphisms are compared in 110 males and 116 females of Atlantic cod using Fishers’ exact test. Six distinct regions are identified with a significant increase in p-values, i.e. above 6SD of the mean. Highest numbers of genotypes with most extreme p-values are found on LG11 (a), whereas reduced numbers with lower p-values are found on LG08, LG14, LG15 and LG17 (b). P-values were calculated using PLINK (v.1.90p) and −log transformed. Unplaced scaffolds have been concatenated for visualization.
Table 2. Linkage groups and regions that contain loci with sex-linked segregation in 226 Atlantic cod specimens.
| LG | Start (bp) | Stop (bp) | Size (bp) | Sex-linked (n) | Total (n) | Max-p |
|---|---|---|---|---|---|---|
| 08a | 1784400 | 1784813 | 413 | 11 | 23 | 47 |
| 08b | 24118524 | 24118864 | 340 | 8 | 14 | 19 |
| 11 | 11864114 | 11918378 | 54264 | 127 | 472 | 67 |
| 14 | 27062221 | 27062388 | 167 | 3 | 8 | 22 |
| 15 | 3678047 | 3679605 | 1558 | 14 | 64 | 41 |
| 17 | 18769676 | 18769822 | 146 | 3 | 6 | 26 |
Start and Stop refer to the location of the first and last locus with a high (greater than six standard deviation from the mean) Fisher’s exact test p-value in a particular region. Size is calculated as the difference between Start and Stop. For each region, the number of sex-linked loci, total number of loci (including those that are not linked to sex, using a MAF of 0.05) and the maximum p-value (Max-p) observed in that region are given.
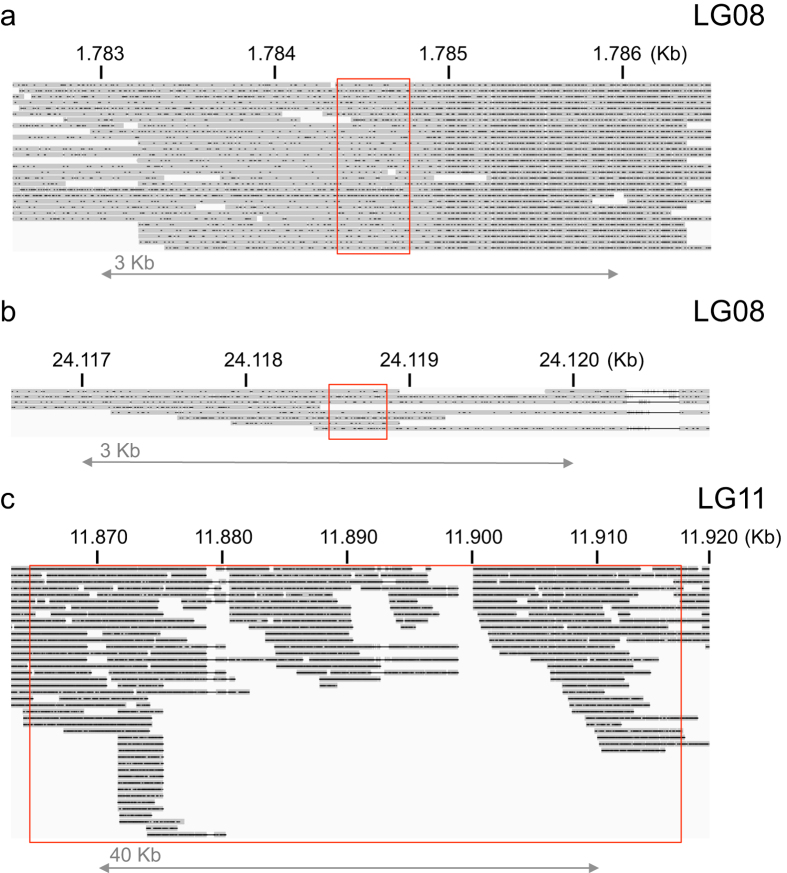
It is possible that assembly errors are causal for the observed pattern of multiple sex-associated regions. We independently assessed the assembly accuracy of these regions by aligning PacBio reads from a female specimen towards the gadMor2 genome30. We obtain long-range coverage for all sex-linked regions (Fig. 2, Supplementary Figures 1–3) and do not observe alignment failures that indicate large-scale assembly errors at the edges of these regions. Nevertheless, within sex-associated region on LG11, no female PacBio alignments can be observed crossing an approximately 1 Kb section around position 11,900,000 bp (Fig. 2C), which can be indicative of assembly error or the presence of larger, female specific genomic rearrangements that cause long-read alignments to fail. Should the latter be the case, we hypothesized that should observe PacBio read alignments crossing this region if we aligned their read ends only, essentially like a paired-end read. By aligning such artificially created “paired-end” PacBio reads, we indeed observe 19 instances of female PacBio reads crossing the region on LG11 (Supplementary Figure 4), further supporting the current orientation of the genome assembly. Overall, we conclude that is unlikely that large-scale assembly errors explain the observed associations with sex over multiple linkage groups.
Figure 2. PacBio read alignments of a female Atlantic cod specimen.
Long read alignments (grey) towards the male gadMor2 reference genome overlap those regions containing sex-associated genotypes (red box) on LG08 (a,b) and LG11 (c). Note the different genomic scale (in Kb) for each of the sub-panels. Small indels (black dots) within the alignments are a typical feature of PacBio read data. Read alignments are visualized using the Integrative Genomics Viewer.
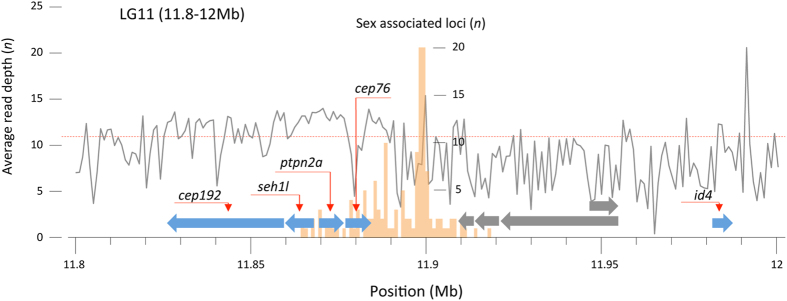
We investigated the locations surrounding the sex-linked regions for presence of genomic features that can provide insight in their function. We do not observe a direct association of the identified regions with any of the candidate genes known to be involved in sex-determination of other fish, or with genes known to be involved in sex-differentiation in Atlantic cod. Instead, the most likely alignments of those candidate genes are found on other linkage groups or are located at least 4 MB away (e.g. dmrt4 on LG17, Table 3). The sex-linked regions on LG14, 15 and 17 occur completely or partly in dispersed repeats (LTR/Copia, data not shown), those on LG8a are associated with an unknown protein model, whereas those on LG8b are not associated with any automated annotation (data not shown). Given that these regions are relatively small and inconsistently sex-linked, we further focus on a 200 Kb region around the sex-linked loci of LG11. Within this region, we find evidence for nine protein coding gene models however; the highest number of sex-linked genotypes is not associated with any of these (Fig. 3). Specifically, the majority of genotypes are located downstream of an annotated gene (cep76) and an unnamed gene model (without evidence for a particular protein homology).
Table 3. Names and genbank ID of sex-determining genes from various teleosts and genes known to be involved in sex-differentiation in Atlantic cod.
| Gene name | Genbank ID | Species | LG | Start | Stop | Alignment score |
|---|---|---|---|---|---|---|
| akap11 | XM_011473624.1 | Oryzias latipes | 20 | 10120287 | 10118045 | 1859 |
| amh | JN802292.1 | Gadus morhua | 12 | 24019786 | 24018572 | 1610 |
| amhy | HM153803.1 | Odontesthes hatcheri | 12 | 24019643 | 24018570 | 473 |
| amhr2 | NM_001280009.1 | Takifugu rubripes | 13 | 9249009 | 9250493 | 574 |
| ar | FJ268742.1 | Gadus morhua | 10 | 19432117 | 19442119 | 3743 |
| cyp19a | DQ402370.1 | Gadus morhua | 14 | 16165860 | 16169755 | 2851 |
| cyp19b | JN802291.1 | Gadus morhua | 9 | 2257039 | 2260649 | 2648 |
| dmrt | AJ506094.1 | Gadus morhua | 6 | 19949821 | 19940091 | 855 |
| dmrt2a | JN802284 | Gadus morhua | 6 | 19911488 | 19908495 | 1650 |
| dmrt3 | JN802285 | Gadus morhua | 6 | 19922948 | 19919278 | 2020 |
| dmrt4 | JN802286 | Gadus morhua | 17 | 14093750 | 14092035 | 2546 |
| dmrt5 | JN802287 | Gadus morhua | 12 | 20988494 | 20990196 | 2537 |
| dmy | NM_001104680.1 | Oryzias latipes | 6 | 19948867 | 19940028 | 382 |
| esr1 | JX178935.1 | Gadus morhua | 21 | 18435483 | 18415167 | 2611 |
| esr2a | JX178936.1 | Gadus morhua | 21 | 8193571 | 8213026 | 3711 |
| esr2b | JK993476.1 | Gadus morhua | 5 | 20297385 | 20297568 | 334 |
| foxl2 | NM_001104888.1 | Oryzias latipes | 1 | 27838167 | 27837834 | 470 |
| gsdf | KC204828.1 | Gadus morhua | 4 | 25085824 | 25083577 | 1040 |
| sdY | NM_001281416.1 | Oncorhynchus mykiss | 7 | 1898620 | 1898410 | 136 |
| sox3 | AB775143.1 | Oryzias dancena | 10 | 17937590 | 17936636 | 1482 |
| sox9a | JN802288.1 | Gadus morhua | 18 | 7659675 | 7661680 | 1722 |
| sox9b | JN802289.1 | Gadus morhua | 2 | 17478629 | 17480857 | 2714 |
| vasa | HM451456.1 | Gadus morhua | 7 | 3034506 | 3016733 | 3812 |
Location (in base pair) on the various linkage groups (LD) was determined by aligning the protein coding sequence to the genome assembly using exonerate 2.2.0 50 with the option –model coding2genome. Alignments with the highest score (–bestn) were selected as the most likely genomic location.
Figure 3. Annotation surrounding the sex-associated region of Atlantic cod on linkage group 11.
Nine gene models (the arrow shows transcriptional direction) have been annotated within a 200 Kb window around the loci with sex-linked genotypic segregation. The histogram (orange) shows the number of sex associated loci (with p-values more than 6 SD from the mean) in windows of 1 Kb. Gene models with cDNA evidence from Atlantic cod (blue) have been annotated with gene names and are in conserved synteny (order and transcriptional direction) with those in three-spined stickleback and spotted gar. Three genes (ptpn2a, cep76 and id4) are in conserved synteny with medaka. Gene models without cDNA evidence (grey) do not have obvious homology with known genes. Average read depth (grey line) is calculated in 1 Kb windows and can be compared to the genome-wide average coverage (red dashed line).
Of the nine gene models, four have no identified orthologs in other species and have no transcription evidence based on Atlantic cod cDNA data. The five genes with transcriptional evidence (annotated cep192, seh1l, ptpn2a, cep76 and id4) based on Atlantic cod cDNA data30, also have high sequence homology towards genes in other species. This region is similarly annotated in the first Atlantic cod genome assembly on GeneScaffold_152331, although that assembly contains substantially more sequence gaps –in particular around the location of the sex-locus. Within the 200 Kb region, the five genes with high sequence homology towards known genes show conserved synteny based on order and transcriptional direction with several other fish species, for instance stickleback (chrXX: 1,45-1,6 Mb) and spotted gar (LG11: 32,28-32,56 Mb). Conserved synteny for three of these (ptpn2a, cep76 and id4) is observed in medaka (chr16: 1.4-1.7 Mb).
Discussion
Here, we identify several genomic regions in Atlantic cod that contain loci with evidence for sex-linked genotypic segregation. Nonetheless, the region on LG 11 shows most evidence for being involved in sex determination. First, this region is an order of magnitude larger than the others, and contains the highest number of sex-linked loci. Second, those loci in this region are the most strongly segregating according to the male-heterogametic sex-determination system that is expected from earlier experimental crosses28. Our data yield no support for a female heterogametic system (i.e., ZZ-ZW system). Finally, based on 13 loci within the region on LG11 that have strict male-heterogametic sex segregation, we can confirm phenotypic sex with genotypic sex in 178 unrelated individuals obtained from geographically separated populations, whereas sex is imperfectly linked in the other genomic regions. Taken together, we conclude that the most likely location for the sex-locus in Atlantic cod is located on LG11 within this 55 Kb window.
The relatively small genomic footprint of the sex-associated regions observed in Atlantic cod limits the type of sequencing strategy that can be used for their detection. Specifically, reduced representation sequencing of genomic DNA (e.g. ddRAD-seq, double digest restriction site – associated DNA sequencing) has been proposed as economically efficient approach for genotyping non-model species32. Using such approach in rockfish, up to 33 sex-specific loci were obtained out of a total of 74,965 loci33. We here identified 166 sex-associated loci out of a total of 55,160,622 variable sites at a rate of 1 in 332,292 loci. Thus, with our observed discovery rate, it is doubtful that a reduced representation sequencing strategy would have yielded sufficient information to characterize the sex-linked regions or deliver reliable sex-linked genotypes. Should similarly sized sex-loci as observed in Atlantic cod be more widespread among teleosts, reduced representation sequencing approaches will likely be of limited use in their detailed characterization.
Automated annotation of the 200 Kb region surrounding the 55 Kb window does not provide strong evidence about the functionality of the sex-locus. We find no sequence homology with candidate SD genes from other teleosts6,7,8,9,10,14 that are all located on different linkage groups. Similarly, no genes reportedly involved in sexual differentiation in Atlantic cod20,21,22 are located on the same linkage group. No ab initio gene model is directly associated with the region that is most strongly associated with sex-specific segregation (Fig. 3). This region’s lack of clear sequence homology and lack of direct association with known candidate genes leaves its functionality unknown. Moreover, based on conserved synteny of five genes (cep192, seh1l, ptpn2a, cep76 and id4) surrounding the Atlantic cod sex-associated region in teleosts like stickleback, spotted gar and medaka, this region is likely of ancient evolutionary origin. In these other species however, no role for sex-determination has been suggested for this location. Therefore, the sex-locus in Atlantic cod appears to be a derived, non-homologous feature. Two scenarios may explain the observed lack of sequence homology; 1) Atlantic cod has recruited a novel type of sex determining locus (with unknown function). 2) Atlantic cod has retained a known locus, yet this subsequently evolved and diverged to such extent that sequence homology analyses fail to detect this locus, whilst maintaining its original functionality. In both scenarios, this observation records extensive evolutionary divergence in the genomic architecture underlying sex-determination. Should the sex-determining locus in Atlantic cod be newly recruited, this adds to a growing body of literature suggesting greater genetic plasticity in the sex-differentiating cascade4 than previously anticipated34.
We note that the sex-associated region is located ~80 kb upstream from id4, which encodes a transcription factor of the inhibitor of DNA binding (ID) protein family. One of the related pathways of id4 is the TGF-β signaling pathway35. Variation in genes of the TGF-β signaling pathway plays a role in the sex determination of a variety of fish. For instance gene duplicates of Anti-Mullerian Hormone (amh) determine sex in Patagonian pejerry10 and Nile Tilapia11, a missense SNP in the Anti-Mullerian Hormone receptor II (amhr2) determines sex in fugu9 and male specific variation in a TGF-β growth factor (gdf6Y) determines sex in killifish13. It may be that male-specific regulation of id4 initiates a downstream cascade through the TGF-β signaling pathway, which eventually results in gender determination in Atlantic cod.
The identification of the Atlantic cod sex-locus has several practical applications. For instance, phenotypic sex is difficult or impossible to obtain if specimens are not large enough for a visual assessment of gonads (i.e. early larval stages) or if fish are in non-spawning condition and preferentially kept alive. Our findings provide the means to retroactively assign genetic sex. Moreover, our finding allows the assignment of the sex phenotype for specimens for which this is impossible to obtain by other means, such as archeological bone material36 or historical specimens37,38 of unknown sex. Finally, this finding eases the identification of masculinized females for the generation of all-female populations28, providing opportunity for a more profitable aquaculture.
Methods
Ethics statement
We always strive to limit the effect of our sampling needs on populations and individuals. Therefore, we collaborate with other sources, like commercial fisheries or aquaculture farms, from which samples can be obtained as a byproduct of conventional business practice. This way, no animals need to be sacrificed to serve our scientific purpose. Samples were taken post-mortem and no scientific experiments have been performed on live animals. All specimens were part of larger hauls, caught by commercial vessels, were euthanized by local fishermen and were intended for human consumption. Sampling in this manner does not fall under any specific legislation in Norway or Iceland. These methods are in accordance with the guidelines set by the ‘Norwegian consensus platform for replacement, reduction and refinement of animal experiments’ (www.norecopa.no).
Sampling and DNA extraction
Atlantic cod specimens (n = 227) were obtained from several locations and sample dates around Norway and Iceland (Supplementary Table 3). Sex was determined through visual assessment of male (n = 112) or female (n = 115) gonads for each individual. DNA was extracted from fin clips, muscle or spleen using DNeasy Blood & Tissue kit (Qiagen), and sheared to an approximate insert size of 350 bp. Over 2 μg of DNA per sample was used to create Illumina compatible sequencing libraries using the TruSeq DNA PCR-Free LT Library Preparation Kit (Illumina). Libraries were individually barcoded, pooled and sequenced together on a HiSeq 2000. After demultiplexing with Illumina RTA (1.18.61.0) & CASAVA (1.8.4), sequencing reads were aligned using the mem algorithm of BWA v.0.7.5a-r40539 to the gadMor2 genome assembly30 available from the European Nucleotide Archive (ENA) LN845748-LN845770. Polymorphic variants were jointly called for all 227 individuals using GATK v. 3.3.040 according to GATK Best Practices recommendation41,42. For the filtered dataset, we selected sites with a minimum quality (FS > 60.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0 || QD<2.0 || MQ<40), allowing no SNPs within 10 base pair (bp) of an indel with BCFTOOLS v. 1.243. After this, we removed all indels and SNPs with more than two alleles, an average read depth higher than 30, a minimum allele frequency (MAF) below 0.05 and SNPs with more than 10% missing data per population using VCFTOOLS v. 0.1.1444. Both datasets –filtered and unfiltered– were used for subsequent analyses.
Identification of putative sex-locus
To identify the location of the putative sex-locus, we use two approaches. First, we analyzed 48 individuals sampled in Lofoten, Norway 2011 (♂ = 21, ♀ = 28, Supplementary Table 1) to identify SNPs and other biallelic polymorphisms with a strict sex-specific segregation (i.e. those either exclusively homozygous or heterozygous depending on sex) using VCFTOOLS v. 0.1.1444. Specifically, we select all polymorphisms with a maximum of two heterozygote genotypes in the homogametic sex, and a maximum of two homozygote genotypes in the heterogametic sex (heterozygote genotypes have a tendency to be lost from low-coverage genome data45), allowing for 10% missing data. Those polymorphisms that agree with these criteria are subsequently used to determine genetic sex in the other 179 specimens by calculating the inbreeding coefficient, F, for each individual using a method of moments using the –het algorithm in VCFTOOLS v. 0.1.1444. Individuals with positive F values (e.g. more homozygous than expected by chance) are classified female, whereas those with negative F values are classified male. We investigated the putative sex determining genomic region using the unfiltered whole genome variants dataset of 110 males and 116 females –excluding a single misclassified individual. We identify sex linked segregation of genotypes by calculating exact p-values using Fishers’ exact test with the options –fisher –model in PLINK v. 1.90p46. This algorithm is general test of association in the 2-by-3 table of genotypes rather than the basic allelic test (which compares frequencies of alleles in cases versus controls). For visualization, p-values were –log transformed.
Alignment of female PacBio reads
Long-range PacBio reads (P6-C4 chemistry) of a female Atlantic cod specimen were aligned to the gadMor2 assembly30 using BLASR v.3.0.147 with the following options: -bestn 2 -clipping subread -affineAlign -noSplitSubreads -nCandidates 20 -minPctIdentity 40 -sdpTupleSize 6. Based on the long-range PacBio data of the female specimen, we created artificial paired-end reads with a length of 300 bp for the forward and reverse reads, using prinseq-lite v.0.20.448. These paired-end reads were subsequently aligned using the mem algorithm of BWA v.0.7.5a-r40539. Aligned full-length PacBio reads and artificial paired-end PacBio reads were visualized using the Integrative Genomics Viewer49.
Location of candidate SD or sex differentiating genes
We identify the location of previously known SD genes or those implicated in sexual differentiation using the following approach. Candidate genes (akap11, amh, amhy, amhr2, ar, cyp19a, cyp19b, dmrt(2a,3,4,5), dmy, esr1, esr2a, esr2b, foxl2, gsdf, sdY, sox3, sox9a, sox9b and vasa) were aligned to the Atlantic cod genome using exonerate 2.2.050 with the option –model coding2genome. Alignments with the highest score were selected as the most likely genomic location in the Atlantic cod genome using the option –bestn.
Protein annotation of sex-determining region
Protein annotation of the Atlantic cod genome (gadMor2) is described in detail elsewhere30. In short, we used MAKER2, v. 2.3151,52 to combine the consolidated evidence from ab initio gene finders and physical evidence (e.g. protein and RNA sequence alignments) in to a set of quality gene models. Evidence from RNA sequence alignments was obtained using a combined RNA-sequence dataset from various sources31,53,54 including a set of PacBio reads55. Gene models were aligned using BLAST56 against the SwissProt/UniProtKG release 2015_12 to obtain putative gene names.
Data availability
The gadMor2 genome assembly is available from the European Nucleotide Archive (ENA) LN845748-LN845770. The annotation of the Atlantic cod genome is available from https://t.co/mdiVe52v4d. All individual read data (including the female PacBio data) associated with linkage groups containing sex-linked genotypes are available from ENA with study accession number PRJEB14672.
Additional Information
How to cite this article: Star, B. et al. Genomic characterization of the Atlantic cod sex-locus. Sci. Rep. 6, 31235; doi: 10.1038/srep31235 (2016).
Supplementary Material
Acknowledgments
We thank three anonymous reviewers for their comments and suggestions that have substantially improved this manuscript. Sequencing was performed by the Norwegian Sequencing Centre, a national technology platform hosted by the University of Oslo (UIO) and supported by the “Functional Genomics” and “Infrastructure” programs of the Research Council of Norway and the Southeastern Regional Health Authorities (www.sequencing.uio.no). Computational intensive analyses were done on the Abel Cluster, owned by the UIO and the Norwegian metacenter for High Performance Computing (NOTUR), and operated by the Department for Research Computing at USIT, the UIO IT-department (http://www.hpc.uio.no/). We thank Dr. Sanne Boessenkool for comments and suggestions. This research was supported by the Norwegian Research Council under project “The Aqua Genome Project (#221734/O30)”. We have adhered to all local, national and international regulations and conventions, and we respected normal scientific ethical practices.
Footnotes
Author Contributions O.K.T. and A.J.N. provided early access to the genome assembly and annotation. O.K.T. and A.J.N. analyzed PacBio read data. C.P. and S.J. provided samples. S.J. supervised laboratory handling and sequencing of samples. K.S.J. provided funding. B.S. designed the study, analyzed and interpreted the data and wrote the manuscript in collaboration with the other authors. All authors have reviewed the manuscript.
References
- Volff J.-N., Nanda I., Schmid M. & Schartl M. Governing sex determination in fish: regulatory putsches and ephemeral dictators. Sex. Dev. 1, 85–99 (2007). [DOI] [PubMed] [Google Scholar]
- Avise J. & Mank J. Evolutionary perspectives on hermaphroditism in fishes. Sex. Dev. 3, 152–163 (2009). [DOI] [PubMed] [Google Scholar]
- Martínez P. et al. Genetic architecture of sex determination in fish: applications to sex ratio control in aquaculture. Front. Genet. 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heule C., Salzburger W. & Böhne A. Genetics of sexual development: an evolutionary playground for fish. Genetics 196, 579–591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I. et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka. Oryzias latipes. PNAS 99, 11778–11783 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M. et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417, 559–563 (2002). [DOI] [PubMed] [Google Scholar]
- Myosho T. et al. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191, 163–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana Y. et al. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 5 (2014). [DOI] [PubMed] [Google Scholar]
- Kamiya T. et al. A Trans-Species Missense SNP in Amhr2 Is Associated with Sex Determination in the Tiger Pufferfish, Takifugu rubripes (Fugu). PLoS Genet 8, e1002798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori R. S. et al. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. PNAS 109, 2955–2959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia. Oreochromis niloticus. PLoS Genet 11, e1005678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46, 253–260 (2014). [DOI] [PubMed] [Google Scholar]
- Reichwald K. et al. Insights into Sex Chromosome Evolution and Aging from the Genome of a Short-Lived Fish. Cell 163, 1527–1538 (2015). [DOI] [PubMed] [Google Scholar]
- Yano A. et al. An immune-related gene evolved into the master sex-determining gene in rainbow trout. Oncorhynchus mykiss. Curr. Biol 22, 1423–1428 (2012). [DOI] [PubMed] [Google Scholar]
- van Doorn G. S. & Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature 449, 909–912 (2007). [DOI] [PubMed] [Google Scholar]
- Tanaka K., Takehana Y., Naruse K., Hamaguchi S. & Sakaizumi M. Evidence for different origins of sex chromosomes in closely related Oryzias fishes: substitution of the master sex-determining gene. Genetics 177, 2075–2081 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. A., Urton J. R., Boland J., Shapiro M. D. & Peichel C. L. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet 5, e1000391 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J. et al. A role for a neo-sex chromosome in stickleback speciation. Nature 461, 1079–1083 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J. & Peichel C. L. Turnover of sex chromosomes and speciation in fishes. Environ Biol Fishes 94, 549–558 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen H. & Andersen Ø. Sex dimorphic expression of five dmrt genes identified in the Atlantic cod genome. The fish-specific dmrt2b diverged from dmrt2a before the fish whole-genome duplication. Gene 505, 221–232 (2012). [DOI] [PubMed] [Google Scholar]
- Johnsen H., Tveiten H., Torgersen J. S. & Andersen Ø. Divergent and sex-dimorphic expression of the paralogs of the Sox9-Amh-Cyp19a1 regulatory cascade in developing and adult atlantic cod (Gadus morhua L.). Mol. Reprod. Dev. 80, 358–370 (2013). [DOI] [PubMed] [Google Scholar]
- Nagasawa K., Presslauer C., Kirtiklis L., Babiak I. & Fernandes J. M. Sexually dimorphic transcription of estrogen receptors in cod gonads throughout a reproductive cycle. J. Mol. Endocrinol. 52, 357–371 (2014). [DOI] [PubMed] [Google Scholar]
- Keyl F., Kempf A. & Sell A. Sexual size dimorphism in three North Sea gadoids. J. Fish Biol 86, 261–275 (2015). [DOI] [PubMed] [Google Scholar]
- Taranger G. L. et al. Control of puberty in farmed fish. Gen. Comp. Endocrinol. 165, 483–515 (2010). [DOI] [PubMed] [Google Scholar]
- Marshall C. T., Needle C. L., Thorsen A., Kjesbu O. S. & Yaragina N. A. Systematic bias in estimates of reproductive potential of an Atlantic cod (Gadus morhua) stock: implications for stock recruit theory and management. Can J Fish Aquat Sci 63, 980–994 (2006). [Google Scholar]
- Morgan M. & Trippel E. Skewed sex ratios in spawning shoals of Atlantic cod (Gadus morhua). ICES J. Mar. Sci. 53, 820–826 (1996). [Google Scholar]
- Whitehead J. A., Benfey T. J. & Martin-Robichaud D. J. Ovarian development and sex ratio of gynogenetic Atlantic cod (Gadus morhua). Aquaculture 324–325, 174–181 (2012). [Google Scholar]
- Haugen T., Andersson E., Norberg B. & Taranger G. The production of hermaphrodites of Atlantic cod (Gadus morhua) by masculinization with orally administered 17-α-methyltestosterone, and subsequent production of all-female cod populations. Aquaculture 311, 248–254 (2011). [Google Scholar]
- Hubert S., Higgins B., Borza T. & Bowman S. Development of a SNP resource and a genetic linkage map for Atlantic cod (Gadus morhua). BMC Genomics 11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tørresen O. K. et al. An improved genome assembly uncovers a prolific tandem repeat structure in Atlantic cod. bioRxiv (2016). [Google Scholar]
- Star B. et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 477, 207–210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson B. K., Weber J. N., Kay E. H., Fisher H. S. & Hoekstra H. E. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PloS one 7, e37135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler B. L. & Buonaccorsi V. P. Genomic characterization of sex-identification markers in Sebastes carnatus and Sebastes chrysomelas rockfishes. Mol. Ecol. (2016). [DOI] [PubMed] [Google Scholar]
- Graves J. A. M. & Peichel C. L. Are homologies in vertebrate sex determination due to shared ancestry or to limited options. Genome Biol 11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M. & Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson W. F. et al. The globalization of naval provisioning: ancient DNA and stable isotope analyses of stored cod from the wreck of the Mary Rose. AD 1545. R. Soc. Open Sci. 2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B. et al. Palindromic sequence artifacts generated during next generation sequencing library preparation from historic and ancient DNA. Plos One 9, e89676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B. et al. Preferential amplification of repetitive DNA during whole genome sequencing library creation from historic samples. STAR 2, 36–45 (2016). [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwera G. A. et al. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 11.10. 11–11.10. 33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Paul J. S., Albrechtsen A. & Song Y. S. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet 12, 443–451 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M. J. & Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13, 238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R. & Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J. T. & Mesirov J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater G. S. & Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6, 31 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L. et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18, 188–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C. & Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe L. et al. Maternal 3′UTRs: from egg to onset of zygotic transcription in Atlantic cod. BMC Genomics 13, 443 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penglase S. et al. Diet affects the redox system in developing Atlantic cod (Gadus morhua) larvae. Redox Biol. 5, 308–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tørresen O. K., Star B., Jentoft S., Jakobsen K. S. & Nederbragt A. J. The improved genome assembly for Atlantic cod reveals a high density of short-tandem-repeats. (In prep.).
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gadMor2 genome assembly is available from the European Nucleotide Archive (ENA) LN845748-LN845770. The annotation of the Atlantic cod genome is available from https://t.co/mdiVe52v4d. All individual read data (including the female PacBio data) associated with linkage groups containing sex-linked genotypes are available from ENA with study accession number PRJEB14672.